Published online May 27, 2023. doi: 10.4240/wjgs.v15.i5.931
Peer-review started: March 13, 2023
First decision: March 28, 2023
Revised: March 29, 2023
Accepted: April 7, 2023
Article in press: April 7, 2023
Published online: May 27, 2023
Processing time: 74 Days and 7.2 Hours
A noninvasive biomarker with high diagnostic performance is urgently needed for the early diagnosis of colorectal cancer (CRC).
To evaluate the diagnostic value of matrix metalloproteinases (MMPs) 2, 7 and 9 in urine for CRC.
Of 59 healthy controls, 47 patients with colon polyps and 82 patients with CRC were included in this study. Carcinoembryonic antigen (CEA) in serum and MMP2, MMP7, and MMP9 in urine were detected. The combined diagnostic model of the indicators was established by binary logistic regression. The receiver operating characteristic curve (ROC) of the subjects was used to evaluate the independent and combined diagnostic value of the indicators.
The MMP2, MMP7, MMP9, and CEA levels in the CRC group differed significantly from levels in the healthy controls (P < 0.05). The levels of MMP7, MMP9, and CEA also differed significantly between the CRC group and the colon polyps group (P < 0.05). The area under the curve (AUC) distinguishing between the healthy control and the CRC patients using the joint model with CEA, MMP2, MMP7 and MMP9 was 0.977, and the sensitivity and specificity were 95.10% and 91.50%, respectively. For early-stage CRC, the AUC was 0.975, and the sensitivity and specificity were 94.30% and 98.30%, respectively. For advanced stage CRC, the AUC was 0.979, and the sensitivity and specificity were 95.70% and 91.50%, respectively. Using CEA, MMP7 and MMP9 to jointly established a model distinguishing the colorectal polyp group from the CRC group, the AUC was 0.849, and the sensitivity and specificity were 84.10% and 70.20%, respectively. For early-stage CRC, the AUC was 0.818, and the sensitivity and specificity were 76.30% and 72.30%, respectively. For advanced stage CRC, the AUC was 0.875, and the sensitivity and specificity were 81.80% and 72.30%, respectively.
MMP2, MMP7 and MMP 9 may exhibit diagnostic value for the early detection of CRC and may serve as auxiliary diagnostic markers for CRC.
Core Tip: Colorectal cancer (CRC) is one of the most common cancers. Early diagnosis and early treatment have become the consensus of CRC diagnosis and treatment. Matrix metalloproteinases (MMPs), as a group of zinc-dependent endopeptidases, participate in the degradation of the extracellular matrix and are secreted and activated outside the cell. We aimed to evaluate the MMP2, MMP7 and MMP9 diagnostic value for early detection of CRC.
- Citation: Peng L, Zhang X, Zhang ML, Jiang T, Zhang PJ. Diagnostic value of matrix metalloproteinases 2, 7 and 9 in urine for early detection of colorectal cancer. World J Gastrointest Surg 2023; 15(5): 931-939
- URL: https://www.wjgnet.com/1948-9366/full/v15/i5/931.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v15.i5.931
Colorectal cancer (CRC) is one of the most common cancers. Although detection and treatment have improved, the incidence and mortality of CRC are on the rise[1]. A large number of clinical studies have confirmed that early diagnosis of CRC can significantly prolong survival, but at present, only approximately 30% to 40% of patients are diagnosed at an early stage. Early diagnosis and early treatment have become the consensus of CRC diagnosis and treatment[2]. At present, the commonly used clinical tests include invasive colonoscopy, noninvasive fecal occult blood testing, fecal DNA testing, carcinoembryonic antigen (CEA), carbohydrate antigen 199 (CA199) levels, and Septin 9 methylation levels, but such tests are limited by their complexity and diagnostic performance[3]. A noninvasive biomarker with high diagnostic performance is urgently needed for the early diagnosis of clinical CRC[4].
Research has demonstrated that cytokines can be used as potential biomarkers for cancer detection and treatment response[5]. Cytokines are soluble peptides that play an important role in inflammation and immune cells signaling as well as in the multistep process of carcinogenesis. Cytokines can bind to their receptor and trigger the production of additional cytokines, leading to high concentrations in blood or other body fluids[6]. Compared with blood or stool, urine represents a better source because of its simple collection method, high patient acceptance and ability to collect repeated samples. Many urine biomarkers for CRC have been studied and have demonstrated potential diagnostic value[7-9]. Matrix metalloproteinases (MMPs), as a group of zinc-dependent endopeptidases, participate in the degrada
The study was approved by the Ethics Committee of Peking University Cancer Hospital & Institute and provided their informed consent. Serum and urine samples were collected from 59 healthy controls, 47 patients with colon polyps and 82 patients with CRC. The age and sex of the healthy control group, colon polyp group and CRC group were matched. Among the 82 patients with CRC, there were 10, 28, 20, and 24 Duke A, B, C and D stage cancers, respectively. The were 38 cases of early CRC and 44 cases of late CRC. The inclusion criteria for the colon polyps and colon cancer groups were as follows: No prior treatment (e.g., surgery, chemotherapy or radiotherapy), histopathological confirmation, no other gastrointestinal diseases and no other major diseases. The age-matched healthy controls all had negative blood biomarker, X-ray, ultrasound, computed tomography (CT), and fecal occult blood tests, and diagnosis was further confirmed by histopathological analysis. CRC sites included the cecum, ascending colon, descending colon, transverse colon, sigmoid colon and rectum. CRC was staged in accordance with Dukes staging criteria. Dukes stage A and B cancers represent early CRC, and Dukes stage C and D cancers represent late CRC. The clinical characteristics of the subjects are presented in Table 1.
| Group | Healthy control | Colon polyps | Colorectal cancer |
| MMP2 | 2128.42 (1635.60, 3119.34) | 15459.62 (12244.16, 18777.56) | 15396.14 (6571.35, 20006.06) |
| MMP7 | 2612.71 (2087.86, 3110.04) | 3237.57 (2513.33, 3915.02) | 4173.63 (3023.82, 6327.17) |
| MMP9 | 8153.00 (5170.05, 11732.83) | 14288.33 (8711.57, 17994.25) | 10324.22 (6005.56, 14932.17) |
| CEA | 1.52 (0.79, 2.26) | 3.05 (1.55, 7.82) | 6.84 (1.86, 14.43) |
Fasting peripheral blood was collected from all subjects included in this study in the morning and centrifuged at 3000 rpm for 10 min, and the supernatants were collected and labeled. All urine samples were collected from the middle section of the second morning urine (10 mL) and centrifuged at 1500 rpm for 10 min; the supernatant was collected, labeled, and stored at -80 °C. CEA and CA199 were detected by chemiluminescence, and a Roche E170 automatic immune analyzer was used for detection. The fasting peripheral blood of the subjects was collected using vacuum collecting tubes, and the serum was separated after centrifugation. CEA and CA199 were detected after the instrument was calibrated using standards. Urine MMP2, MMP7, and MMP9 were detected by a modular collection rapid detection system and using the antigen-antibody combination luminescence principle, which can detect trace MMP9 levels.
The data of this study were analyzed using SPSS 20.0. The CEA, CA199, MMP2, MMP7, and MMP9 levels are expressed as medians (25%, 75%). Differences in marker levels among the healthy control group, colon polyp group and CRC group were tested using one-way ANOVA for significance. The combined diagnostic model of the three indicators was established using binary logistic regression. The receiver operating characteristic curve (ROC) was used to evaluate the independent and combined diagnostic value of the indicators. P < 0.05 was defined as a significant difference.
The MMP2, MMP7 MMP9, and CEA levels in the three groups were detected and compared. As shown in Table 1, MMP2, MMP7 MMP9, and CEA levels in the healthy controls were 2128.42 (1635.60, 3119.34), 2612.71 (2087.86, 3110.04) and 8153.00 (5170.05, 11732.83), respectively. In the colon polyp group, the MMP2, MMP7 MMP9, and CEA levels were 15459.62 (12244.16, 18777.56), 3237.57 (2513.33, 3915.02), 14288.33 (8711.57, 17994.25), and 3.05 (1.55, 7.82). In the CRC group, the MMP2, MMP7 MMP9, and CEA levels were 15396.14 (6571.35, 20006.06), 4173.63 (3023.82, 6327.17), 8324.22 (4005.56, 11932.17), and 6.84 (1.86, 14.43), respectively. The MMP2, MMP7, MMP9, and CEA levels in the CRC group differed significantly from those in the healthy control group (P < 0.05). The MMP7, MMP9, and CEA levels in the CRC group differed significantly from the levels in the colon polyp group (P < 0.05). MMP2 levels did not differ significantly between the colon polyp and CRC groups.
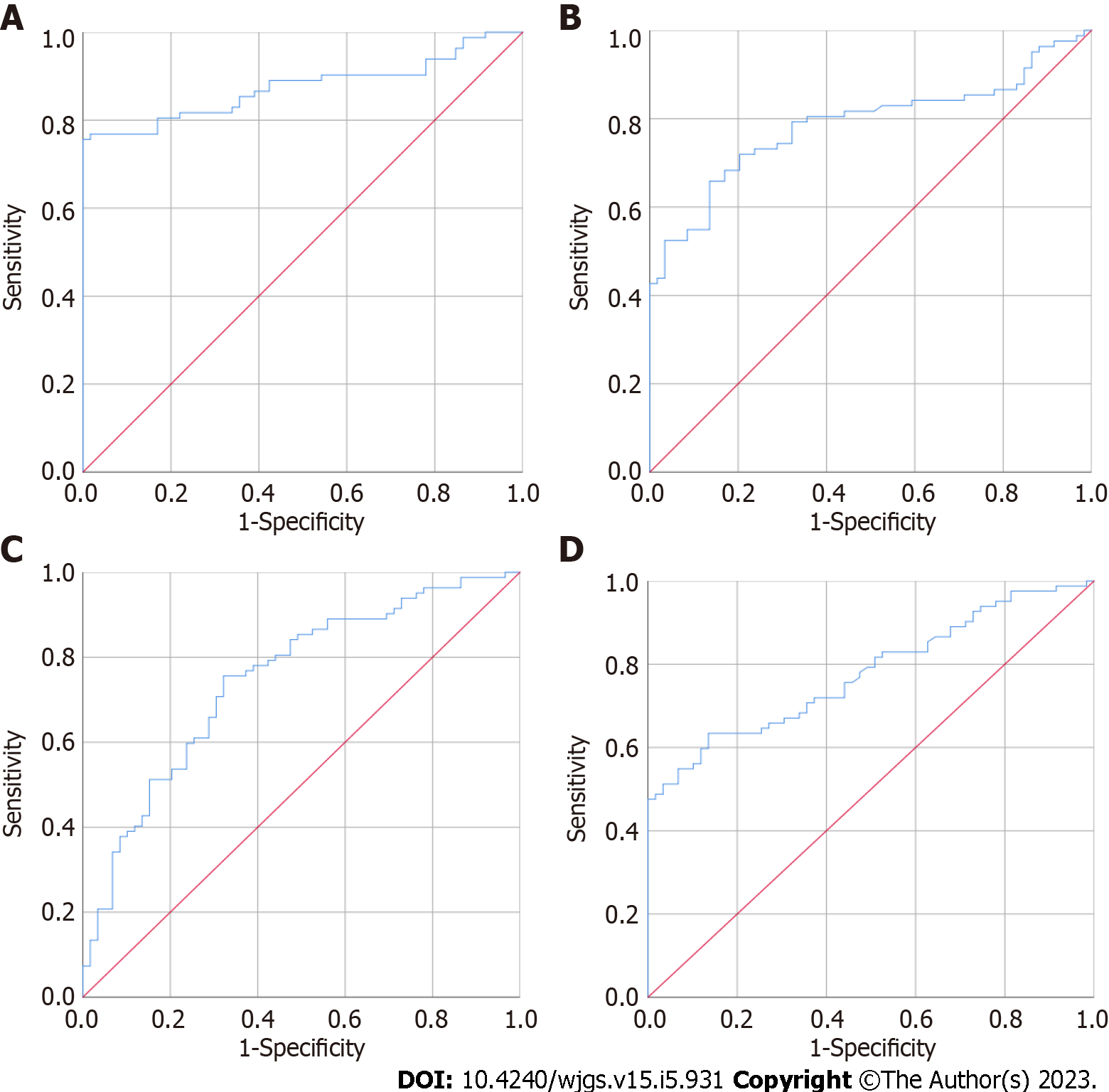
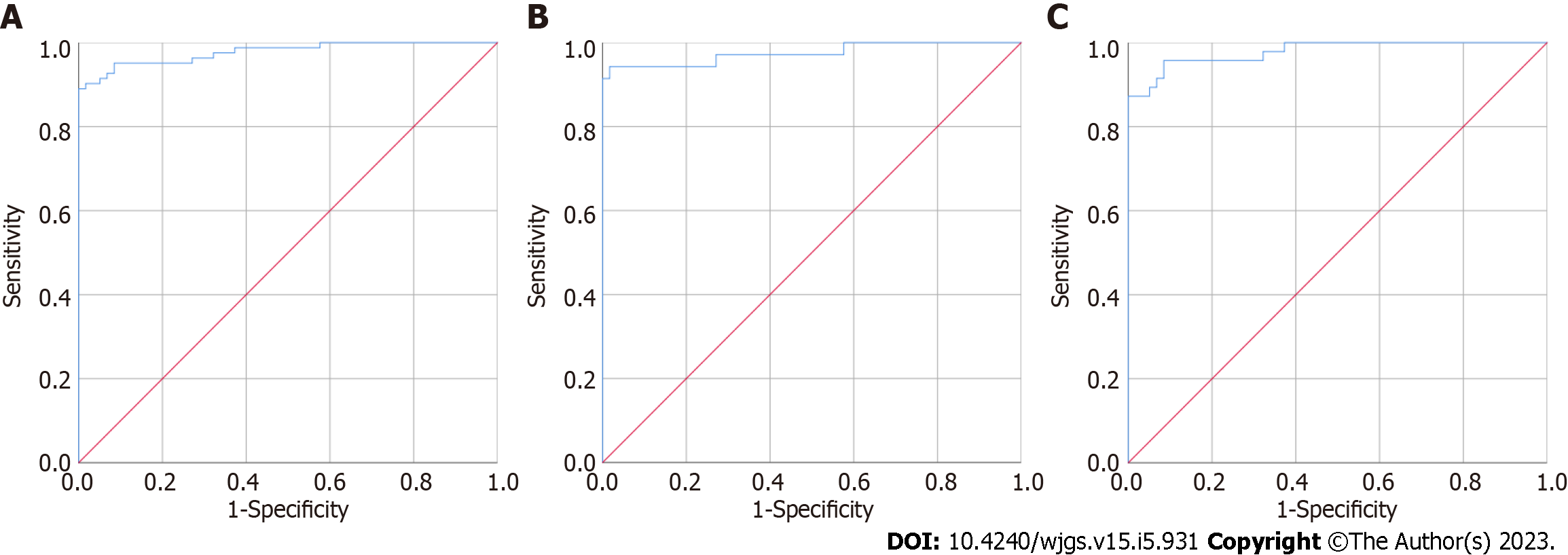
CEA, MMP2, MMP7 and MMP9 were separately used to distinguish the 59 healthy controls from the 82 CRC patients; the results are presented in Figure 1 and Table 2, respectively. The area under the curve (AUC) of urine MMP2 was the highest, 0.875 [95% confidence interval (CI): 0.815-0.935], followed by MMP7, CEA, and MMP9. The AUCs were 0.786 (95%CI: 0.711-0.862), 0.779 (95%CI: 0.704-0.853) and 0.748 (95%CI: 0.667-0.830), respectively. As shown in Figure 2, when CEA, MMP2, MMP7 and MMP9 levels were combined to established a model to distinguish the 59 healthy controls and 82 CRC patients, the AUC was 0.977 (95%CI: 0.957-0.998), and the sensitivity and specificity were 95.10% and 91.50%, respectively. When this combined model was used to distinguish 59 healthy controls and 38 patients with early CRC, the AUC was 0.975 (95%CI: 0.940-1.000), and the sensitivity and specificity were 94.30% and 98.30%, respectively. When used to distinguish 59 healthy controls and 47 patients with advanced CRC, the AUC was 0.979 (95%CI: 0.956-1.000), and the sensitivity and specificity were 95.70% and 91.50%, respectively.
| AUC | Standard error | P value | 95%CI | ||
| Lower | Upper | ||||
| MMP2 | 0.875 | 0.031 | < 0.001 | 0.815 | 0.935 |
| MMP7 | 0.786 | 0.039 | < 0.001 | 0.711 | 0.862 |
| MMP9 | 0.748 | 0.042 | < 0.001 | 0.667 | 0.830 |
| CEA | 0.779 | 0.038 | < 0.001 | 0.704 | 0.853 |
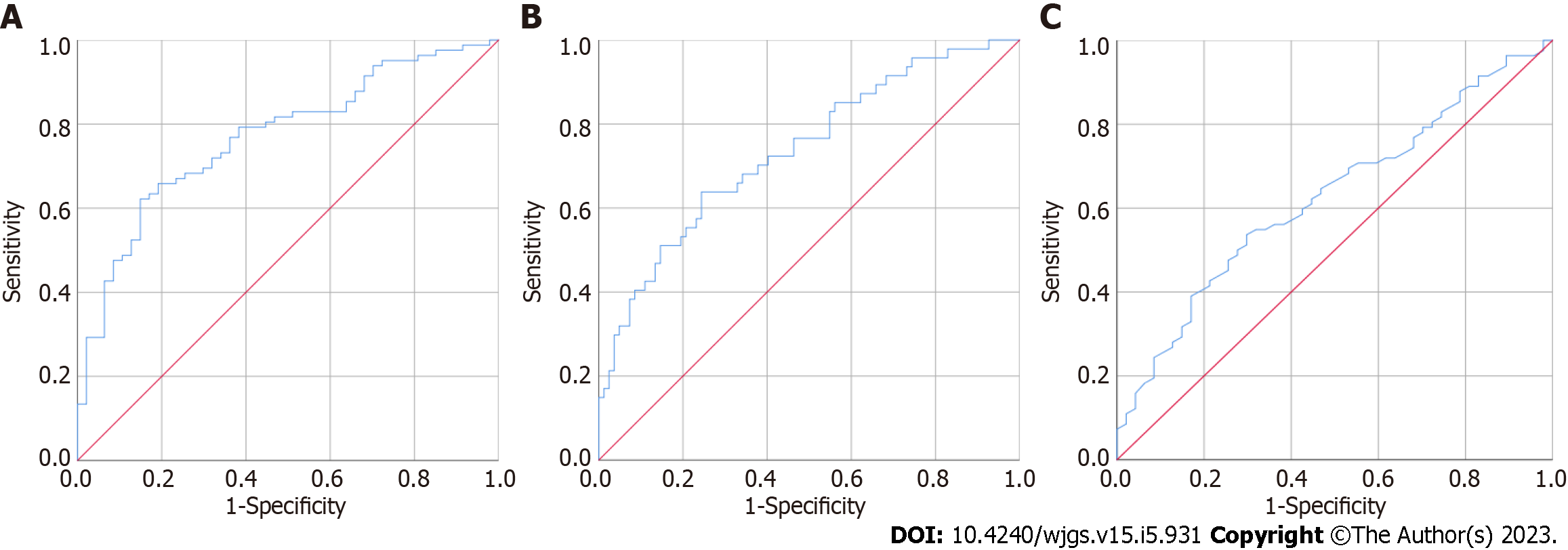
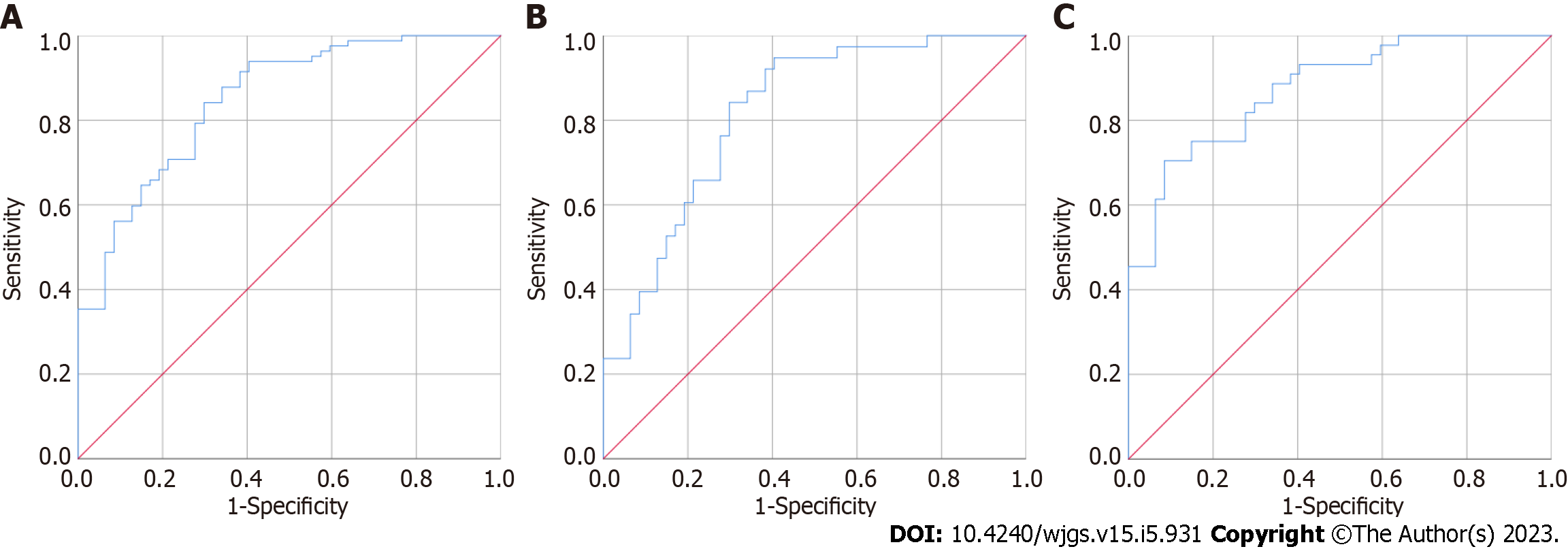
CEA, MMP7 and MMP9 were separately used to distinguish the 47 patients with colorectal polyps from the 82 CRC patients; the results are presented in Figure 3 and Table 3. The AUC of urine MMP7 was the highest, 0.769 (95%CI: 0.688-0.851), followed by MMP9 and CEA. The AUCs for MMP9 and CEA were 0.737 (95%CI: 0.647-0.827) and 0.626 (95%CI: 0.528-0.723), respectively. As shown in Figure 4, when CEA, MMP7 and MMP9 were combined to establish a model to distinguish the 47 colorectal polyp patients from the 82 CRC patients, the AUC was 0.849 (95%CI: 0.781-0.916), and the sensitivity and specificity were 84.10% and 70.20%, respectively. When this combined model was used to distinguish the 47 colorectal polyp patients and 38 patients with early CRC, the AUC was 0.818 (95%CI: 0.730-906), and the sensitivity and specificity were 76.30% and 72.30%, respectively. When used to distinguish 47 colorectal polyp patients and 47 patients with advanced CRC, the AUC was 0.875 (95%CI: 0.806-944), and the sensitivity and specificity were 81.80% and 72.30%, respectively.
| Indicator | AUC | Standard error | P value | 95%CI | |
| Lower | Upper | ||||
| MMP2 | 0.532 | 0.050 | 0.550 | 0.433 | 0.630 |
| MMP7 | 0.769 | 0.042 | < 0.001 | 0.688 | 0.851 |
| MMP9 | 0.737 | 0.046 | < 0.001 | 0.647 | 0.827 |
| CEA | 0.626 | 0.050 | 0.018 | 0.528 | 0.723 |
CRC is one of the most common cancers worldwide and a leading cause of death. Detection of CRC at an early stage can significantly reduce CRC mortality. Early diagnosis is particularly important for improving the survival and quality of life of CRC patients[14]. Colonoscopy is recognized as the gold standard for CRC screening due to its high sensitivity and specificity[15]. However, colonoscopy requires experienced endoscopic doctors and patient cooperation. With the development of molecular biotechnology, the detection and treatment of tumors has improved. At present, protein, DNA (mutation and methylation), RNA (primarily microRNA), volatile organic compounds, and intestinal microflora have been identified as potential early diagnostic markers of CRC[16-18]; however, there remain some limitations for their use in the early diagnosis of CRC[19]. A highly sensitive and specific, easily collected, and noninvasive or minimally invasive method is urgently needed for the early diagnosis of CRC.
CEA has been used in diagnosis, disease monitoring and treatment response of various gastrointestinal tumors, and patients with positive and negative serum CEA expression prior surgery exhibit significant differences in the incidence of lymph node metastasis, nerve invasion and TNM staging[20]. The rates of CEA positivity were 24%, 44%, 56% and 87% in patients stage I to IV CRC patients, respectively. Measurements of serum CEA levels can predict the disease status of CRC[21], including the tumor stage and presence of lymph node metastasis, and can provide guidance for clinical treatment and prognosis. In addition, CEA, as a common marker of CRC, can be used in combination with multiple indicators to improve the detection of CRC[22]. In this study, CEA levels in CRC increased significantly, which demonstrates its diagnostic value for CRC.
MMP9 is upregulated in macrophages of various types of tumors, primarily in the subpopulations of macrophages located at the edge of tumors, indicating that the specific expression of MMP9 in macrophages is directly related to cancer invasion. In addition, MMP9 has been reportedly linked to the development and progression of cancer, including but not limited to cancer invasion, metastasis and angiogenesis[23]. Furthermore, the value of MMP9 for tumor diagnosis, treatment response and disease progression has been studied in a variety of tumors[24,25]. When MMP9 is used alone as a biomarker, it may lack sufficient specificity for clinical application. The combination of biomarkers can improve the specificity of biomarkers. To achieve high specificity, MMP9 can be used in combination with other cancer biomarkers. With the development of statistical methods, bioinformatics and interdisciplinary research, a variety of multiparameter diagnostic models have been widely used in clinical diagnosis[26], and their diagnostic efficiency is superior to that of single indicator detection[27].
This study has limitations. First, our research team evaluated the diagnostic value of serum MMP9 for the detection of early-stage CRC. Compared with urine MMP, serum MMP9 exhibits less diagnostic value for early CRC, and the relationship between serum and urine MMP9 requires further study. Second, although a diagnostic model based on three indicators has been established, the model has not yet been verified using a large sample size. Third, although the diagnostic value of this model is superior to the conventional indicators CEA or CA199, our model may be combined with other potential biomarkers or artificial intelligence to establish a multi-indicator model with improved diagnostic value.
Compared with the commonly used indicator CEA, the diagnostic performance of a model combining CEA, MMP2, MMP7 and MMP9 levels was significantly improved and can be used as a potential diagnostic method for CRC.
Early diagnosis and early treatment are critical to improved colorectal cancer (CRC) diagnosis and treatment.
A noninvasive biomarker with high diagnostic performance is urgently needed for the early clinical diagnosis of CRC.
To evaluate the diagnostic value of matrix metalloproteinases (MMPs) 2, 7 and 9 for the early detection of CRC.
Serum carcinoembryonic antigen (CEA) and urine MMP2, MMP7, and MMP9 levels were measured in 59 healthy controls, 47 patients with colon polyps and 82 patients with CRC. The independent and combined diagnostic values of the indicators for the detection of CRC were compared.
A model for CRC detection using CEA, MMP2, MMP7 and MMP9 exhibited an area under the curve (AUC) of 0.977. For early-stage and advanced-stage CRC, the model AUCs were 0.975 and 0.979, respectively. To distinguish the colorectal polyp patients from the CRC patients, the model using CEA, MMP7 and MMP9 levels produced an AUC of 0.849. For early-stage and advanced-stage CRC, the AUCs were 0.818 and 0.875, respectively.
Compared with CEA alone, the diagnostic performance of a model combining CEA, MMP2, MMP7 and MMP9 established in this study was significantly improved.
Validation of the model built in our study using a larger sample size should be performed.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lunkka P, Finland; Topi S, Italy S-Editor: Wang JJ L-Editor: A P-Editor: Zhao S
| 1. | Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M, Saad A. Colorectal Cancer Epidemiology: Recent Trends and Impact on Outcomes. Curr Drug Targets. 2021;22:998-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 154] [Article Influence: 38.5] [Reference Citation Analysis (2)] |
| 2. | Chan SCH, Liang JQ. Advances in tests for colorectal cancer screening and diagnosis. Expert Rev Mol Diagn. 2022;22:449-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 42] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 3. | Betesh AL, Schnoll-Sussman FH. Colorectal Cancer Screening in the Elderly. Clin Geriatr Med. 2021;37:173-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 4. | Ladabaum U, Dominitz JA, Kahi C, Schoen RE. Strategies for Colorectal Cancer Screening. Gastroenterology. 2020;158:418-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 402] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 5. | Loo SW, Pui TS. Cytokine and Cancer Biomarkers Detection: The Dawn of Electrochemical Paper-Based Biosensor. Sensors (Basel). 2020;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 6. | Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat Rev Cancer. 2004;4:11-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1023] [Cited by in RCA: 1061] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 7. | Lech G, Słotwiński R, Słodkowski M, Krasnodębski IW. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J Gastroenterol. 2016;22:1745-1755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 234] [Cited by in RCA: 285] [Article Influence: 31.7] [Reference Citation Analysis (8)] |
| 8. | Li A, Wang WC, McAlister V, Zhou Q, Zheng X. Circular RNA in colorectal cancer. J Cell Mol Med. 2021;25:3667-3679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 9. | Lu DC, Zhang QF, Li L, Luo XK, Liang B, Lu YH, Hu BL, Jiang HX. Methylated Septin9 has moderate diagnostic value in colorectal cancer detection in Chinese population: a multicenter study. BMC Gastroenterol. 2022;22:232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 10. | Böckelman C, Beilmann-Lehtonen I, Kaprio T, Koskensalo S, Tervahartiala T, Mustonen H, Stenman UH, Sorsa T, Haglund C. Serum MMP-8 and TIMP-1 predict prognosis in colorectal cancer. BMC Cancer. 2018;18:679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 11. | Sun DW, Zhang YY, Qi Y, Zhou XT, Lv GY. Prognostic significance of MMP-7 expression in colorectal cancer: a meta-analysis. Cancer Epidemiol. 2015;39:135-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 12. | Tong WH, Mu JF, Zhang SP. LINC00346 accelerates the malignant progression of colorectal cancer via competitively binding to miRNA-101-5p/MMP9. Eur Rev Med Pharmacol Sci. 2020;24:6639-6646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 13. | Yueh TC, Wu CN, Hung YW, Chang WS, Fu CK, Pei JS, Wu MH, Lai YL, Lee YM, Yen ST, Li HT, Tsai CW, Bau DT. The Contribution of MMP-7 Genotypes to Colorectal Cancer Susceptibility in Taiwan. Cancer Genomics Proteomics. 2018;15:207-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 14. | Issa IA, Noureddine M. Colorectal cancer screening: An updated review of the available options. World J Gastroenterol. 2017;23:5086-5096. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 434] [Cited by in RCA: 391] [Article Influence: 48.9] [Reference Citation Analysis (11)] |
| 15. | Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1570] [Cited by in RCA: 3004] [Article Influence: 500.7] [Reference Citation Analysis (3)] |
| 16. | Jung G, Hernández-Illán E, Moreira L, Balaguer F, Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17:111-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 542] [Cited by in RCA: 516] [Article Influence: 103.2] [Reference Citation Analysis (0)] |
| 17. | Zhang N, Hu X, Du Y, Du J. The role of miRNAs in colorectal cancer progression and chemoradiotherapy. Biomed Pharmacother. 2021;134:111099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 91] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 18. | Liu W, Zhang R, Shu R, Yu J, Li H, Long H, Jin S, Li S, Hu Q, Yao F, Zhou C, Huang Q, Hu X, Chen M, Hu W, Wang Q, Fang S, Wu Q. Study of the Relationship between Microbiome and Colorectal Cancer Susceptibility Using 16SrRNA Sequencing. Biomed Res Int. 2020;2020:7828392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 19. | Eng C, Jácome AA, Agarwal R, Hayat MH, Byndloss MX, Holowatyj AN, Bailey C, Lieu CH. A comprehensive framework for early-onset colorectal cancer research. Lancet Oncol. 2022;23:e116-e128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 104] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 20. | Duffy MJ, Lamerz R, Haglund C, Nicolini A, Kalousová M, Holubec L, Sturgeon C. Tumor markers in colorectal cancer, gastric cancer and gastrointestinal stromal cancers: European group on tumor markers 2014 guidelines update. Int J Cancer. 2014;134:2513-2522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 248] [Article Influence: 22.5] [Reference Citation Analysis (1)] |
| 21. | Grotowski M. [Antigens (CEA and CA 19-9) in diagnosis and prognosis colorectal cancer]. Pol Merkur Lekarski. 2002;12:77-80. [PubMed] |
| 22. | Fakih MG, Padmanabhan A. CEA monitoring in colorectal cancer. What you should know. Oncology (Williston Park). 2006;20:579-87; discussion 588, 594, 596 passim. [PubMed] |
| 23. | Zhao C, Yuan G, Jiang Y, Xu J, Ye L, Zhan W, Wang J. Capn4 contributes to tumor invasion and metastasis in gastric cancer via activation of the Wnt/β-catenin/MMP9 signalling pathways. Exp Cell Res. 2020;395:112220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Xu Y, Yang Q, Fang Z, Tan X, Zhang M, Chen W. TRIM66 Promotes Malignant Progression of Non-Small-Cell Lung Cancer Cells via Targeting MMP9. Comput Math Methods Med. 2022;2022:6058720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 25. | Chen Y, Jiang T, Mao A, Xu J. Esophageal cancer stem cells express PLGF to increase cancer invasion through MMP9 activation. Tumour Biol. 2014;35:12749-12755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Muinao T, Deka Boruah HP, Pal M. Multi-biomarker panel signature as the key to diagnosis of ovarian cancer. Heliyon. 2019;5:e02826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 27. | Thrift AP, Garcia JM, El-Serag HB. A multibiomarker risk score helps predict risk for Barrett's esophagus. Clin Gastroenterol Hepatol. 2014;12:1267-1271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |