Published online Mar 27, 2023. doi: 10.4240/wjgs.v15.i3.346
Peer-review started: December 12, 2022
First decision: January 5, 2023
Revised: January 17, 2023
Accepted: February 16, 2023
Article in press: February 16, 2023
Published online: March 27, 2023
Processing time: 105 Days and 4.4 Hours
The relationship between hepatitis B surface antigen (HBsAg)-positive carrier status and liver cancer has been extensively studied. However, the epigenetic changes that occur during progression from HBsAg-positive carrier status or cirrhosis to liver cancer are unknown. The epigenetic modification of DNA hydroxymethylation is critical in tumor development. Further, 5-hydroxymethylcytosine (5hmC) is an important base for DNA demethylation and epigenetic regulation. It is also involved in the assembly of chromosomes and the regulation of gene expression. However, the mechanism of action of 5hmC in HBsAg-positive carriers or patients with cirrhosis who develop liver cancer has not been fully elucidated.
To investigate the possible epigenetic mechanism of HBsAg-positive carriers and hepatocellular carcinoma (HCC) progression from cirrhosis.
Forty HBsAg-positive carriers, forty patients with liver cirrhosis, and forty patients with liver cancer admitted to the First People's Hospital of Yongkang between March 2020 and November 2021 were selected as participants. Free DNA was extracted using a cf-DNA kit. cfDNA was extracted by 5hmC DNA se
A total of 16455 hydroxymethylated genes were identified. Sequencing results showed that 32 genes had significant 5hmC modification differences between HBsAg carriers and liver cancer patients, of which 30 were upregulated and 2 downregulated in patients with HCC compared with HBsAg-positive carriers. Significant 5hmC modification differences between liver cirrhosis and liver cancer patients were identified in 20 genes, of which 17 were upregulated and 3 were downregulated in patients with HCC compared with those with cirrhosis. These genes may have potential loci that are undiscovered or unelucidated, which contribute to the development and progression of liver cancer. Analysis of gene ontology enrichment and Kyoto Encyclopedia of Genes and Genomes showed that the major signaling pathways involved in the differential genes were biliary secretion and insulin secretion. The analysis of protein interactions showed that the important genes in the protein-protein interaction network were phosphoenolpyruvate carboxykinase and solute carrier family 2.
The occurrence and development of liver cancer involves multiple genes and pathways, which may be potential targets for preventing hepatitis B carriers from developing liver cancer.
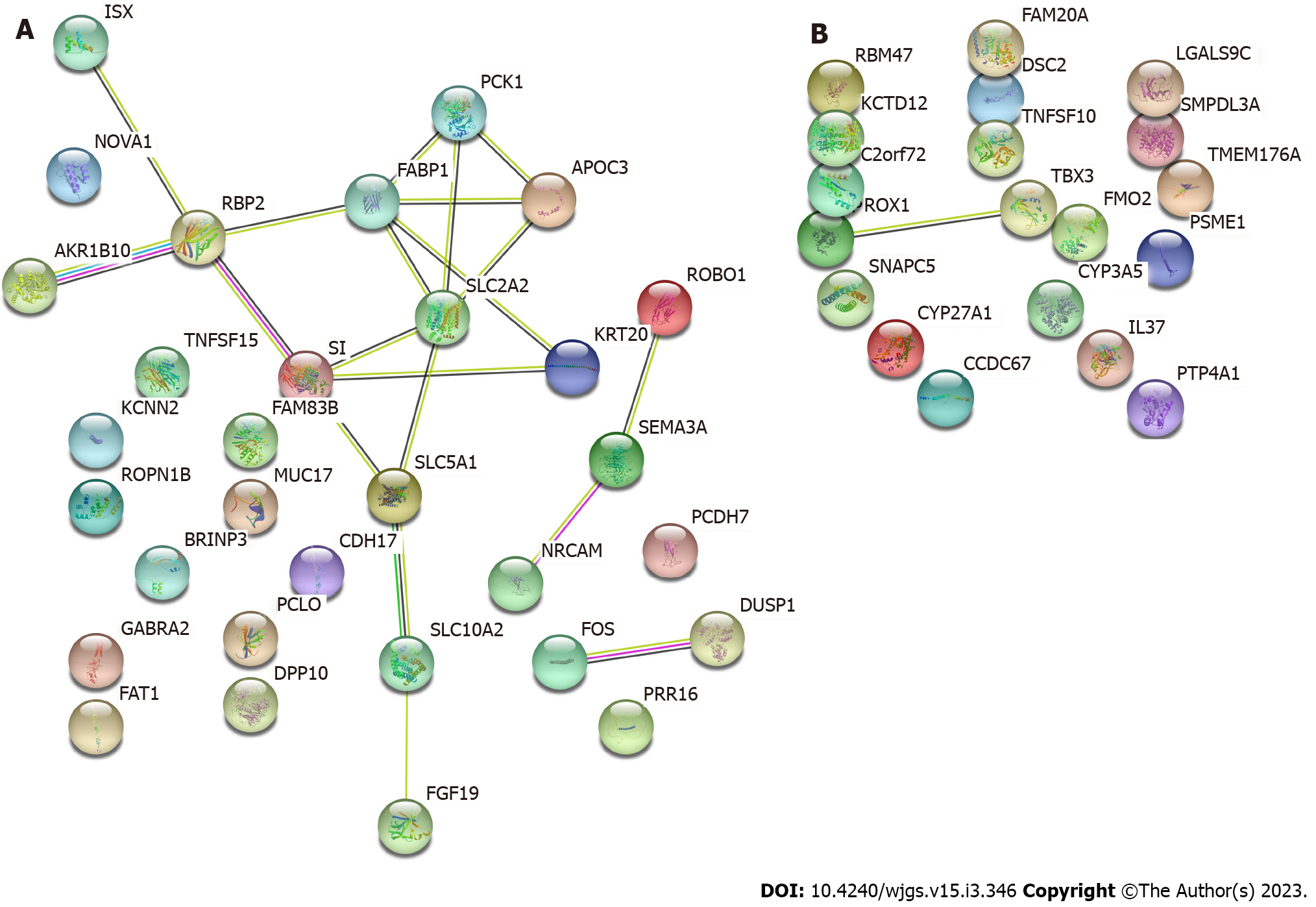
Core Tip: Major signaling pathways involved in differentially expressed genes are biliary secretion and insulin secretion. Abnormal secretion of bile and insulin in tumor cells may promote or symbolize the occurrence and development of liver cancer. SLC2A2 and PCK1 are the central nodes of the differential genes, which may be most closely related to the occurrence of liver cancer. FABP1, APOC3, SI, KRT20, SLC5A1, SLC10A2, RBP2, and AKR1B10 may be key genes in the protein regulatory network, and these genes may play regulatory roles after modification by 5-hydroxymethylcytosine (5hmC). Therefore, we suggest that the difference in 5hmC modification levels is related to the occurrence and progression of liver cancer.
- Citation: Li YC, Hu WY, Li CH, Zhang LL, Xu XW, Li J, Luo HX. Differential expression and significance of 5-hydroxymethylcytosine modification in hepatitis B virus carriers and patients with liver cirrhosis and liver cancer. World J Gastrointest Surg 2023; 15(3): 346-361
- URL: https://www.wjgnet.com/1948-9366/full/v15/i3/346.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v15.i3.346
Primary liver cancer, mainly hepatocellular carcinoma (HCC), is a solid tumor generated from the malignant transformation of hepatocytes or intrahepatic bile duct cells and is one of the most common malignancies in China[1]. Additionally, the number of hepatitis B virus (HBV) carriers in China is high. If patients develop an infection, chronic HBV infection, liver cirrhosis, and even HCC may occur. The relationship between hepatitis B surface antigen (HBsAg)-positive carriers and liver cancer has been reported in many studies[2]. There is consensus that liver cirrhosis can progress to liver cancer. Although great progress has been made in the prevention and treatment of hepatitis B in China, including a reduction in the transmission rate of HBV, effective treatments for HBV carriers are still lacking. Drugs such as nucleotide analogs, including entecavir, cannot rapidly reduce HBV titers in the short term. Additionally, administration of these drugs to HBV carriers remains controversial in clinical practice[3]. At present, the mechanisms underlying the transformation from the HBsAg-positive state or liver cirrhosis to liver cancer is not entirely clear, especially the potential mechanism of epigenetic changes in the occurrence and progression of these diseases. Therefore, the differences in epigenetic modifications between the HBsAg-positive state, liver cirrhosis, and liver cancer must be explored.
DNA hydroxymethylation is a process in which 5-methylcytosine is oxidized to 5-hydroxymethylcytosine (5hmC) by ten-eleven translocation enzymes during DNA methylation[4]. Further, 5hmC is an important base in DNA demethylation as well as in epigenetic regulation. Some studies suggest that 5hmC is the sixth genome base and is involved in the assembly of chromosomes and regulation of gene expression[5]. Studies have shown that the level of hydroxymethylation in tumors and other diseases is significantly different from that in the normal state, and the difference gradually increases with the progression of diseases, indicating that the epigenetic modification of hydroxymethylation is essential in the development of tumors[6]. However, the mechanism of 5hmC in HBsAg-positive carriers or patients with cirrhosis that develops into liver cancer has not yet been fully elucidated. 5hmC sequencing is a new technology that constructs free nucleic acid fragments containing 5hmC using polymerase chain reaction (PCR) technology and detects the 5hmC sequence using second-generation DNA sequencing technology. Because the number of 5hmC modifications is far less than the number of DNA methylations, 5hmC sequencing has a higher cost performance, efficiency, accuracy, and economy, as well as extensive clinical application value. In this study, we analyzed differentially expressed genes (DEGs) in HBsAg-positive carriers, patients with liver cirrhosis, and patients with liver cancer using 5hmC sequencing technology and further annotated the function of DEGs to explore the similarities and differences in gene hydroxymethylation between HBsAg-positive carriers, patients with liver cirrhosis, and patients with liver cancer and the regulatory role of signaling pathways.
Forty HBsAg-positive carriers, forty patients with liver cirrhosis, and forty patients with liver cancer admitted to the First People's Hospital of Yongkang between March 2020 and November 2021 were selected as participants. This study was approved by the medical ethics committee of the hospital. The diagnostic criteria for liver cirrhosis were based on the International Guidelines for the Diagnosis and Treatment of Liver Cirrhosis updated in 2021[7]. There were no differences in baseline data, such as sex and age, between the three groups of patients. Inclusion criteria for primary liver cancer were as follows[8]: Body mass index of 20–30 kg/m2 in men and 19–34 kg/m2 in women; complete results of general biochemical indexes such as liver function; imaging data such as tumor computed tomography/magnetic resonance imaging and serological indexes; a history of hepatitis B; no history of diabetes or other systemic diseases; no history of other infectious diseases; and no pregnancy or lactation.
cfDNA extraction: Blood samples from HBsAg-positive carriers, patients with liver cirrhosis, and patients with liver cancer were collected at the time of diagnosis and sent to Zhongke Jinzhen Co., Ltd. for DNA extraction, pyrolysis of specimen precipitate after centrifugation, and extraction of cfDNA from plasma using the Quick-cfDNA Serum & Plasma Kit (Zymo).
DNA quality test and results: Qubit accurately quantified the DNA concentration, and Q-sep analyzed the size, distribution, and relative quantification of DNA fragments. After the sample was quantified, the DNA of the sample was first repaired, a tail was added to the 3' end, and the sequencing joint was connected. Biotins were connected by a transglycosylation reaction and click chemistry. The DNA fragment containing 5hmC was captured using streptavidin beads, and PCR amplification was performed to complete the entire library construction. After the library quality inspection, different libraries were sequenced using Nova6000 according to the effective concentration and target data volume. After obtaining the original sequence (sequenced reads), the original gene fragments were returned to the correct human reference genome position to calculate the gene expression of each sample.
Principal component analysis (PCA) is an unsupervised multivariate statistical analysis method that can generally reflect the overall differences between samples and the variation between samples within the group. It is commonly used to evaluate sample differences between groups and consistency within groups. PCA was used for dimensionality reduction of all 5hmC gene sequencing results. After data visualization, outlier samples or sample clusters with high similarity were identified.
Through the analysis of the significant difference in the 5hmC modification expression matrix of all samples, functional gene modification differences among HBsAg-positive carriers, patients with liver cirrhosis, and patients with liver cancer were found. In this study, the differences in 5hmC expression were analyzed using normalization, discrete estimation, and significance tests. Normalization and discrete estimation mainly remove the influence of sample sequencing depth differences and reduce the false-positive rate through homogenization. DEGs were further detected using DEseq2 and PossionDis algorithms. DEGs were screened according to the log2FoldChange ≥ 0.26, gene 5hmC modified expression value, and adjusted P value ≤ 0.05. The sample classification of this analysis included three groups: HBsAg-positive carriers, patients with liver cirrhosis, and patients with liver cancer, with 40 patients in each group. In the three types of samples, the differences between the liver cirrhosis group and liver cancer group, HBsAg-positive carrier group, and liver cancer patient group were analyzed. Lists of all differential genes are in the original data.
Based on the results of gene ontology (GO) and gene pathway annotation, the DEGs were enriched and classified according to the aforementioned functions and pathways. GO enrichment analysis was used for functional classification, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was used for pathway classification. GO enrichment analysis was performed from three aspects: cellular component (CC), biological process (BP), and molecular function (MF). KEGG is a database used for analyzing gene functions, linking multiple genes with specific pathways and functions, and classifying genes according to the pathways involved. STRING (https://string-db.org/) is currently the largest protein interaction database. DEGs were imported into the STRING database, and the homology, literature, and co-expression relationship of known proteins in the database were used to determine the interaction between the imported encoded proteins to construct a protein-protein interaction (PPI) network analysis.
Pearson correlation analysis was performed between each pair of samples using the Cor function in R software; P < 0.05 was considered statistically significant. The expression of 5hmC modified genes obeyed a Poisson distribution after standardization. The P value was calculated using the Wald test and corrected using multiple hypothesis tests. Statistical significance was set at P < 0.05.
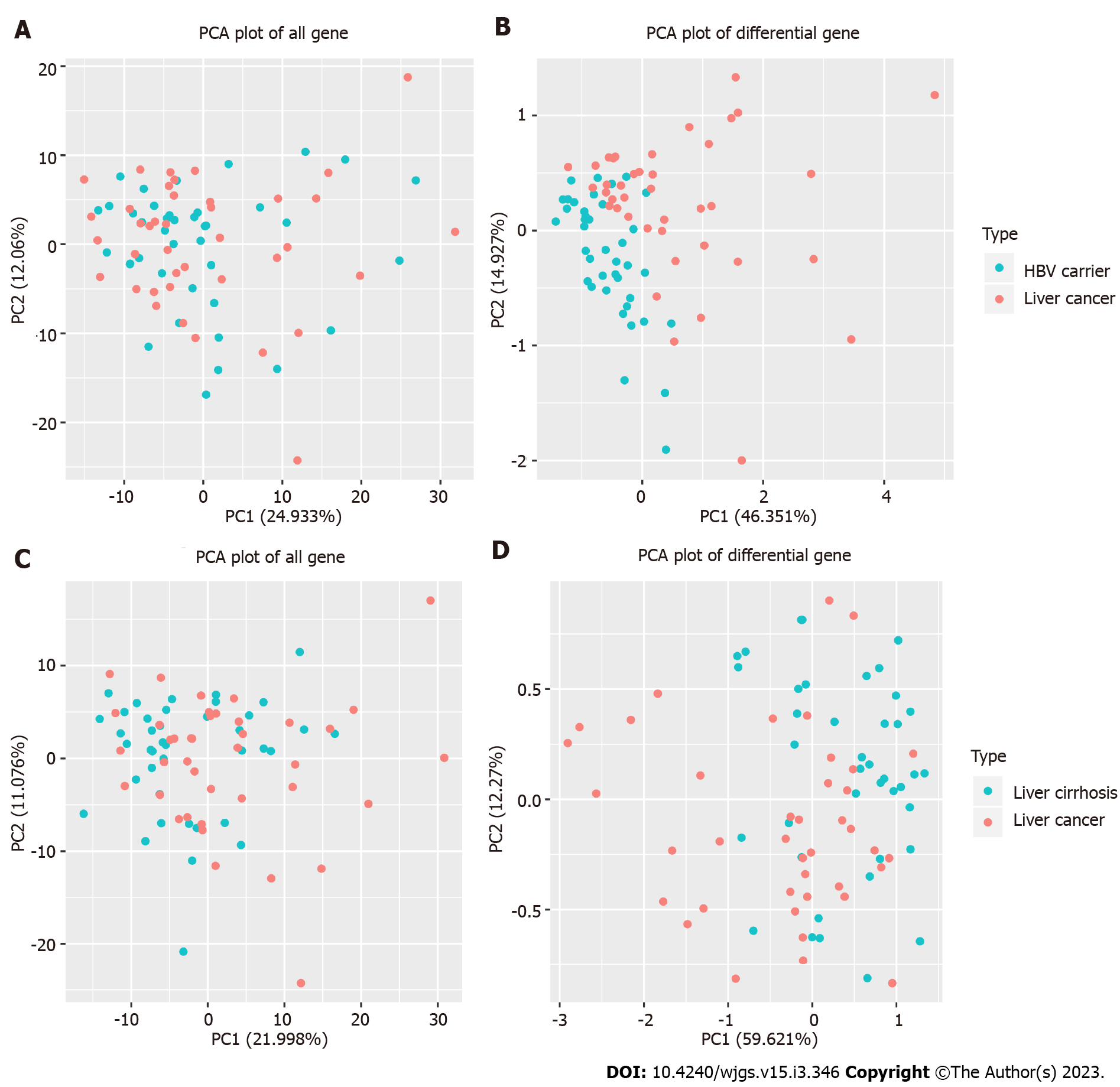
The Nova6000 platform was used to measure samples from the three groups of patients. Each group contained 40 patients, and 16455 genes were detected. The percentages of principal components (PC) 1 and 2 in the PCA plot of all genes in HBsAg-positive carriers and patients with liver cancer were 24.933% and 12.06% (Figure 1A), respectively. The percentages of PC1 and PC2 in the PCA plot of differential genes were 46.351% and 14.927% (Figure 1B), respectively, suggesting that differential genes can distinguish HBsAg-positive carriers from patients with liver cancer.
Similarly, when detecting the same number of genes, the percentages of PC1 and PC2 in the PCA plot of all genes in patients with liver cirrhosis and liver cancer were 21.998% and 11.076% (Figure 1C), respectively, and the percentages of PC1 and PC2 in the PCA plot of differential genes were 59.621% and 12.27% (Figure 1D), respectively, suggesting that differential genes have the ability to distinguish between liver cirrhosis and liver cancer.
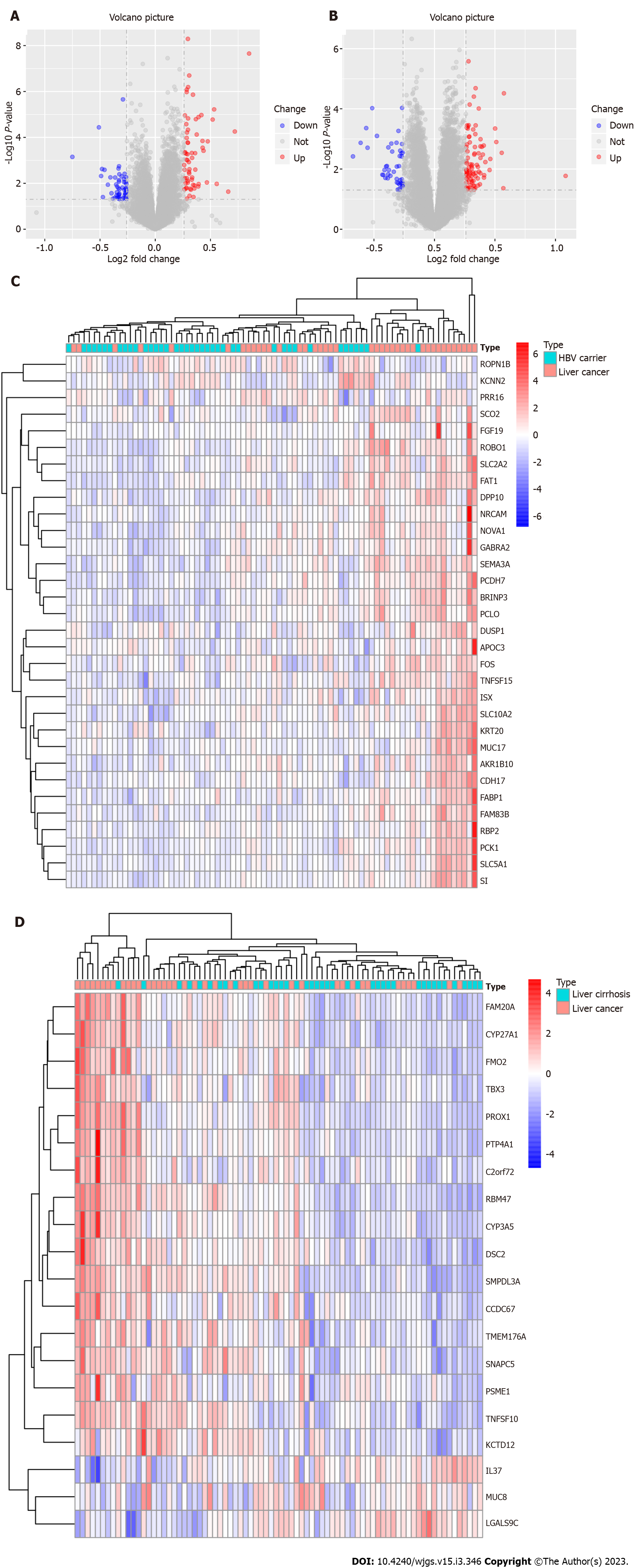
Differences between patients with liver cirrhosis and those with liver cancer and differences between HBsAg-positive carriers and patients with liver cancer were analyzed. According to the levels of 5hmC modified genes in each sample, DEGs between HBsAg-positive carriers and patients with liver cirrhosis or liver cancer were screened. The results showed 32 DEGs between HBsAg-positive carriers and patients with liver cancer, of which 30 were upregulated and 2 genes were downregulated by 5hmC modification. The distribution of DEGs is shown by a volcanic and thermal map in Figure 2A and C. All DEGs upregulated and downregulated in HBsAg-positive carriers and liver cancer groups are shown in Table 1. There were 20 DEGs between patients with liver cirrhosis and those with liver cancer, of which 17 were upregulated and 3 genes were downregulated by 5hmC modification. The distribution of the DEGs is shown by a volcanic and thermal map (Figure 2B and D). All DEGs that were upregulated and downregulated in patients with liver cirrhosis and liver cancer are shown in Table 2.
| Symbol | Ensembl | log2FoldChange | padj |
| SI | ENSG00000090402 | 0.849003917 | 0.000172596 |
| FAM83B | ENSG00000168143 | 0.532487323 | 0.00425853 |
| PCK1 | ENSG00000124253 | 0.519299173 | 0.006848676 |
| KCNN2 | ENSG00000080709 | -0.510730422 | 0.011078186 |
| FAT1 | ENSG00000083857 | 0.47499449 | 0.02542581 |
| FGF19 | ENSG00000162344 | 0.428605733 | 0.036854977 |
| FABP1 | ENSG00000163586 | 0.427372957 | 0.025268146 |
| SLC10A2 | ENSG00000125255 | 0.417559665 | 0.005504835 |
| ISX | ENSG00000175329 | 0.391889896 | 0.005936182 |
| APOC3 | ENSG00000110245 | 0.374978715 | 0.017711634 |
| NOVA1 | ENSG00000139910 | 0.367147827 | 0.006848676 |
| KRT20 | ENSG00000171431 | 0.361001798 | 0.049694505 |
| DUSP1 | ENSG00000120129 | 0.341097702 | 0.001523335 |
| RBP2 | ENSG00000114113 | 0.330491105 | 0.049694505 |
| PCLO | ENSG00000186472 | 0.307272826 | 0.000520372 |
| BRINP3 | ENSG00000162670 | 0.297534011 | 0.001107995 |
| SLC2A2 | ENSG00000163581 | 0.297032232 | 0.006630228 |
| AKR1B10 | ENSG00000198074 | 0.296134756 | 0.012660193 |
| SEMA3A | ENSG00000075213 | 0.295498456 | 7.92E-05 |
| SLC5A1 | ENSG00000100170 | 0.295343232 | 0.027331533 |
| ROBO1 | ENSG00000169855 | 0.294408057 | 0.026805139 |
| ROPN1B | ENSG00000114547 | -0.292505301 | 0.002291301 |
| PRR16 | ENSG00000184838 | 0.291799543 | 0.049694505 |
| TNFSF15 | ENSG00000181634 | 0.286998557 | 0.01533013 |
| CDH17 | ENSG00000079112 | 0.284940571 | 0.001204498 |
| MUC17 | ENSG00000169876 | 0.284637524 | 0.022168549 |
| PCDH7 | ENSG00000169851 | 0.284173648 | 0.00131498 |
| FOS | ENSG00000170345 | 0.28075648 | 0.008754834 |
| GABRA2 | ENSG00000151834 | 0.27973066 | 0.049694505 |
| NRCAM | ENSG00000091129 | 0.270536808 | 0.047702973 |
| DPP10 | ENSG00000175497 | 0.268657512 | 0.005504835 |
| Symbol | Ensembl | log2FoldChange | padj |
| MUC8 | NA | -0.61023 | 0.048721 |
| PROX1 | ENSG00000117707 | 0.510971 | 0.047662 |
| TBX3 | ENSG00000135111 | 0.380857 | 0.04644 |
| DSC2 | ENSG00000134755 | 0.366222 | 0.021397 |
| PSME1 | ENSG00000092010 | 0.340336 | 0.036054 |
| C2orf72 | ENSG00000204128 | 0.339959 | 0.015473 |
| PTP4A1 | ENSG00000112245 | 0.33768 | 0.043273 |
| KCTD12 | E0NSG00000178695 | 0.325745 | 0.016818 |
| FMO2 | ENSG00000094963 | 0.320229 | 0.040782 |
| LGALS9C | ENSG00000171916 | -0.29776 | 0.040782 |
| CYP3A5 | ENSG00000106258 | 0.292488 | 0.048557 |
| SNAPC5 | ENSG00000174446 | 0.290892 | 0.020989 |
| TMEM176A | ENSG00000002933 | 0.285556 | 0.04535 |
| SMPDL3A | ENSG00000172594 | 0.283335 | 0.036054 |
| TNFSF10 | ENSG00000121858 | 0.281624 | 0.009552 |
| CYP27A1 | ENSG00000135929 | 0.275649 | 0.038131 |
| FAM20A | ENSG00000108950 | 0.275255 | 0.040338 |
| CCDC67 | NA | 0.268828 | 0.036054 |
| IL37 | ENSG00000125571 | -0.26789 | 0.021397 |
| RBM47 | ENSG00000163694 | 0.267243 | 0.038605 |
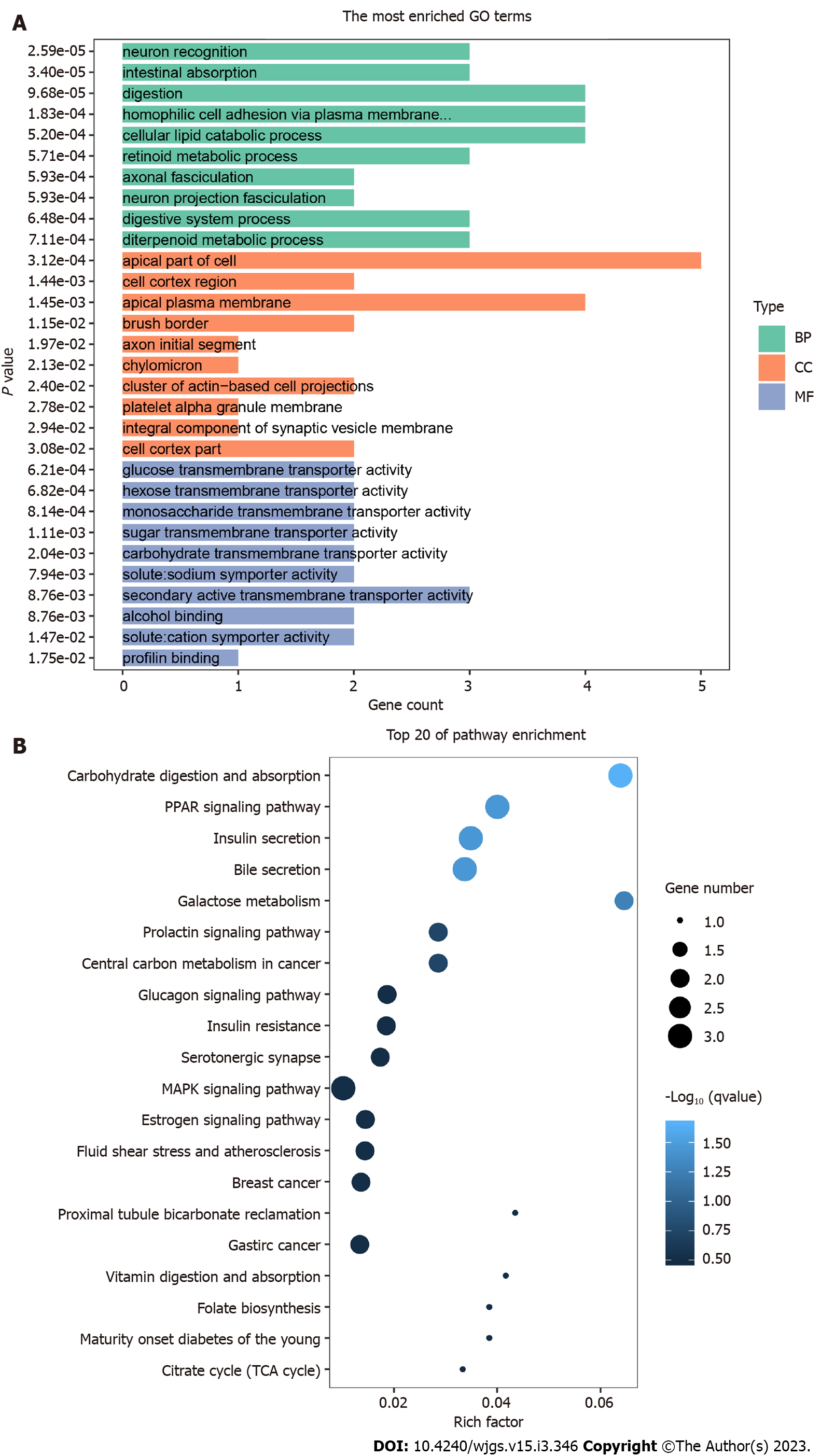
According to the results of DEG detection, GO enrichment analysis was used for the functional annotation of the 5hmC sequences. After the difference analysis between HBsAg carrier and liver cancer groups, the top 10 enriched GO terms related to BP, CC, and MF were used as the main functions of differential hydroxymethylation genes between HBsAg carriers and patients with liver cancer. The main pathways involved are shown in Figure 3A. The numbers of genes involved in the GO BP of TOP3 were 4, 4, and 4, respectively, which were mainly related to digestion, homophilic cell adhesion via plasma membrane adhesion membrane, and cellular lipid catabolic processes. The numbers of genes included in the MF of TOP3 were 3, 2, and 2, respectively, which were related to the activities of secondary active transmembrane transporter activity, alcohol binding, and solute:cation symporter activity. The numbers of genes in TOP3 cell components were 5, 4, and 2, respectively, which were mainly related to the apical part of the cell, apical plasma membrane, and cell cortex region. These three kinds of gene function together suggest that the main differences between HBsAg-positive carriers and patients with liver cancer may be digestive function and cell information transmission.
From the perspective of pathway enrichment, 92 pathways were found to be closely related to KEGG enrichment in HBsAg carriers and patients with liver cancer. The main pathways are shown in Figure 3B. Among them, the number of pathways related to digestive secretion, such as carbohydrate digestion and absorption, insulin and bile secretion, galactose metabolism, and the glucagon signaling pathway, were most significant. The peroxisome proliferator-activated receptor and mitogen-activated protein kinase (MAPK) signaling pathways were also significantly enriched in differentially hydroxymethylated genes. These results indicate that the main difference between HBsAg carriers and patients with liver cancer lies in their digestive function. Soluble carrier family 2 member 2 (SLC2A2) and Fos proto-oncogene, AP-1 transcription factor subunit (FOS) were involved in the most enriched pathways. SLC2A2 is mainly enriched in insulin and bile secretion, the prolactin signaling pathway, and central carbon metabolism in cancer. FOS is mainly enriched in the prolactin, MAPK, and B-cell receptor signaling pathways and the PD-L1/PD-1 pathway, suggesting that SLC2A2 is related to liver-digestive function and FOS is related to immune signal transduction.
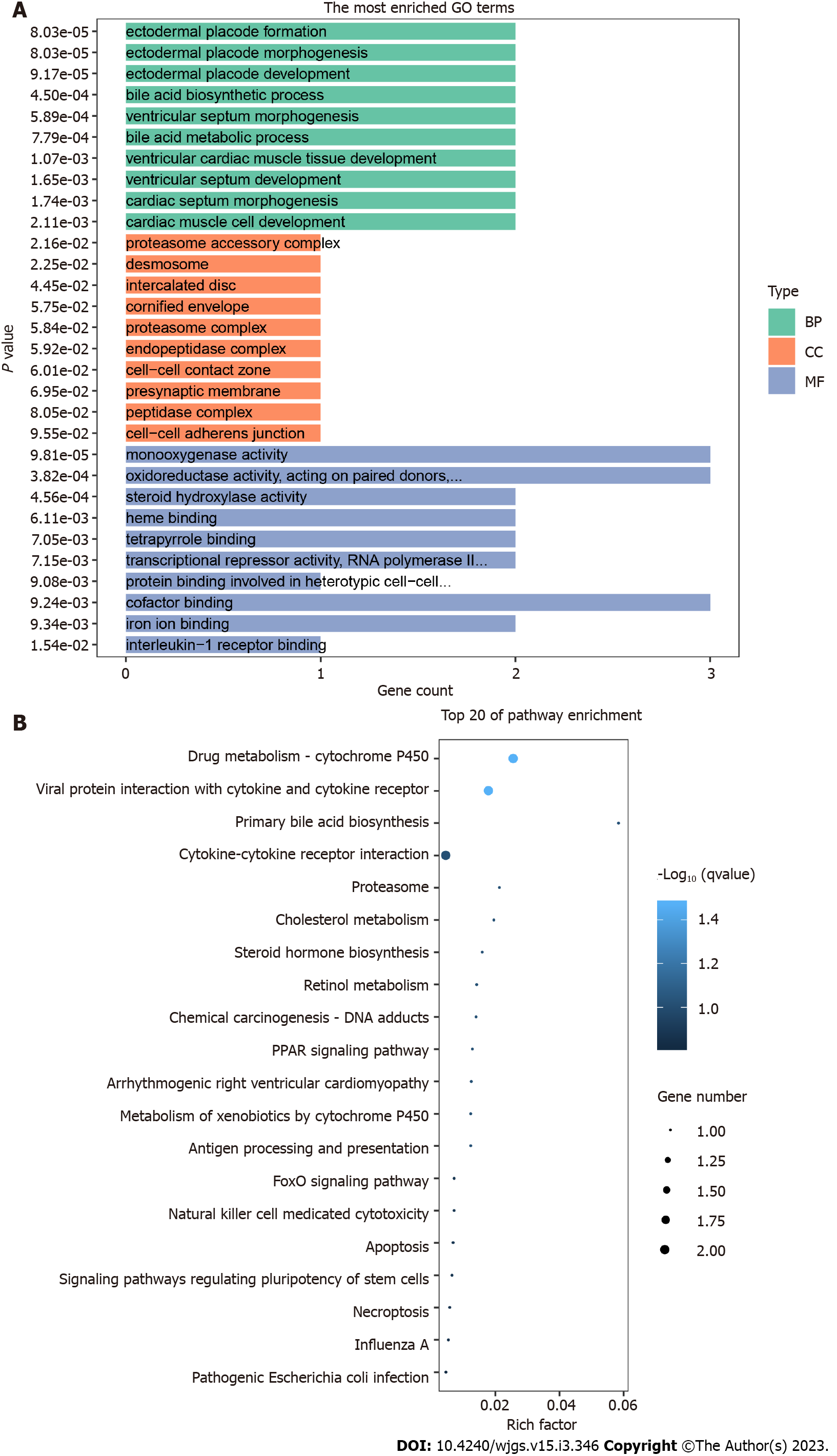
After the difference analysis between the liver cirrhosis and liver cancer groups, the main pathways involved in the GO enrichment analysis were determined, as shown in Figure 4A. Among them, the numbers of genes involved in the GO BP of TOP3 were 2, 2, and 2, respectively, which were mainly related to the bile acid biosynthetic process, ectodermal placode development, and ventricular cardiac muscle tissue development. The numbers of genes included in the MF of TOP3 were 3, 3 and 3, respectively, which were related to monooxygenase activity, oxidoreductase activity, acting on paired donors, and cofactor binding. The numbers of genes contained in the TOP3 cell components were 1, 1, and 1, respectively, which were mainly related to the hydrolysis of short peptides, ubiquitination and protein synthesis. These three types of gene functions jointly suggest that the main differential functions between liver cirrhosis and liver cancer may be bile acid metabolism and oxidoreductase activity.
The KEGG enrichment results of the liver cirrhosis and liver cancer groups differed from those of the above enrichment pathways. The results showed that 22 pathways were closely related; the main pathways involved are shown in Figure 4B. Among them, pathways related to the metabolism of various biological components were enriched, but specific pathways were different, such as primary bile acid biosynthesis, cholesterol metabolism, retinol metabolism, and drug metabolism (cytochrome P450), as well as cell death-related signaling pathways such as cytotoxicity, apoptosis, and necrotizing apoptosis mediated by natural killer cells, which were also significantly enriched in differentially hydroxymethylated genes. These results indicate that the main difference between liver cirrhosis and liver cancer is first reflected in the metabolic disorders of various active substances, and that the death behavior of tumor cells is different from that of non-tumor cells.
Proteins usually perform biological functions by binding to one another. According to the https://string-db.org/ STRING protein interaction database, DEGs were analyzed using PPI, and the results are shown in Figure 5A, which shows the PPI network of DEGs in HBsAg carriers and patients with liver cancer. PCK1 and SLC2A2 were most closely related to each protein. Protein interactions were mainly concentrated between genes such as FABP1, APOC3, SI, KRT20, SLC5A1, SLC10A2, RBP2, and AKR1B10; Figure 5B displays the PPI network of patients with liver cirrhosis and those with liver cancer. It shows that only PROX1 and TBX3 have potential protein interactions, and the relationship between other proteins is unclear.
China has the most frequent occurrences of liver cancer, accounting for 47% of cases globally. Liver cancer ranks third among malignant tumors in China. In 2018, the number of newly diagnosed patients with liver cancer in China was 390000, and the number of deaths was 360000. Therefore, it remains a major threat to the health of the Chinese population[9]. Although many basic and clinical studies have explored the mechanisms of liver cancer formation and development, it remains unclear, and there are limited studies on epigenetic modifications in the occurrence and development of liver cancer, especially on the regulation of hydroxymethylation. To further define the potential relationship between HBsAg carrier status, liver cirrhosis, and liver cancer in the epigenetic modification of hydroxymethylation, serum samples from 40 HBsAg carriers, 40 patients with liver cirrhosis, and 40 patients with liver cancer were collected in our hospital. The bioinformatics method of 5hmC sequencing was used to conduct an in-depth analysis of these datasets, and DEGs were screened for 5hmC; the genes and pathways closely related to them were identified through bioinformatic analysis.
In this study, 16455 hydroxymethylated genes were identified. Sequencing showed that 32 genes had significant differences in 5hmC modifications between HBsAg carriers and patients with liver cancer, and 20 genes had significant differences in 5hmC modifications between patients with liver cirrhosis and liver cancer. These genes may have potential loci that have not been discovered or clearly studied, leading to the occurrence and development of liver cancer, which is congruent with the findings of previous studies on epigenetic modifications[10]. Studies have shown that multiple DEGs of 5hmC are upregulated and downregulated in HBsAg carriers, patients with liver cirrhosis, and patients with liver cancer, involving multiple cellular signaling pathways and biological processes[11].
GO enrichment analysis showed that 32 DEGs were enriched in both HBsAg carriers and patients with liver cancer, indicating that liver cancer progression is the result of multiple pathways. For example, in GO BP analysis, the digestion and cellular lipid catabolic processes are enriched; digestion is one of the most important functions of the liver, and lipid catabolism is part of the digestive link and is responsible for one of the most basic biological processes[12]. In GO MF, the enrichment of secondary active transmembrane transporters and cation symporter activity was the most obvious, indicating that transmembrane transporters and ion transporters are involved in the digestive function of liver cells and may affect the progression of liver cancer[13]. In GO CC analysis, the apical part of the cell and apical plasma membrane were enriched, suggesting that the interaction between cells mainly occurs on the cell membrane[14]. Interestingly, in the GO BP analysis of the liver cirrhosis and liver cancer groups, the most obvious enrichment was bile acid biosynthesis and metabolic processes. Bile acid can increase the contact area of lipase by emulsification of fat, improve the activity of pancreatic and lipoprotein lipases, and promote intestinal fat transport to promote fat metabolism[15]. In the GO MF analysis, oxidation-related enzymes were enriched, indicating that the redox function of liver cirrhosis was further affected after liver cancer progression, which is consistent with the results of Chen et al[16]. In GO CC analysis, the enrichment of the proteasome complex further supports the results of these molecular functions. Minor et al[17] also pointed out that HBV proteins can promote the occurrence and development of HCC by forming a ubiquitinated proteasome complex.
KEGG analysis showed that the DEGs between HBsAg carriers and liver cancer groups had the largest number of genes enriched in the series of digestive and metabolic pathways, and the genes enriched in liver digestive function-related pathways accounted for the largest proportion of DEGs. Most genes are involved in digestive processes such as insulin and bile secretion, galactose metabolism, and the glucagon signaling pathway. In addition to these pathways, DEGs in the liver cirrhosis and liver cancer groups were also related to death processes, such as apoptosis and necrotizing apoptosis. Normal liver cells often develop abnormal metabolic behavior during their gradual development into tumor cells. This abnormal behavior affects the normal function of the liver, which can be considered a progressive feature of liver cancer. However, most previous studies concluded that no obvious pathological changes appeared in the liver of HBV carriers without disease. A few studies pointed out that the secretion of digestive juice in HBsAg-positive carriers might be related to the occurrence and development of liver cancer. An increasing number of researchers believe that the normal transaminase levels of HBsAg-positive carriers do not indicate fibrosis or inflammatory necrosis in the liver, which may also show different degrees of hepatocyte degeneration, necrosis, and even fibrosis[18]. In the liver tissue of HBV carriers with normal alanine aminotransferase (ALT) levels and without treatment, the proportion of liver inflammation grade G2-3 was 25.0%, which was higher than that in the group with a slight increase in ALT, and HBV carriers were accompanied by different degrees of liver fibrosis[18]. Studies have shown that insulin inhibits HBsAg expression in hepatocytes[19]. Moreover, the presence of insulin resistance-related diseases during entecavir treatment is associated with a slower decline in HBsAg expression[20]. Researchers have found that, while lovastatin is used to block the p21Ras signaling pathway of insulin, it also inhibits the secretion of HBsAg in Hep3B cells, which may be caused by the instability of lipid rafts due to the depletion of cholesterol from the membrane[21]. Therefore, it is reasonable to believe that changes in liver digestive function-related pathways are closely related to the progression of liver cancer.
PPI analysis showed that protein interactions were mainly concentrated among genes with PCK1 and SLC2A2 as the central nodes, and protein interactions were mainly concentrated among genes such as FABP1, APOC3, SI, KRT20, SLC5A1, SLC10A2, RBP2, and AKR1B10. Most of them are involved in the regulation of cellular insulin, glucocorticoids, glucagon, and other signaling pathways and play a key role in glucose metabolism and gluconeogenesis. The PCK1 gene, encoding cytosolic phosphoenolpyruvate carboxykinase (PEPCK-C), functions as a glucoamylase in the liver and kidneys[22]. Studies have found that the type 2 diabetes phenotypes, such as obesity, fat malnutrition, fatty liver, and even death in serious circumstances, appear in mice after knockout of systemic and tissue-specific PCK1 genes[23]. In this study, PCK1 was highly expressed in liver cancer tissues (log2FC = 0.519299173). Considering that the 5hmC modification of PCK1 may affect the transcription and translation of the PEPCK-C isoenzyme, it is necessary to verify the effect of 5hmC on its protein expression and explore the relationship between PCK1 and the occurrence and development of liver cancer. SLC2A2 is a membrane glycoprotein that encodes liver cells, pancreatic β cells, and intestinal and renal epithelial cells and mediates easy bidirectional glucose transport. Owing to its low affinity for glucose, it is a glucose sensor rather than the main transporter of glucose. This gene mutation is reportedly related to susceptibility to Fanconi–Bickel syndrome and non-insulin-dependent diabetes[24]. It participates in many processes, such as cell proliferation, differentiation, and apoptosis, by regulating glucose metabolism[25,26]. This study found that the 5hmC modification level of SLC2A2 was slightly increased (log2FC = 0.297032232); however, whether the 5hmC modification affects the function of SLC2A2 requires further confirmation. The valuable information obtained in the PPI network construction of liver cirrhosis and liver cancer groups is limited, and we can only infer that PROX1-TBX3 may have molecular interactions and a role in the progression of liver cirrhosis to liver cancer.
This study showed that the 5hmC modification of genes in the liver digestion-related pathway may be closely related to the occurrence and development of liver cancer. Abnormal bile and insulin secretion by tumor cells may promote or symbolize the occurrence and development of liver cancer. SLC2A2 and PCK1 are the central nodes of the DEGs, which may be most closely related to the occurrence of liver cancer. FABP1, APOC3, SI, KRT20, SLC5A1, SLC10A2, RBP2, and AKR1B10 may be the key genes in the protein regulatory network. 5hmC modification of these genes alters their transcription and other functions and plays a regulatory role. These differences in the 5hmC modification levels of DEGs provide further insights into the development and progression of liver cancer. We plan to verify the 5hmC modification levels of these genes to further explore the mechanisms of liver cancer.
As an important base in DNA demethylation, 5-hydroxymethylcytosine (5hmC) is also involved in epigenetic regulation, specifically in the assembly of chromosomes and the regulation of gene expression. However, the mechanism of action of 5hmC in hepatitis B surface antigen (HBsAg)-positive carriers and during the transition from cirrhosis to liver cancer remains unclear.
This study investigated the relationship between HBsAg-positive carriers, patients with cirrhosis, and patients with hepatocellular carcinoma using their serum samples and to identify potential genes and signaling pathways of DNA hydroxymethylation in order to understand the possible developmental mechanisms.
Using 5hmC sequencing technology, we analyzed the differentially expressed genes (DEGs) of HBsAg-positive carriers, patients with liver cirrhosis, and patients with liver cancer. The function of DEGs was further elucidated to explore the differences and similarities in gene hydroxymethylation and the regulatory role of signaling pathways in these groups.
Using 5hmC DNA sequencing technology, we detected the expression profile of the samples, and the DEGs modified by DNA hydroxymethylation were screened. Bioinformatic analysis was used to enrich the DEGs.
The 5hmC DNA sequencing results showed that 30 genes were upregulated and 2 genes were downregulated in patients with hepatocellular carcinoma compared with HBsAg positive carriers. Further, 17 genes were upregulated and 3 genes were downregulated in hepatocellular carcinoma, compared with cirrhosis. Abnormal secretion of bile and insulin in tumor cells may promote or symbolize the occurrence and development of liver cancer. SLC2A2 and PCK1 are the central nodes of the differential genes, which may be most closely related to the occurrence of liver cancer. FABP1, APOC3, SI, KRT20, SLC5A1, SLC10A2, RBP2, and AKR1B10 may be the key genes in the protein regulatory network that play a regulatory role through 5hmC modification.
The occurrence and development of liver cancer are related to several 5hmC-modified pathway genes and metabolic pathways, which may be potential therapeutic targets to prevent the progression of liver cancer in hepatitis B carriers.
The differences and similarities in gene hydroxymethylation in HBsAg positive carriers, patients with cirrhosis, and patients with liver cancer and the regulatory role of signaling pathways were revealed by 5hmC sequencing technology.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Heij LR, Germany; Na-Bangchang K, Thailand S-Editor: Wang JL L-Editor: A P-Editor: Wang JL
| 1. | Tümen D, Heumann P, Gülow K, Demirci CN, Cosma LS, Müller M, Kandulski A. Pathogenesis and Current Treatment Strategies of Hepatocellular Carcinoma. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 71] [Article Influence: 23.7] [Reference Citation Analysis (1)] |
| 2. | Yu CX, Chen YS, Ge ZJ, Zhang YH, Xu X, Tian T, Wen Y, Zhu J, Song C, Chen JG, Hu ZB. Dietary habits and risk of hepatocellular carcinoma among hepatitis B surface antigen carriers: A prospective cohort study in China. J Dig Dis. 2020;21:406-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 3. | Long Q, Yao YQ, Yan CG. [Research advances in necessity of antiviral therapy for chronic hepatitis B virus carriers]. Zhonghua Gan Zang Bing Za Zhi. 2016;24:465-468. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 4. | Skvortsova K, Stirzaker C, Taberlay P. The DNA methylation landscape in cancer. Essays Biochem. 2019;63:797-811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 200] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 5. | Fu S, Wu H, Zhang H, Lian CG, Lu Q. DNA methylation/hydroxymethylation in melanoma. Oncotarget. 2017;8:78163-78173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 6. | Rustad SR, Papale LA, Alisch RS. DNA Methylation and Hydroxymethylation and Behavior. Curr Top Behav Neurosci. 2019;42:51-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 7. | Chinese Society of Hepatology; Chinese Medical Association. [Chinese guidelines on the management of liver cirrhosis]. Zhonghua Gan Zang Bing Za Zhi. 2019;27:846-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 8. | Bureau of Medical Administration; National Health Commination of the People's Republic of China. [Standardization for diagnosis and treatment of primary hepatic carcinom (2019 edition)]. Zhongguo Shiyong Waike Zazhi. 2020;40:121-138. [DOI] [Full Text] |
| 9. | Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301-1314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2800] [Cited by in RCA: 4102] [Article Influence: 586.0] [Reference Citation Analysis (6)] |
| 10. | Dhar D, Baglieri J, Kisseleva T, Brenner DA. Mechanisms of liver fibrosis and its role in liver cancer. Exp Biol Med (Maywood). 2020;245:96-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 278] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 11. | Cai J, Chen L, Zhang Z, Zhang X, Lu X, Liu W, Shi G, Ge Y, Gao P, Yang Y, Ke A, Xiao L, Dong R, Zhu Y, Yang X, Wang J, Zhu T, Yang D, Huang X, Sui C, Qiu S, Shen F, Sun H, Zhou W, Zhou J, Nie J, Zeng C, Stroup EK, Chiu BC, Lau WY, He C, Wang H, Zhang W, Fan J. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut. 2019;68:2195-2205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 199] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 12. | Solhi R, Lotfinia M, Gramignoli R, Najimi M, Vosough M. Metabolic hallmarks of liver regeneration. Trends Endocrinol Metab. 2021;32:731-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 13. | Paulusma CC, Lamers WH, Broer S, van de Graaf SFJ. Amino acid metabolism, transport and signalling in the liver revisited. Biochem Pharmacol. 2022;201:115074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 77] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 14. | Li YH, Liu Y, Li YD, Liu YH, Li F, Ju Q, Xie PL, Li GC. GABA stimulates human hepatocellular carcinoma growth through overexpressed GABAA receptor theta subunit. World J Gastroenterol. 2012;18:2704-2711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 41] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 15. | Garrido A, Kim E, Teijeiro A, Sánchez Sánchez P, Gallo R, Nair A, Matamala Montoya M, Perna C, Vicent GP, Muñoz J, Campos-Olivas R, Melms JC, Izar B, Schwabe RF, Djouder N. Histone acetylation of bile acid transporter genes plays a critical role in cirrhosis. J Hepatol. 2022;76:850-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 16. | Chen W, Yang A, Jia J, Popov YV, Schuppan D, You H. Lysyl Oxidase (LOX) Family Members: Rationale and Their Potential as Therapeutic Targets for Liver Fibrosis. Hepatology. 2020;72:729-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 175] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 17. | Minor MM, Slagle BL. Hepatitis B virus HBx protein interactions with the ubiquitin proteasome system. Viruses. 2014;6:4683-4702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 18. | An ZW, Wang B, Dai SM, Jiang L, Yan ZC. [Study on the relationship between serum Tim-3 level and the disease process in different state of hepatitis B virus]. Zhongguo Shiyan Zenduanxue. 2017;21:1901-1903. [DOI] [Full Text] |
| 19. | Senoymak MC, Ozkan H. Evaluation of the Relationship between Insulin Resistance and HBV DNA Level in Patients with HBeAg-negative Chronic HBV Infection (Natural Course Phase 3). Euroasian J Hepatogastroenterol. 2020;10:85-91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 20. | Lin TC, Liu WC, Hsu YH, Lin JJ, Chiu YC, Chiu HC, Cheng PN, Chen CY, Chang TT, Wu IC. Insulin Resistance Associated Disorders Pivoting Long-Term Hepatitis B Surface Antigen Decline During Entecavir Therapy. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 21. | Lin YL, Shiao MS, Mettling C, Chou CK. Cholesterol requirement of hepatitis B surface antigen (HBsAg) secretion. Virology. 2003;314:253-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 22. | Xu D, Wang Z, Xia Y, Shao F, Xia W, Wei Y, Li X, Qian X, Lee JH, Du L, Zheng Y, Lv G, Leu JS, Wang H, Xing D, Liang T, Hung MC, Lu Z. The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature. 2020;580:530-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 242] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 23. | Hu Q, Chen H, Zuo Y, He Q, He X, Simpson S Jr, Huang W, Yang H, Zhang H, Lin R. Role of PCK1 gene on oil tea-induced glucose homeostasis and type 2 diabetes: an animal experiment and a case-control study. Nutr Metab (Lond). 2019;16:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Mägi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparsø T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proença C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O'Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Böttcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jørgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martínez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orrù M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvänen AC, Tanaka T, Thorand B, Tichet J, Tönjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Ríos M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators, Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105-116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1722] [Cited by in RCA: 1721] [Article Influence: 114.7] [Reference Citation Analysis (0)] |
| 25. | Hoppstädter J, Ammit AJ. Role of Dual-Specificity Phosphatase 1 in Glucocorticoid-Driven Anti-inflammatory Responses. Front Immunol. 2019;10:1446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 88] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 26. | Jin Q, Li R, Hu N, Xin T, Zhu P, Hu S, Ma S, Zhu H, Ren J, Zhou H. DUSP1 alleviates cardiac ischemia/reperfusion injury by suppressing the Mff-required mitochondrial fission and Bnip3-related mitophagy via the JNK pathways. Redox Biol. 2018;14:576-587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 252] [Cited by in RCA: 380] [Article Influence: 47.5] [Reference Citation Analysis (0)] |