Published online Aug 27, 2024. doi: 10.4240/wjgs.v16.i8.2386
Revised: May 16, 2024
Accepted: June 5, 2024
Published online: August 27, 2024
Processing time: 150 Days and 2.2 Hours
Hepatocellular carcinoma (HCC) presents challenges due to its high recurrence and metastasis rates and poor prognosis. While current clinical diagnostic and prognostic indicators exist, their accuracy remains imperfect due to their biol
Core Tip: Hepatocellular carcinoma (HCC), ranking as the third leading cause of cancer-related mortality globally, is characterized by high rates of recurrence and metastasis. Long non-coding RNAs related to genomic instability emerge as promising biomarkers for HCC prognosis. Here, we discuss their clinical significance as prognostic models and offer ins
- Citation: Xing XW, Huang X, Li WP, Wang MK, Yang JS. Clinical application value of long non-coding RNAs signatures of genomic instability in predicting prognosis of hepatocellular carcinoma. World J Gastrointest Surg 2024; 16(8): 2386-2392
- URL: https://www.wjgnet.com/1948-9366/full/v16/i8/2386.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v16.i8.2386
Hepatocellular carcinoma (HCC), also known as the "king of cancer", ranks fifth in incidence and third in mortality in China, underscoring the critical importance of early screening and prognosis assessment. With the recognition of long non-coding RNAs (lncRNAs) as potential prognostic factors in various cancers including HCC, exploration into lncRNAs related to genomic instability (GI) has surged. In a recent study published in the World Journal of Gastrointestinal Surgery, Duan et al[1] identified a GI-derived lncRNA signature (GI-LncSig) by integrating lncRNA expression and somatic mu
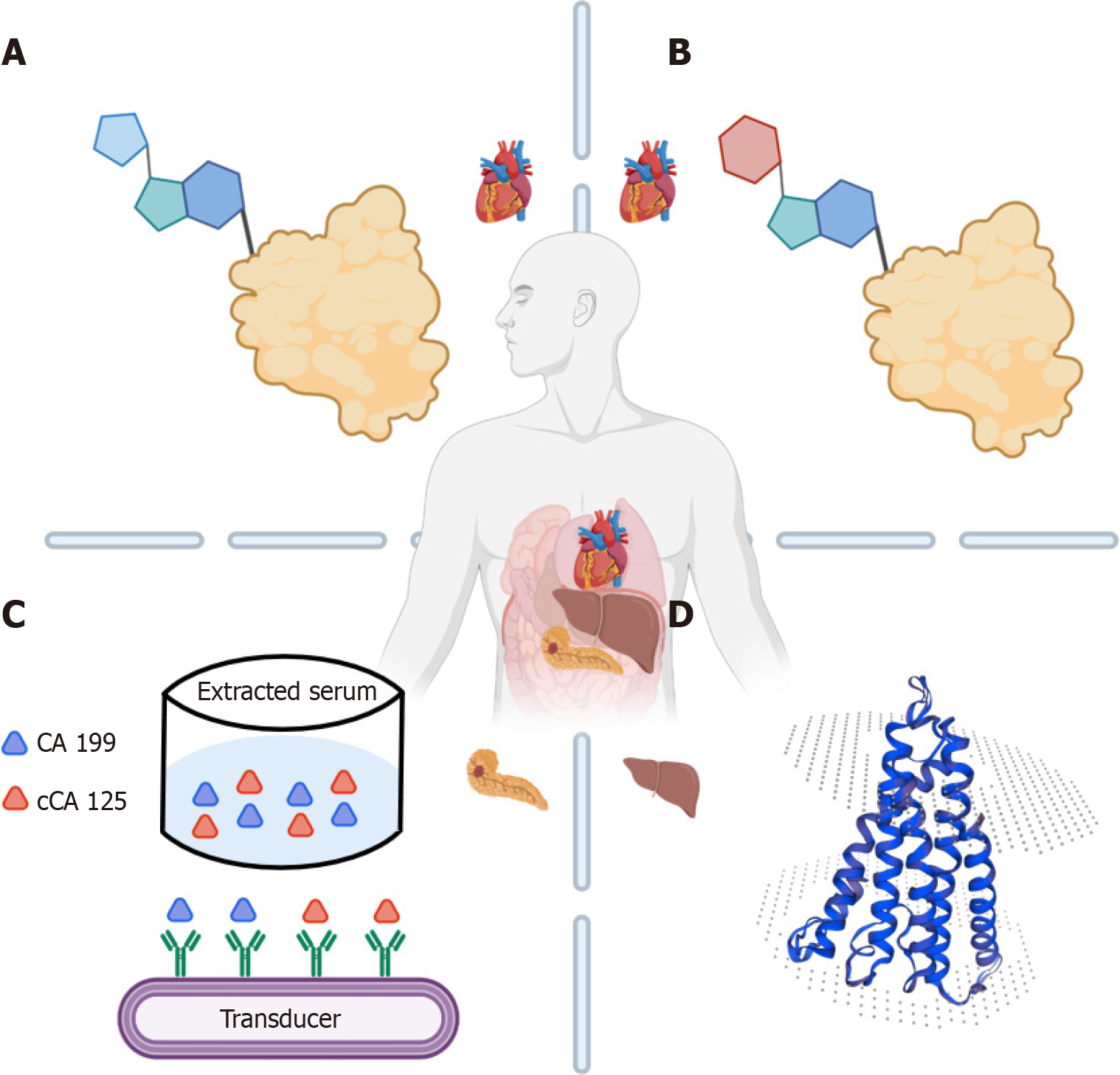
Five common HCC markers are routinely employed in clinical settings: Alpha-fetoprotein (AFP), carbohydrate antigen (CA) 199, cancer-derived CA 125 (cCA125), AFP anisoplasts (AFP-L3), and abnormal prothrombin (PIVKA-II) (Figure 1). The following sections provide a brief description of the representative significance, detection scope, and prognostic value of each marker.
AFP, primarily synthesized by HCC, exhibits elevated levels in 60%-70% of HCC patients, making it the most frequently utilized tumor marker. Both the United States National Comprehensive Cancer Network Guidelines and the Guidelines for Diagnosis and Treatment of Primary Liver Cancer in China (2022 edition)[2] recommend AFP as a standard tumor marker for HCC screening, aiming to enhance early detection rates. Normally, serum AFP concentration remains below 20 ng/mL. Routine screening for HCC involves ultrasound with or without AFP assessment every 6 months. The combination of ultrasound and AFP has shown marginal improvements in detection (6%-8% higher than ultrasound alone)[3]; however, this may also increase false-positive results, which limits the specificity of AFP. Despite its strong prognostic significance in patients with HCC undergoing systemic therapy, elevated AFP levels were also observed in various other conditions including acute and chronic hepatitis, cirrhosis, viral and neonatal hepatitis, pregnancy and germ cell tumors, gastrointestinal tumors, liver injury, and telangiectasia. Additionally, certain patients with HCC were negative for AFP (AFP < 20 ng/mL)[4], indicating AFP’s limited sensitivity and specificity for HCC.
AFP-L3, a subfraction of AFP originating from malignant hepatocytes, serves as a valuable indicator for HCC. Given that AFP is negative in approximately 30% of patients with HCC, AFP-L3 acts as a complementary marker for AFP[5]. The ratio of AFP-L3 to total AFP aids in distinguishing between non-malignant hepatic disease and HCC. In 2005, the United States Food and Drug Administration approved AFP-L3 for HCC diagnosis. Normally, the serum AFP-L3 to AFP ratio remains below 10%; however, even with low AFP levels, an AFP-L3 ratio exceeding 10% suggests HCC occurrence. In a prospective study, AFP-L3 (AFP bound to lens culinaris agglutinin) and des-γ-carboxyprothrombin (DCP) biomarkers exhibited strong predictive capabilities for early HCC recurrence, surpassing AFP alone and effectively reducing false-negatives and false-positives[6]. In China, AFP, AFP-L3, and DCP have been included in the "13th Five-Year Plan" for infectious disease prevention and control, which is expected to become a common diagnostic criterion for HCC globally. Nonetheless, the relationship between pre-treatment serum AFP-L3% levels and tumor invasion, metastasis, and other clinicopathological parameters (such as tumor grade, stage, and cirrhosis) reported in some studies lacks reliability, hindering the estimation of their impact on overall survival (OS) or disease-free survival (DFS)[7]. Conflicting data have also emerged regarding the ability of pre-treatment serum AFP-L3% to predict DFS and OS in HCC, thereby reducing the confidence of AFP-L3% for HCC patient prognosis.
CA199 and cCA125, two glycoprotein macromolecules, are commonly utilized markers for adenocarcinoma, notably elevated in lung, pancreatic, colorectal, endometrial, ovarian, and other cancers[8]. Approximately 10% of HCC cases originate from bile duct epithelial cells or rare tumor types, wherein AFP is negative while CA199 is elevated. Fu
PIVKA-II arises in the presence of glutamyl carboxylase and vitamin K deficiency. When hepatocytes fail to synthesize normal vitamin K-dependent clotting factors, abnormal serum prothrombin concentrations are elevated. Since 2015, China's "Guidelines for the Prevention and Treatment of Chronic Hepatitis B" have recommended PIVKA-II as a crucial indicator for HCC diagnosis, serving as a complementary marker to AFP to enhance early detection rates of primary liver cancer[12]. Under normal conditions, PIVKA-II concentrations are below 40 mAU/mL, with a diagnostic rate of 74% for early-stage liver cancer[13]. PIVKA-II holds significant diagnostic value in preoperative diagnosis and postoperative monitoring of liver cancer, with levels typically decreasing post-surgery. A rise in PIVKA-II levels post-surgery indicates tumor recurrence. However, the pathological mechanism underlying the elevation of PIVKA-II in HCC remains incompletely understood, rendering it a serological marker with clinically significant associations. The clinical sensitivity of PIVKA-II-positive HCC stands at 55%, only positioning it as a reference marker in clinical diagnosis[14].
The elevation of tumor markers often correlates with tumor occurrence and progression, albeit influenced by benign diseases, inflammation, physiological changes, lifestyle habits, and other factors. Frequently, a single tumor marker alone may not conclusively indicate cancer; rather, multiple markers and detection methods are required for accurate identification. Cancer is characterized by abnormal and uncontrolled cell growth due to genetic mutations, a trait often referred to as GI[15].
Zhou et al[2] validated the GI-LncSig model, constructed using five GI-lncRNAs, which was established at the genetic level related to pathogenesis, thus circumventing environmental and individual differences affecting changes in HCC markers. Utilizing the risk score derived from this model, patients with HCC in the database were categorized into high-risk and low-risk groups. A comparison of the 5-year survival rates between these groups revealed a survival rate of 9.3% for high-risk patients and 19.8% for low-risk patients. The prognostic performance of GI-LncSig was assessed via receiver operating characteristic curve analysis, yielding an area under the curve (AUC) of 0.736, surpassing that of GulncSig (AUC = 0.664) or WulncSig (AUC = 0.725). These findings indicate that GI-LncSig exhibits superior prognostic per
Directly using lncRNA expression profiles and somatic cell mutation profiles at the molecular level enables the pr
Current screening methods for cancer include ultrasound imaging and serum antigen detection, despite their limited sensitivity (ranging from 47% to 84%) and specificity (from 67% to over 90%)[17]. However, their quickness and con
The current arsenal of serum biomarkers for predicting HCC prognosis remains insufficient, characterized by low sensitivity and heterogeneous specificity. Currently, apart from AFP and those mentioned above, new biomarkers have yet to be integrated into routine clinical practice. Therefore, researchers are diligently exploring alternative biomarkers for early diagnosis, personalized treatment approaches, and post-treatment prognosis using proteomics, metabolomics, genomics, and other novel technologies such as microbiome analysis[19].
Currently, researchers have mined genetic information associated with HCC-related processes, including cell senescence, cuproptosis, cell necrosis, cell-free DNA, natural killer cells, basement membrane, and cell cycles, to identify biomarkers that accurately assess patient prognosis. Integrating proteomic studies with gene-editing models enables the analysis of HCC patient prognosis, shedding light on post-translational modifications and complex pathological pro
| Species | Name | Feature | Ref. |
| Genomics | PDXK | Cuproptosis-related gene signature | Chen et al[21] |
| m6A/m5C/m1A | Poor prognosis and immune microenvironment in HCC | Li et al[22] | |
| CANT1 | Histologic grade | Liu et al[23] | |
| CTSA | The most critical basement membrane-related genes | Sun et al[24] | |
| Mutation Capsule Plus | Multiple analyses of a cfDNA sample to obtain its whole genome information | Wang et al[25] | |
| IL18RAP, CHP1, VAMP2, PIK3R1, PRKCD | 5-NKRLSig, associated with natural killer cells | Xi et al[26] | |
| CDCA8, CENPA, SPC25, TTK | Four central genes involved in cellular senescence | Zhang et al[27] | |
| PHF19 | As a crucial constituent part of Polycomb repressive complex 2, PHD finger protein 19 plays a pivotal role in epigenetic regulation | Zhu et al[28] | |
| Proteomics | Lysine crotonylation | Higher crotonylation in HCC cells facilitated cell invasiveness | Zhang et al[29] |
| SLC1A4 | SLC1A4 inhibited cell proliferation, migration, and cell cycle progression, and promoted cell apoptosis in HCC | Peng et al[30] | |
| Metabolomics | MG (monoacylglyceride) | It might accumulate in patients with advanced HCC due to the deficit of MGLL | Lin et al[31] |
| NrLR | Neutrophil times γ-glutamyl transpeptidase to lymphocyte ratio | Wu et al[32] | |
| IL-6 | Interleukin-6 promotes the growth of the HCC microenvironment | Dalbeni et al[33] | |
| Others | Gut microbiome | Strong diagnosis potential for early HCC and even advanced HCC | Ren et al[34] |
| Intratumor microbiome | It can affect HCC patients' prognosis by modulating the cancer stemness and immune response | Song et al[35] | |
| LI-RADS | Liver imaging reporting and data system | Ronot et al[36] | |
| A combined clinicoradiological MR-based model integrating radiomics features | This model was shown to be associated with recurrence-free survival | Song et al[37] |
This literature review provides an overview of biomarkers utilized in the diagnosis and prognosis of patients with HCC in clinical practice, comparing their detection time and cost with those of the GI-LncSig prognostic model. AFP, AFP-L3, CA199, cCA125, PIVKA-II, and other indicators exhibit varying degrees of deficiencies and inaccuracies in terms of accuracy, sensitivity, applicability, as well as representativeness. In contrast, the GI-LncSig model effectively addresses these limitations and demonstrates superior alignment with the database, resulting in enhanced prognostic accuracy. However, the current implementation of GI-LncSig is hindered by cumbersome detection sampling, high costs, and prolonged detection times. If these issues can be resolved, GI-LncSig technology will offer higher accuracy than tra
| 1. | Duan BT, Zhao XK, Cui YY, Liu DZ, Wang L, Zhou L, Zhang XY. Construction and validation of somatic mutation-derived long non-coding RNAs signatures of genomic instability to predict prognosis of hepatocellular carcinoma. World J Gastrointest Surg. 2024;16:842-859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 2. | Zhou J, Sun H, Wang Z, Cong W, Zeng M, Zhou W, Bie P, Liu L, Wen T, Kuang M, Han G, Yan Z, Wang M, Liu R, Lu L, Ren Z, Zeng Z, Liang P, Liang C, Chen M, Yan F, Wang W, Hou J, Ji Y, Yun J, Bai X, Cai D, Chen W, Chen Y, Cheng W, Cheng S, Dai C, Guo W, Guo Y, Hua B, Huang X, Jia W, Li Q, Li T, Li X, Li Y, Li Y, Liang J, Ling C, Liu T, Liu X, Lu S, Lv G, Mao Y, Meng Z, Peng T, Ren W, Shi H, Shi G, Shi M, Song T, Tao K, Wang J, Wang K, Wang L, Wang W, Wang X, Wang Z, Xiang B, Xing B, Xu J, Yang J, Yang J, Yang Y, Yang Y, Ye S, Yin Z, Zeng Y, Zhang B, Zhang B, Zhang L, Zhang S, Zhang T, Zhang Y, Zhao M, Zhao Y, Zheng H, Zhou L, Zhu J, Zhu K, Liu R, Shi Y, Xiao Y, Zhang L, Yang C, Wu Z, Dai Z, Chen M, Cai J, Wang W, Cai X, Li Q, Shen F, Qin S, Teng G, Dong J, Fan J. Guidelines for the Diagnosis and Treatment of Primary Liver Cancer (2022 Edition). Liver Cancer. 2023;12:405-444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 195] [Reference Citation Analysis (0)] |
| 3. | Force M, Park G, Chalikonda D, Roth C, Cohen M, Halegoua-DeMarzio D, Hann HW. Alpha-Fetoprotein (AFP) and AFP-L3 Is Most Useful in Detection of Recurrence of Hepatocellular Carcinoma in Patients after Tumor Ablation and with Low AFP Level. Viruses. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 4. | Lim DH, Casadei-Gardini A, Lee MA, Lonardi S, Kim JW, Masi G, Chon HJ, Rimini M, Kim I, Cheon J, Hwang JE, Kang JH, Lim HY, Yoo C. Prognostic implication of serum AFP in patients with hepatocellular carcinoma treated with regorafenib. Future Oncol. 2022;18:3021-3030. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 5. | Kawahara I, Fukuzawa H, Urushihara N, Kosaka Y, Kuroda Y, Fujieda Y, Takeuchi Y, Uemura K, Iwade T, Samejima Y, Morita K, Maeda K. AFP-L3 as a Prognostic Predictor of Recurrence in Hepatoblastoma: A Pilot Study. J Pediatr Hematol Oncol. 2021;43:e76-e79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 6. | Norman JS, Li PJ, Kotwani P, Shui AM, Yao F, Mehta N. AFP-L3 and DCP strongly predict early hepatocellular carcinoma recurrence after liver transplantation. J Hepatol. 2023;79:1469-1477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 71] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 7. | Cheng J, Wang W, Zhang Y, Liu X, Li M, Wu Z, Liu Z, Lv Y, Wang B. Prognostic role of pre-treatment serum AFP-L3% in hepatocellular carcinoma: systematic review and meta-analysis. PLoS One. 2014;9:e87011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 8. | Lin S, Wang Y, Peng Z, Chen Z, Hu F. Detection of cancer biomarkers CA125 and CA199 via terahertz metasurface immunosensor. Talanta. 2022;248:123628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 9. | Gao Y, Wang J, Zhou Y, Sheng S, Qian SY, Huo X. Evaluation of Serum CEA, CA19-9, CA72-4, CA125 and Ferritin as Diagnostic Markers and Factors of Clinical Parameters for Colorectal Cancer. Sci Rep. 2018;8:2732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 200] [Article Influence: 28.6] [Reference Citation Analysis (1)] |
| 10. | Huang Y, Zeng J, Liu T, Lin X, Guo P, Zeng J, Zhou W, Liu J. Prognostic Significance of Elevated Preoperative Serum CA125 Levels After Curative Hepatectomy for Hepatocellular Carcinoma. Onco Targets Ther. 2020;13:4559-4567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 11. | Zhang J, Qin SD, Li Y, Lu F, Gong WF, Zhong JH, Ma L, Zhao JF, Zhan GH, Li PZ, Song B, De Xiang B. Prognostic significance of combined α-fetoprotein and CA19-9 for hepatocellular carcinoma after hepatectomy. World J Surg Oncol. 2022;20:346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 12. | You H, Wang F, Li T, Xu X, Sun Y, Nan Y, Wang G, Hou J, Duan Z, Wei L, Jia J, Zhuang H; Chinese Society of Hepatology, Chinese Medical Association; Chinese Society of Infectious Diseases, Chinese Medical Association. Guidelines for the Prevention and Treatment of Chronic Hepatitis B (version 2022). J Clin Transl Hepatol. 2023;11:1425-1442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 82] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 13. | Feng H, Li B, Li Z, Wei Q, Ren L. PIVKA-II serves as a potential biomarker that complements AFP for the diagnosis of hepatocellular carcinoma. BMC Cancer. 2021;21:401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 100] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 14. | Fujita K, Kinukawa H, Ohno K, Ito Y, Saegusa H, Yoshimura T. Development and evaluation of analytical performance of a fully automated chemiluminescent immunoassay for protein induced by vitamin K absence or antagonist II. Clin Biochem. 2015;48:1330-1336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 15. | Martínez-Jiménez F, Muiños F, Sentís I, Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, Mularoni L, Pich O, Bonet J, Kranas H, Gonzalez-Perez A, Lopez-Bigas N. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20:555-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 385] [Cited by in RCA: 736] [Article Influence: 147.2] [Reference Citation Analysis (0)] |
| 16. | Caruso S, Calatayud AL, Pilet J, La Bella T, Rekik S, Imbeaud S, Letouzé E, Meunier L, Bayard Q, Rohr-Udilova N, Péneau C, Grasl-Kraupp B, de Koning L, Ouine B, Bioulac-Sage P, Couchy G, Calderaro J, Nault JC, Zucman-Rossi J, Rebouissou S. Analysis of Liver Cancer Cell Lines Identifies Agents With Likely Efficacy Against Hepatocellular Carcinoma and Markers of Response. Gastroenterology. 2019;157:760-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 137] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 17. | Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, Waljee AK, Singal AG. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients With Cirrhosis: A Meta-analysis. Gastroenterology. 2018;154:1706-1718.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 638] [Cited by in RCA: 809] [Article Influence: 115.6] [Reference Citation Analysis (0)] |
| 18. | Qiu M, Yu C, Zhu S, Liu S, Peng H, Xiong X, Chen J, Jiang X, Du H, Li Q, Zhang Z, Yang C. RNA sequencing reveals lncRNA-mediated non-mendelian inheritance of feather growth change in chickens. Genes Genomics. 2022;44:1323-1331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 19. | Piñero F, Dirchwolf M, Pessôa MG. Biomarkers in Hepatocellular Carcinoma: Diagnosis, Prognosis and Treatment Response Assessment. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 328] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 20. | Yang C, Huang X, Liu Z, Qin W, Wang C. Metabolism-associated molecular classification of hepatocellular carcinoma. Mol Oncol. 2020;14:896-913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 150] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 21. | Chen Y, Tang L, Huang W, Abisola FH, Zhang Y, Zhang G, Yao L. Identification of a prognostic cuproptosis-related signature in hepatocellular carcinoma. Biol Direct. 2023;18:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 50] [Reference Citation Analysis (0)] |
| 22. | Li D, Li K, Zhang W, Yang KW, Mu DA, Jiang GJ, Shi RS, Ke D. The m6A/m5C/m1A Regulated Gene Signature Predicts the Prognosis and Correlates With the Immune Status of Hepatocellular Carcinoma. Front Immunol. 2022;13:918140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 79] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 23. | Liu T, Li ZZ, Sun L, Yang K, Chen JM, Han XY, Qi LM, Zhou XG, Wang P. Upregulated CANT1 is correlated with poor prognosis in hepatocellular carcinoma. BMC Cancer. 2023;23:1007. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 24. | Sun W, Wang J, Wang Z, Xu M, Lin Q, Sun P, Yuan Y. Combining WGCNA and machine learning to construct basement membrane-related gene index helps to predict the prognosis and tumor microenvironment of HCC patients and verifies the carcinogenesis of key gene CTSA. Front Immunol. 2023;14:1185916. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 25. | Wang P, Song Q, Ren J, Zhang W, Wang Y, Zhou L, Wang D, Chen K, Jiang L, Zhang B, Chen W, Qu C, Zhao H, Jiao Y. Simultaneous analysis of mutations and methylations in circulating cell-free DNA for hepatocellular carcinoma detection. Sci Transl Med. 2022;14:eabp8704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 26. | Xi D, Wang J, Yang Y, Ji F, Li C, Yan X. A novel natural killer-related signature to effectively predict prognosis in hepatocellular carcinoma. BMC Med Genomics. 2023;16:211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 27. | Zhang S, Zheng Y, Li X, Zhang S, Hu H, Kuang W. Cellular senescence-related gene signature as a valuable predictor of prognosis in hepatocellular carcinoma. Aging (Albany NY). 2023;15:3064-3093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 28. | Zhu ZY, Tang N, Wang MF, Zhou JC, Wang JL, Ren HZ, Shi XL. Comprehensive Pan-Cancer Genomic Analysis Reveals PHF19 as a Carcinogenic Indicator Related to Immune Infiltration and Prognosis of Hepatocellular Carcinoma. Front Immunol. 2021;12:781087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 29. | Zhang XY, Liu ZX, Zhang YF, Xu LX, Chen MK, Zhou YF, Yu J, Li XX, Zhang N. SEPT2 crotonylation promotes metastasis and recurrence in hepatocellular carcinoma and is associated with poor survival. Cell Biosci. 2023;13:63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 30. | Peng X, Chen R, Cai S, Lu S, Zhang Y. SLC1A4: A Powerful Prognostic Marker and Promising Therapeutic Target for HCC. Front Oncol. 2021;11:650355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 31. | Lin Z, Li H, He C, Yang M, Chen H, Yang X, Zhuo J, Shen W, Hu Z, Pan L, Wei X, Lu D, Zheng S, Xu X. Metabolomic biomarkers for the diagnosis and post-transplant outcomes of AFP negative hepatocellular carcinoma. Front Oncol. 2023;13:1072775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 32. | Wu Q, Zeng J, Zeng J. Inflammation-Related Marker NrLR Predicts Prognosis in AFP-Negative HCC Patients After Curative Resection. J Hepatocell Carcinoma. 2023;10:193-202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 33. | Dalbeni A, Natola LA, Garbin M, Zoncapè M, Cattazzo F, Mantovani A, Vella A, Canè S, Kassem J, Bevilacqua M, Conci S, Campagnaro T, Ruzzenente A, Auriemma A, Drudi A, Zanoni G, Guglielmi A, Milella M, Sacerdoti D. Interleukin-6: A New Marker of Advanced-Sarcopenic HCC Cirrhotic Patients. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 34. | Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, Xie H, Chen X, Shao L, Zhang R, Xu S, Zhang H, Cui G, Chen X, Sun R, Wen H, Lerut JP, Kan Q, Li L, Zheng S. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68:1014-1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 542] [Cited by in RCA: 508] [Article Influence: 84.7] [Reference Citation Analysis (0)] |
| 35. | Song Y, Xiang Z, Lu Z, Su R, Shu W, Sui M, Wei X, Xu X. Identification of a brand intratumor microbiome signature for predicting prognosis of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2023;149:11319-11332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 36. | Ronot M, Chernyak V, Burgoyne A, Chang J, Jiang H, Bashir M, Fowler KJ. Imaging to Predict Prognosis in Hepatocellular Carcinoma: Current and Future Perspectives. Radiology. 2023;307:e221429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 51] [Reference Citation Analysis (0)] |
| 37. | Song W, Yu X, Guo D, Liu H, Tang Z, Liu X, Zhou J, Zhang H, Liu Y, Liu X. MRI-Based Radiomics: Associations With the Recurrence-Free Survival of Patients With Hepatocellular Carcinoma Treated With Conventional Transcatheter Arterial Chemoembolization. J Magn Reson Imaging. 2020;52:461-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |