Published online Nov 27, 2023. doi: 10.4240/wjgs.v15.i11.2382
Peer-review started: June 26, 2023
First decision: August 24, 2023
Revised: August 30, 2023
Accepted: September 27, 2023
Article in press: September 27, 2023
Published online: November 27, 2023
Processing time: 153 Days and 21.9 Hours
The earliest and most accurate detection of the pathological manifestations of hepatic diseases ensures effective treatments and thus positive prognostic outcomes. In clinical settings, screening and determining the extent of a pathology are prominent factors in preparing remedial agents and administering approp
Core Tip: Computerized imaging has a vital role in modern liver disease diagnosis and therapeutic intervention, including surgery. The scheme generally involves four elements, i.e., preprocessing, segmentation, modeling and simulation, and software development. This paper describes and discusses how this progressive multidisciplinary technology assists physicians, radiologists, and surgeons in carrying out their tasks effectively and efficiently, hence improving the posttherapeutic outcomes of patients diagnosed with liver diseases.
- Citation: Horkaew P, Chansangrat J, Keeratibharat N, Le DC. Recent advances in computerized imaging and its vital roles in liver disease diagnosis, preoperative planning, and interventional liver surgery: A review. World J Gastrointest Surg 2023; 15(11): 2382-2397
- URL: https://www.wjgnet.com/1948-9366/full/v15/i11/2382.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v15.i11.2382
It is estimated that there are 20 million new cancer cases worldwide and 10 million cancer-related deaths every year[1]. Among these cases, liver cancer is the third leading cause of cancer death. In 2020, 905700 people globally were diagnosed with liver cancer, and 830200 people died from the disease. Scientists have estimated that in 2040, approximately 1.4 million people will be diagnosed with the disease, while 1.3 million people will die from it[2].
The vital function of the liver is filtering blood flow from the digestive tract before circulating the blood back to the rest of the body. Consequently, the liver is subject to various diseases, e.g., fascioliasis, cirrhosis, hepatitis, and alcoholic liver disease[3]. In particular, cancer is associated with increases in both the number and size of abnormal cells. If diagnosed early, it can be treated by interventional radiology, chemotherapy, radiation therapy, or a combination thereof. Among these treatments, liver surgery removing the tumors is efficient in preventing their recurrence and prolonging the life expectancy of the patient, especially those in primary and secondary stages[4]. Liver surgery is a complex and challenging procedure that requires comprehensive knowledge of the liver anatomy, blood supply, and tumor locations and characteristics. Consequently, preoperative imaging is necessary for its planning.
In recent years, there have been significant advancements in diagnostic and interventional imaging technologies, including the use of software equipped with artificial intelligence (AI), to enhance the accuracy of preoperative imaging. This comprehensive review, therefore, aims to offer an in-depth exploration of the latest progress and applications of imaging techniques. In particular, it highlights their pivotal role in improving the outcomes of diagnoses, preoperative planning, and interventional liver surgeries. The main topics discussed in the remainder of this paper cover liver segmentation, diagnostic imaging, preoperative planning and simulation, surgical and therapeutic intervention, and finally software as a medical device.
The functional anatomy of a liver is considered in terms of its dual blood supply, as well as its venous and biliary drainage systems. It is divided into four sectors by the three hepatic veins, each of which drains into the inferior vena cava (IVC) and runs within its scissurae. This nomenclature system was famously described by Couinaud in 1957 and later amended at the Brisbane meeting in 2000. Its primary advantage is enabling anatomical resection of this seemingly almost asymmetrical organ. With this classification, each liver subdivision is self-contained in its artery and portal venous supply and biliary drainage[5,6].
According to the Couinaud classification[5], the liver is divided into eight functionally independent segments, each of which has its own vascular in- and outflows, as well as biliary drainage (Figure 1). There are three major planes that divide the liver vertically: The right hepatic vein plane divides the right hepatic lobe into anterior and posterior segments; the middle hepatic vein divides the liver into right and left lobes; and the umbilical plane running from the falciform ligament to the IVC divides the left lobe into medial and lateral parts. The portal vein divides the liver into superior and inferior segments.
Diagnosis, treatment, and prognosis of diseases of the liver involve defining its anatomical boundary as well as characterizing its pathologies, typically from computed tomography (CT) and magnetic resonance (MR) images. On an image plane, a liver is described by closed contours separating itself from the background. Conventionally, physicians must mentally reconstruct the whole liver and relevant structure in 3-dimension (3D) while navigating through its tomographic planes. However, recent advances in computing technology have enabled virtual 3D reconstruction, modeling, and simulation of the organ in vivo[7,8].
With these technologies, physicians can accurately calculate the hepatic volumetry. In addition, visualization of a 3D liver can also help locate its arteries, veins, and biliary tracts and, hence, determine its functional segments. In surgical planning, the spatial relationship between tumors and hepatic vasculature with a 3D model increases the precision of proposal resection over that with a 2D counterpart by up to 31%[9]. Last but not least, the use of such a model also reduces time and strain during surgical planning and intervention[10].
Modeling a liver from medical images first involves delineating its boundary from other connective tissues and adjacent organs[11]. One of the key challenges is that, as a complex organ, the liver comprises not only its parenchyma but also an extensive vascular network, as well as lesions in pathological cases. In addition, despite recent advances in tomographic imaging[12], liver images remain contaminated with noise of various distributions, depending on the modality. These problems call for the development of effective preconditioning and robust image analysis algorithms. This section, therefore, investigates state-of-the-art methods, as well as their features, limitations, and challenges. These approaches include data preprocessing and fully automatic and semiautomatic segmentation methods.
Medical images are often degraded by noise and artifacts during acquisition. Depending on their model assumptions, various noise reduction strategies are applied prior to image analyses[13]. The perturbation function due to noise is normally random and hence unknown, except for only their distribution. Therefore, in medical imaging, the most frequently assumed distribution models include Gaussian, Poisson, and Rician distributions for charge couple device, X-ray, CT, and MR images, respectively. However, directly applying an inverse filter to reduce noise and possibly other inherent artifacts could adversely affect smaller features or abate anatomical boundaries, such as vasculature, calcification, and connective tissue. Instead, several applications adopt structural adaptive anisotropic[14], spatial frequency or wavelet[15], blind deconvolution[16], regularized diffusion[17] filters or, much more recently, those based on machine learning (ML) or AI models, e.g., convolutional neural networks (CNNs)[18,19].
On measuring their performance, the peak signal-to-noise ratio (PSNR) and structural similarity (SSIM) are often considered. However, a recent study[20] revealed that although visual quality is clearly improved with advanced filters, correlations between PSNR and SSIM and application-specific performance, such as classification (i.e., based on area under the curve), are not clearly present. In fact, fine tuning neural network parameters to a particular noise model is recommended.
In practice, to balance complexity and intended analysis, a trivial anisotropic diffusion filter has been applied to denoise a T1-weighted MR image of a liver while enhancing its border[21] prior to 3D surface generation. Meanwhile, in some other studies, the least commitment principle[22] has been adopted with no preprocessing of an image other than adjusting its windows and levels, but taking noise into account during subsequent analyses[23]. Once preconditioned, a series of cross-sectional images proceeds to the next stage, in which the liver, its peripherals, and lesions are separated.
Since liver pixels appear very similar to those of other nearby organs, existing automatic schemes thus rely on auxiliary information, e.g., data-driven appearance models or empirical understanding of its morphology[23]. Accordingly, its main advantage is low inter- and intraobserver variability due to manual intervention. Recent extensive surveys on the topic are found in the literature[24,25]. Several early works were developed and validated based on a public dataset called SLIVER07[26]. They contained 3D CT liver images of 20 and 10 subjects for training and testing, respectively. These images had 512 × 512 pixels at 0.56 to 0.86 mm2 in-plane resolution and covered 64 to 502 slices with spacing between 0.7 mm and 5 mm. Another recent dataset is 3D-IRCADb[27]. It contains CT images of 20 patients, three thirds of which contain hepatic tumors. Their voxel size and resolution are similar to those of its predecessor. Furthermore, each image is labeled with not only pathologies but also segmentation challenges. Based on these datasets, a number of computerized methods were proposed to delineate a liver and benchmarked. Examples of the recent results are summarized in Table 1[28-34]. Note that these studies may employ different accuracy metrics, e.g., volumetric overlap error or dice similarity.
| Ref. | Key techniques | Dataset | Accuracy |
| Kumar et al[29], 2013 | Region growing | Proprietary | 98% ± 1% |
| Chen et al[30], 2012 | AAM, graph cut | SLIVER07 | 93.5% ± 1.8% |
| Huang et al[31], 2016 | Template matching (SBLDA) | 3D-IRCADb | 92.16% ± 2.95% |
| Wu et al[32], 2016 | Linear clustering, graph cut | SLIVER07 | 75.2%-71.4% |
| Mohamed et al[33], 2017 | Bayesian model | Proprietary | 95.5% |
| Zheng et al[34], 2022 | DL (CNN, C-LSTM) | SLIVER07 | 82.5% ± 7.7% |
The most promising approach in this category is one based on modeling from pretrained data, both statistically[26,30,33,35-37] and using CNNs[34,38-40]. The former iteratively deforms a liver model to fit underlying imaging features while imposing anatomically plausible constraints found in the training, e.g., the active appearance model. The latter learns from some segmented livers, their spatial architecture, and the relationship among their convolutional features, cascaded through a deep network, and fuses them with weighted nonlinear functions. Subsequently, pixels of an unseen image proceed through the same network and are labeled accordingly, resulting in the final segmentation. However, these methods require sufficiently large prelabeled samples and hence a substantial amount of computing power for model learning.
It is evident that various factors, e.g., nearby organs such as the stomach, pancreas, duodenum, and heart, as well as artifacts due to implants, could have adverse effects on segmentation quality. Currently, it remains challenging to incorporate computable elements to automatically address this issue. Therefore, user interaction is often involved but kept to a minimum, i.e., at initialization[23,41], during the process[42,43], in final adjustment[23], or with a combination thereof. The recent works are summarized in Table 2[21,28,41,43-46].
| Ref. | Key techniques | Dataset | Accuracy |
| Chen et al[44], 2009 | Quasi-Monte Carlo | Proprietary | NA |
| Yang et al[41], 2014 | Level set | SLIVER07 | 78.9% |
| Liao et al[45], 2016 | Graph cut | SLIVER07 | 94.2% ± 3.3% |
| Lu et al[46], 2017 | 3D CNN, graph cut | 3D-IRCADb | 90.64% ± 3.34% |
| Chartrand et al[43], 2017 | Deformable model | SLIVER07 | 92.38% ± 1.35% |
| Le et al[23], 2021 | Mixture model, graph cut | SLIVER07 | 92.2% ± 1.5% |
At varying degrees of interaction, many methods can achieve reasonable accuracy without previously trained liver data. For instance, trivial thresholding with K-means clustering has been applied to CT angiography to separate the liver from the kidneys and ribs[47]. Similar methods automatically set these thresholds by learning the pattern of abdominal histograms[48,49] or that of textures[50]. However, they often require prior knowledge of the anatomy for initialization and postprocessing, e.g., manual editing or morphological operators, to remove oversegmented regions. Instead of defining pixel membership by thresholds, many researchers have expanded a region of interest accumulatively from seeding points. They then have exploited different strategies to control new inclusions, e.g., convex hulls[51], binary morphology and anatomical constraints[52], significant differences in boundaries[44,53], and anatomical priors[42].
In addition to region-based approaches, delineating contours around a liver has also attracted considerable interest. Initially, the active contour model and its variants were explored[54,55] based on gradient and curvature and later extended to the level set[43,56-59]. By these methods, starting contours were specified by a user or estimated by other segmentations. They were then implicitly driven by gradient and embedding surface curvature. Unlike its counterparts, any aberration would be regularized by geometric continuity on the hypersurface. Empirical and anatomical knowledge, e.g., distance to centroid, nominal contrast, and segmental and anatomical markers, were translated to computable conditions to assert the evolution of these contours.
Another approach poses the segmentation problem as that of graph optimization[60]. In liver imaging, the deviation of intensity at a pixel from a predefined distribution and its gradient strength are formulated as region and boundary cost functions, respectively[61]. Similar functions are obtained from texture images and supervoxels[32] or constrained by statistically trained shape[45] and intensity[32] models or initialized by CNN[46]. It was shown that automation of the remaining steps was possible if anatomical constraints (e.g., vena cava and tumor) were imposed for postprocessing[62].
It has been discussed previously that although the liver and its vasculature are clearly presented to a radiologist in medical images, mentally extracting them requires substantial knowledge and expertise regarding hepatic anatomy and physiology. Therefore, automating this process with algorithmic codes remains an open research challenge. Similarly, diagnosing a hepatic disease involves assessing liver damage and characterizing its lesions based on their vascularity and composition, as well as their implications on adjacent vessels[63,64]. A range of radiological and computerized imaging techniques can be utilized and are summarized as follows.
Arterial anatomy: Hepatobiliary surgery, liver transplantation procedures, and endovascular treatments can all benefit from information on anatomical variations in hepatic arteries. The existence of such variations may call for adjustment of surgical procedures to avoid unintentional vascular damage, hemorrhage, and biliary problems.
The Michels classification[65] and its modification by Hiatt et al[66] are the most frequently used categories for describing hepatic arterial variations in the literature. Approximately 55%-60% of people have the classic pattern of the common hepatic artery branching from the celiac artery, with the hepatic artery normally splitting off into the right and left hepatic arteries to supply the entire liver. Replaced and accessory left or right hepatic arteries are the most often found anatomical variants. In cases of transarterial embolization of traumatic liver injury or embolization of liver tumors, if the bleeding point or arterial feeders cannot be demonstrated on conventional hepatic angiogram, searching for these possible anatomical variants is crucial. Another example is when left hepatectomy is performed in a patient with a replaced or accessory left hepatic artery, ligation of the left hepatic artery at its origin in the left gastric artery is needed.
Although the anatomical variant classification has been widely accepted, not all variants are surgically significant. Furthermore, the course of the hepatic artery and its topographic relationship to the surrounding structures, such as the portal vein and bile ducts, are not taken into consideration[67].
Portal vein anatomy: There are numerous variations in the portal vein branching patterns. The classic anatomy, which is found in approximately 65% of patients, consists of the main portal vein branching into the right and left portal veins at the porta hepatis. The right portal vein later subdivides into anterior and posterior branches. Found in approximately 35% of patients, the two most common variants are trifurcation of the portal vein trunk and a right posterior branch as the first branch of the portal vein trunk, with the latter being more common and known as the Z-type pattern[68,69].
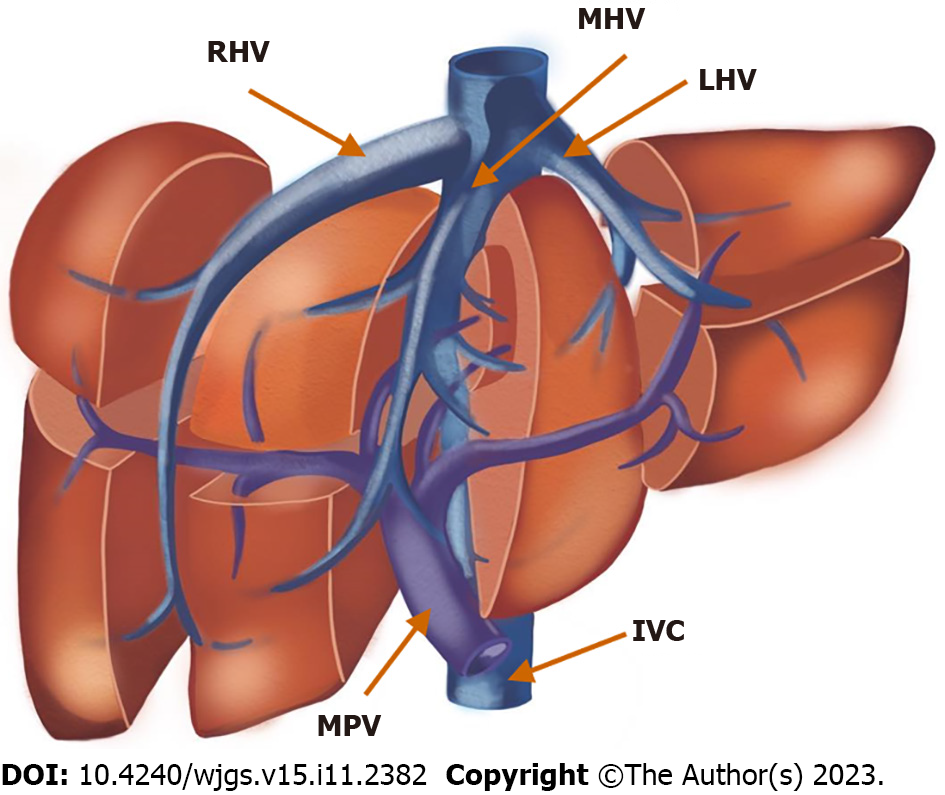
Hepatic vein anatomy: Accurate perception of the hepatic vein anatomy before liver surgery is crucial. Inadvertent injury of the hepatic veins leads to a higher risk of bleeding and functional loss of the hepatic segment with a compromised venous outflow. Generally, there are three hepatic veins: The right hepatic vein drains segments V, VI, and VII, the middle hepatic vein drains segments IV, V, and VIII, and the left hepatic vein drains segments II and III. The classic anatomy of the hepatic veins, which form a common trunk between the left and middle hepatic veins, is found in approximately 65%-85% of patients[70,71].
Biliary anatomy: The normal biliary anatomy found in approximately 58% of the population comprises the right hepatic duct draining the right hepatic lobe and the left hepatic duct draining the left hepatic lobe. The right hepatic duct divides into the right posterior sectional duct, coursing in a horizontal plane and draining segments VI and VII; the right anterior sectional duct, coursing in a vertical plane, drains segments V and VIII. The left hepatic duct divides into the left superior sectional duct and drains segment IVa, and the left inferior sectional duct drains segments II, III, and IVb[72]. The caudate lobe usually drains into the proximal left or right hepatic duct.
Generally, 20%-30% of patients have synchronous hepatic disease, while hepatic metastasis occurs in more than 50% of colorectal cancer patients[73]. Primary liver tumors, such as hepatocellular carcinoma (HCC), mass-forming cholangiocarcinoma, hepatic adenoma, or focal nodular hyperplasia, as well as liver metastasis, have distinct cellular components and, hence, unique imaging appearances. As such, they can be characterized by means of CT and MRI.
To date, the only treatment associated with long-term survival for both HCC and colorectal liver metastasis is surgical resection. Imaging studies are essential for identifying potential surgical candidates. Specifically, for the best outcome in the resection procedure, all lesions need to be removed, while a sufficient functioning liver must be preserved. One of the major challenges is that because metachronous hepatic metastasis can occur in over 50% of patients with colorectal cancer[74,75], the imaging sensitivity should be sufficiently high to detect these lesions. Although CT is available worldwide and enables evaluation of extrahepatic disease and vascular structures, the modality has some limitations. These include an inferior ability to delineate the tumor margin, to perform tissue characterization, and to detect and characterize small lesions and associated radiation. Alternatively, MRI with hepatocyte-specific agents is currently the most accurate imaging modality to identify hepatic disease in patients with colorectal cancer[76-78]. Despite its sensitivity, additional metastatic foci can be found intraoperatively in up to 25% of patients after MRI[79,80]. Another drawback of MRI is that in patients with coexisting benign focal liver lesions, such as hemangioma, an ill-defined heterogeneous echoic nodule could lead to confusion during surgery. To resolve the ambiguity, contrast-enhanced ultrasound has increasingly been adopted intraoperatively as a complement. Table 3 summarizes the existing research related to the sensitivity of focal liver tumor detection[63,78,81-83].
The anatomy of both the liver and its peripherals has been extensively explored in the medical literature, and the most common patterns have been firmly established. Despite the highly deformable structure and large intersubject variability of this organ, it has been continually demonstrated that computerized methods can be applied to extract relevant objects with reasonable degrees of accuracy[24,26]. Thus far, pathological manifestations can result in irregular appearances of the interconnecting parts, undermining their merits in clinical and surgical practice[4]. In fact, with the recent advances in ML and AI, research focus has now been particularly directed toward identifying, delineating, and characterizing lesions from tomographic images. Prominent works in the field are summarized and discussed here.
ML algorithms have been widely employed in segmenting the tumoral liver. After a seed point was estimated within a lesion, fuzzy C-means (FCM) was used to expand the coverage toward its margin[29]. Likewise, a watershed was applied to CT images to extract supervoxels with similar characteristics. Subsequently, tumors were identified from the liver and other objects by merging those subregions with FCM and K-means clustering using their textural information[50], i.e., pixel intensities, directional derivative, local binary pattern, and local differences. Based on 22 trained and 22 tested instances, the highest classification accuracies of 95.64% to 98.88% were reported. K-means clustering was applied to approximate liver contours, which were later refined by Graph-Cut[62]. Once the vena cava had been detected and other segments had been discarded by anatomical templates, tumors were extracted by cavity filling. Note, however, that this assumption failed to identify those on the liver boundary. For percutaneous radiofrequency ablation (RFA) to remove inoperable primary or metastatic tumors, the ablation zone was first determined by max-flow min-cuts of a 3D spherical graph expanded from a seed point[84]. It was later automated by using FCM to extract the ablation zone and then cyclic morphology to refine one[85].
Meanwhile, with rapid development in AI and CNNs in particular, a number of network architectures have been adopted for diagnosing tumoral livers. Li et al[38] used 2D and 3D DenseUNets to extract within-slice features and to learn their spatial relationships between slices, respectively. The results from both networks were finally fused to produce labels of both liver and tumor pixels. Despite relatively high benchmark scores (i.e., 93.7%), their models took 30 h in total for training with only limited series. Another example[86] applied a simple 3D U-Net to first extract the liver. Super pixel blocks of tumors were localized by a multiscale candidate generation method. The exact regions of these candidates were defined and then refined by a 3D fractal residual network and active contour, which reached a 67% accuracy during evaluation. It was pointed out in another study that a main drawback of data-driven AI is the imbalanced proportion between healthy livers and those with pathologies[87]. As a result, many existing dice loss (DL) models tend to predict lesions as part of the liver or backgrounds. To address this issue, they tried to assemble cascade U-ResNets, each trained with a different loss function, i.e., weighted cross entropy, DL, weighted DL, Teverskry loss (TL), and weighted TL. With ensemble learning, tumors could be segmented with a 75% accuracy, compared to the approximately 65%-70% accuracy obtained by competing networks. The same accuracy of 74.5% was achieved by a 2.5D fully CNN whose loss function consisted of cross-entropy, a similarity coefficient, and a novel boundary loss function[88]. The latter was prescribed based on the boundary between segmented objects by means of logical morphology. Alternatively, using a two-stage densely connected network, where a liver was first localized by an encoder-decoder CNN, tumors were detected with attention modules at a 72.5% accuracy[89].
Since these methods recognize tumors by their features, implicitly learned by examples, the irregularities found on the boundaries of lesions are not precisely traced, while adjacent lesions are sometimes merged. Thus, postprocessing by another empirical model or manual processing by radiologists is often needed. In fact, some studies have shown that, as a baseline, even skilled human raters could achieve only a 78% accuracy.
Conducting a virtual liver resection prior to the live procedure is highly beneficial. For example, the right portal pedicle divides into anterior and posterior branches, each of which further splits into two segments (i.e., V and VIII, and VI and VII, respectively). The left portal pedicle has a longer and more horizontal extrahepatic course. This allows the surgeon access and exposure to the relevant areas, for instance, during biliary system reconstruction. Moreover, segmental branches arise from the left portal pedicle supplying segments II, III, and IV. The ligamentum venosum, a fibrous remnant of the ductus venosum in the fetus, which connects the left portal vein to the left hepatic vein at the IVC, serves as a distinctive landmark for gaining access to the left portal pedicle and the left hepatic vein, whose terminating discharge is often merged with the middle hepatic vein and thus can be challenging to identify and control during a left hepatectomy.
Upon estimating potential risks of the surgical procedure, liver cirrhosis and portal hypertension (which are usually associated with liver cancer, particularly HCC), must be diagnosed preoperatively. To this end, multidisciplinary teams consider any evidence of advanced cirrhosis or inadequate liver function while devising an appropriate management plan, e.g., by using the Child-Pugh scoring system[90,91]. Specifically, for patients having cirrhosis and meeting certain criteria, liver resection and transplantation address an underlying field change that predisposes the parenchyma to tumor recurrence. Options for patients who are not candidates for those procedures include RFA, microwave ablation, trans-arterial chemoembolization, and other locoregional therapies[92].
Surgical resection is a primary curative treatment for patients with primary and metastatic liver tumors. Unfortunately, fewer than 25% of patients are suitable for surgery[93,94]. With a better understanding of hepatic anatomy and surgical technique refinements, the extent of liver sections that can be surgically removed is expanding. However, a tendency toward more aggressive liver resections in patients with preexisting liver disease requires thorough evaluation of hepatic function, especially the amount and quality of the postoperative future liver remnant (FLR).
It has been established that an inadequate liver volume following surgery is a robust, independent predictor of postoperative hepatic dysfunction and complications[95]. Generally, the FLR per total liver volume (TLV) ratio must be 25%-30% to minimize postoperative complications[96,97]. Patients with hepatotoxic chemotherapy or hepatic steatosis should have an FLR ratio of greater than 30%, whereas those with cirrhosis should maintain an above 40% ratio. Likewise, in living donor transplantation, the donor’s liver volume has to be 30%-35% more than that of the recipient[98], or 40% in cases with hepatic disease[99].
Volumetric CT has currently become the gold standard for determining whether hepatectomy can be performed[100]. To this end, computer software is employed to reconstruct a 3D liver and estimate the ratio of FLR to nontumorous TLV. Normally, the latter is measured directly by CT. Alternatively, it may be estimated from the patient’s body surface area[101]. These methods are called mTLV and eTLV, respectively. It was found in some studies[4,102] that eTLV could identify cases where mTLV was previously underestimated. In addition, there have been a number of recent advancements in automated liver volumetry by medical image computing.
Unlike image segmentation, where whole liver boundaries are traced on an underlying volumetric image, functional segmentation or resection involves estimating its composition of independently functional segments, each of which has its own vascular in- and outflow, according to Couinaud’s scheme. MeVis LiverAnalyzer™ (MeVis Medical Solution, Germany) and Synapse Vincent™ (Fujifilm, Japan) are currently standard software programs for virtual liver resection that rely on the surgeon’s judgment[103]. Common practice involves the user’s interactive tracing of the segments with respect to their major vascular tracks. Not only are individual experiences and skills needed, but the process is also tedious and time-consuming, as well as inducing large inter- and intraobserver variability. Meanwhile, many novel imaging algorithms have also been continuously developed to assist or complement this task and have rapidly become an emerging area of investigation.
Provided that portal and hepatic veins are extracted, liver segments are defined with respect to voxel distances to specific branches[104,105], voxel projections onto vascular intersections[106], or categorical search by Voronoi diagram[107-109]. However, these methods suffer from computationally intensive voxel sorting. Moreover, the topology of voxel aggregation is neither validated nor rectified. Alternatively, to accelerate computation and ensure surgically plausible resection, an extracted liver volume is first converted to a surface and subdivided, based on vasculature and salient anatomical landmarks, by differential geometry[110-115]. Thus far, manual correction is often inevitable. Otherwise, additional anatomical constraints, e.g., from a statistically trained deformable atlas[116], are necessary.
Unfortunately, not every method was able to segment all eight Couinaud segments nor was it always validated on the same liver dataset. Thus, a recent study[115] compared some prominent algorithms only according to their volumetric ratios, i.e., at lobe and sectional levels. The average values are listed in Table 4[11,106,108,112,113,115]. Since these methods rely on accurate extraction of the hepatic vasculature and liver boundary, future directions worth exploring are advanced pattern recognition (PR) of the gastrointestinal structures and integration of other imaging modalities.
Once the resection is made, removal of pathological segments could be planned, and FLR could hence be estimated. One of the most widely utilized 3D software programs in preoperative liver surgery is Synapse Vincent™ (Fujifilm, Japan)[103]. It helps automate liver segmentations and their volumetric assessment. However, with recent surgical techniques, liver resection is no longer limited to only right hepatectomy. Several surgical plans have been devised or tailored for an individual, i.e., patient-specific strategies. Therefore, FLR should be resilient to variations in such planning. In addition, other volumetric assessments are also equally important posttherapy, e.g., graft regeneration after transplantation and responses to cancer treatment[117].
In addition to resected volumetry, liver function also needs to be evaluated. In fact, technical limitations of resection and its safety have been exceeded by continually developed procedures, aiming at increasing FLR in patients with insufficient liver volume by utilizing its regeneration in response to blood flow, also known as flow modulation. Among the most often chosen procedures is portal vein embolization (PVE), where the portal vein on the opposite side of the FLR has a catheter radiologically inserted and is then embolized with vascular plugs, coils, particles, or glue[117]. Consequences of a diseased liver parenchyma in terms of liver function may be analyzed by biopsy, performed on the living donor liver prior to transplantation[118]. In patients who may need PVE to enhance the FLR ratio, these anatomical and functional criteria are also relevant, given the proper context. Comprehensive assessment of liver conditions is required prior to any therapy because livers with such as cirrhosis and steatosis, for instance, demand a significantly greater FLR than a healthy liver[119].
The indocyanine green (ICG) clearance test, asialoglycoprotein receptor scintigraphy using 99mTc-galactosyl human serum albumin, and serum hyaluronic acid level assessment are prominent methods that can be used to evaluate the residual liver's ability to function[120,121]. Injection of ICG, a tricarbocyanine dye, causes it to bond with albumin and be carried throughout the body via the circulatory system. Elimination of ICG occurs solely through biliary excretion. Thus, serum blood tests or an optical sensor on the finger can reveal the excretion level. ICG levels in the blood should be below 10% at 15 min after injection (ICG-R15). Therefore, blood samples taken at 5-min intervals postinjection can be analyzed to determine the plasma disappearance rate of ICG (ICGK), which is calculated by using linear regression of the plasma ICG concentration[122].
Although there is accurate automated anatomical liver volumetry and identification of its biomarkers, there is currently no computer software with biomarker mapping that can precisely delineate the area of residual functioning liver. A multidisciplinary approach is therefore recommended to determine both the liver volume and function. The selection of the surgical plane, feasibility of resection, visualization of the tumor and its extent, etc., all rely on maintaining an ongoing interaction between the surgeon and radiologist, as well as reliable, though probably not the most precise, imaging software.
The risk of postoperative hepatic failure is substantially associated with the extent of liver resection. Although this is logical and simple to assess, the volume of the liver that remains present is more indicative and must be precisely determined. Additionally, because the segmental anatomy and its volume significantly vary among patients, only determining the segmental numbers is inconclusive. Specifically, the right side of the liver accounts for more than half of the TLV in most people, but its variations extend from 49% to 82%, while those of the left side range from 17% to 49%[123]. Therefore, a formal radiologic volumetric assessment is necessary to ensure accurate FLR, especially when planning a major liver resection.
CT or MRI is typically performed to characterize lesions and devise preoperative planning[21]. Consequently, FLR is estimated from the planned resection outlines by sequential marking on respective cross-sectional images, given slice thickness and voxel dimensions. It has been shown that both the intended and actual FLR as well as their actual surface and volumes are highly correlated radiographically[124,125]. As such, additional tests are not required during the procedure.
For patients with an insufficient FLR who are being considered for hepatic resection, FLR augmentation by PVE via interventional radiography is most widely used. With this procedure, the portal vein (with or without segment IV branches) is embolized. Usually, the procedure is performed with percutaneous vascular access[117]. Subsequently, those who exhibit more than 2.0% growth per week on repeated volumetry have no risk of liver failure during the perioperative phase following hepatectomy[126].
An anatomic resection involves the removal of a Couinaud segment by selective ligation of the main HPV and portal triad. With this approach, there is a higher chance of obtaining disease-free margins because it resects areas distal to the tumor that are at risk for vascular micrometastasis. Alternatively, nonanatomic resection or parenchymal transection disregards those segmental planes; it is often employed for benign tumors, debulking treatment, or when attempting to preserve the residual parenchyma. A microscopic margin negative (R0) resection must be performed to minimize local recurrence. It has been shown that a resection margin of 1 cm or smaller is safe[127].
Standard anatomic hepatectomy involves controlling both vascular inflow and outflow prior to parenchymal transection. Accordingly, removal can be performed without affecting adjacent hepatic segments. Generally, intraoperative ultrasonography is utilized to determine the presence of vascular structures and to assess the location and size of the tumor as well as their relation to the surrounding vasculature.
The development of computerized imaging techniques to further enhance minimally invasive liver surgery has rapidly progressed[128,129]. For instance, near-infrared fluorescence is adopted in laparoscopic and robotic camera systems, allowing the identification of different preoperatively injected dyes (e.g., indocyanine green). This contrast agent propagates through the biliary tree while illuminating the structure after being metabolized mostly by hepatocytes. Recently, this modality has been exploited to differentiate between well- and inadequately-perfused hepatic parenchyma to guide parenchymal dissection following vascular control[130-132].
It has been established that computer-based 3D reconstruction of liver tumors could improve the accuracy of their localization and the precision of surgical planning[9]. Thus far, 2D/3D image reviewing during surgery on a traditional picture archiving and communication system in the operating room has been found to be distracting. Therefore, real-time localization of lesions and the identification of arteries and biliary structures by using intraoperative ultrasonography are usually preferred. Nevertheless, similar to what was pointed out in another survey on tumor surgery[133], the need for additional port sites to interpret 2D images and hence to mentally recreate the 3D anatomy with respect to the orientations of ultrasound probes has restricted its wider adoption in minimally invasive surgery.
Currently, an augmented reality (AR) endoscopic overlay of the patient-specific anatomy with associated virtual reality (VR) models has attracted considerable attention as it could increase the surgical efficiency in real-time with intelligent operative guidance[134-136]. With this approach, 3D reconstructed data can be precisely overlaid onto the operated area. Effectively, cognitive strain conventionally imposed on the surgeon could be lessened. For uterine myomectomy, it has been shown that spatial recognition based on AR could improve the localization accuracy[137].
To adopt VR and AR in hepatobiliary surgery, one has to confront the technical challenges of continuously coregistering the computer-generated models to a mobile liver with significant tissue deformation. To address this issue, a recent study[28], for example, applied conformal parameterization to an extracted liver surface. With this technique, the triangle mesh of genus-0 of the surface was mapped onto its topological homeomorphism[138]. Given a set of landmarks on a liver surface, representing the resection paths according to Couinaud’s definition, a deformation that bijectively maps a liver and its section onto another instance with minimal distortion could be realized. However, since the liver is morphologically diverse, it was suggested that localized alignment should be the focus. In fact, to ensure physiologically plausible correspondence within or across subjects, statistical deformable models[30,43] are incorporated. Additionally, clinical management aspects, e.g., tumor board evaluations, preoperative strategy, and intraoperative access, also need to be considered.
Since the early 1990s, Digital Imaging and Communication in Medicine (DICOM) developed by the American College of Radiology and the National Electrical Manufacturers Association has been a gold standard for archiving, transferring, and presenting imaging data among acquiring and processing devices[139] in radiological practices[140]. It features a unique structural content[141], consisting of not only an imaging matrix and its encoding but also relevant medical data, e.g., patient information and study details, as well as a scanning protocol. In a typical liver examination, for instance, its DICOM structure contains a series of multislice CT images in the axial direction, covering the upper abdomen, and each image is numbered and labeled with physical geometry, thickness, and resolution, and perhaps suggested window-level settings. This information is vital for accurately reconstructing a whole 3D liver for diagnostic and intervention purposes. Hence, most current medical image computing software does support this standard by default. In addition, the Neuroimaging Informatics Technology Initiative (NIfTI)[142] has been specifically designed and developed by neuroimaging scientists to resolve physical orientation objects within a brain image. Nonetheless, this data format is also adopted in other fields, where geometric information is needed.
Presently, computer software has increasingly been integrated into digital platforms that serve medical purposes. Software that is a medical device in its own right is called Software as a Medical Device (SaMD)[143]. It is to be distinguished from software in a medical device and that used in manufacturing or maintaining a medical device. Specifically, the International Medical Device Regulators Forum (IMDRF) defines SaMD as “software intended to be used for one or more medical purposes (e.g., to make clinical decisions) that performs these purposes without being part of a hardware medical device.” With its unique features, a working group by IMDRF as the representative of regulators worldwide developed a common framework aiming to support innovation and timely access by both patients and providers to safe and effective SaMD.
In liver imaging, SaMD, regardless of its computing platforms, may be used for diagnoses both in vivo and in vitro, prevention, screening or monitoring, and treatment or alleviation of liver diseases. A manufacturer who intends to make SaMD available for use under their name would be subject to regulations, not only throughout its software engineering life cycle (e.g., ISO/IEC 14764:2006 Software Engineering) but also postmarket surveillance and any subsequent updates, in which risk identification and countermeasures are established[144].
To maintain regulatory compliance, the roles of an SaMD and its deployment in clinical environments must be declared. Its recommendation for intended uses (i.e., diagnosing or treating a disease and informing or driving clinical management) with potential adverse consequences (i.e., critical, serious, and nonserious situations or conditions) must be classified. Most importantly, software evaluation (according to established protocols)[145], clinical evaluations, and relevant evidence must be attached. Finally, its linguistic design and instructions must conform to standard medical terms. Other considerations include technology and sociotechnical system, environment, and information security with respect to safety.
This paper has provided an extensive review on computerized imaging in both current and emerging clinical practices and when integrated with state-of-the-art algorithms. The vital roles of this modality include the diagnosis of liver disease and its curative planning, treatment, and surgical intervention. It has been demonstrated in the recent literature that, depending on the data condition, prior knowledge, and amount of user interaction involved, various computer algorithms yield reasonable diagnostic and simulation accuracies. Nonetheless, it is worth noting that while these algorithms perform particularly well for functional segment classification of normal or slightly pathological livers, their performance on hepatic lesion characterization remains to be much further improved.
Although ML and AI strategies have rapidly become the main players in liver imaging and thus far have exhibited promising results, it remains challenging to acquire sufficiently large and heterogeneous datasets with labeled ground truth for training. This issue has been partly addressed in many less critical applications by using, for instance, big and crowdsourced data.
Finally, with advances in medical imaging, many computer algorithms will be adopted and implemented in SaMD. Therefore, researchers, digital health manufacturers, and physicians should be made aware of relevant regulatory requirements and guidelines to ensure the safety of patients.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Radiology, nuclear medicine and medical imaging
Country/Territory of origin: Thailand
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Li HL, China; Neri V, Italy; Sugimoto M, Japan; Tchilikidi KY, Russia S-Editor: Yan JP L-Editor: Wang TQ P-Editor: Yu HG
| 1. | PAHO. World Cancer Day 2023: Close the care gap. Feb 4, 2023. [cited 25 June 2023]. Available from: https://www.paho.org/en/campaigns/world-cancer-day-2023-close-care-gap. |
| 2. | Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, Laversanne M, McGlynn KA, Soerjomataram I. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol. 2022;77:1598-1606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 1071] [Article Influence: 357.0] [Reference Citation Analysis (0)] |
| 3. | Machicado C, Machicado JD, Maco V, Terashima A, Marcos LA. Association of Fasciola hepatica Infection with Liver Fibrosis, Cirrhosis, and Cancer: A Systematic Review. PLoS Negl Trop Dis. 2016;10:e0004962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 4. | Martel G, Cieslak KP, Huang R, van Lienden KP, Wiggers JK, Belblidia A, Dagenais M, Lapointe R, van Gulik TM, Vandenbroucke-Menu F. Comparison of techniques for volumetric analysis of the future liver remnant: implications for major hepatic resections. HPB (Oxford). 2015;17:1051-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 5. | Couinaud C. Le foie: études anatomiques et chirurgicales. Paris: Masson & Cie, 1957: 1-530. |
| 6. | Pang YY. The Brisbane 2000 terminology of liver anatomy and resections. HPB 2000; 2:333-39. HPB (Oxford). 2002;4:99; author reply 99-99; author reply100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 259] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 7. | Nakayama K, Oshiro Y, Miyamoto R, Kohno K, Fukunaga K, Ohkohchi N. The Effect of Three-Dimensional Preoperative Simulation on Liver Surgery. World J Surg. 2017;41:1840-1847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 8. | Yeo CT, MacDonald A, Ungi T, Lasso A, Jalink D, Zevin B, Fichtinger G, Nanji S. Utility of 3D Reconstruction of 2D Liver Computed Tomography/Magnetic Resonance Images as a Surgical Planning Tool for Residents in Liver Resection Surgery. J Surg Educ. 2018;75:792-797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 9. | Lamadé W, Glombitza G, Fischer L, Chiu P, Cárdenas CE Sr, Thorn M, Meinzer HP, Grenacher L, Bauer H, Lehnert T, Herfarth C. The impact of 3-dimensional reconstructions on operation planning in liver surgery. Arch Surg. 2000;135:1256-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 120] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 10. | Agha RA, Fowler AJ. The role and validity of surgical simulation. Int Surg. 2015;100:350-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 141] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 11. | Umetsu S, Shimizu A, Watanabe H, Kobayake H, Nawano S. An Automated Segmentation Algorithm for CT Volumes of Livers with Atypical Shapes and Large Pathological Lesions. IEICE Trans Inf Syst. 2014;E97.D:951-963. [DOI] [Full Text] |
| 12. | Potigailo V, Kohli A, Pakpoor J, Cain DW, Passi N, Mohsen N. Recent Advances in Computed Tomography and MR Imaging. PET Clin. 2020;15:381-402. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 13. | Gravel P, Beaudoin G, De Guise JA. A method for modeling noise in medical images. IEEE Trans Med Imaging. 2004;23:1221-1232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 82] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 14. | Greenberg S, Kogan D. Improved structure-adaptive anisotropic filter. Pattern Recogn Lett. 2006;27:59-65. [RCA] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 15. | Relin Francis Raj J, Vijayalakshmi K, Priya SK. Medical image denoising using multi-resolution transforms. Measurement. 2019;145:769-778. [DOI] [Full Text] |
| 16. | Michailovich O, Tannenbaum A. Blind deconvolution of medical ultrasound images: a parametric inverse filtering approach. IEEE Trans Image Process. 2007;16:3005-3019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 51] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | Chung H, Lee ES, Ye JC. MR Image Denoising and Super-Resolution Using Regularized Reverse Diffusion. IEEE Trans Med Imaging. 2023;42:922-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 26] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 18. | Zhang K, Zuo W, Chen Y, Meng D, Zhang L. Beyond a Gaussian Denoiser: Residual Learning of Deep CNN for Image Denoising. IEEE Trans Image Process. 2017;26:3142-3155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3624] [Cited by in RCA: 1327] [Article Influence: 165.9] [Reference Citation Analysis (0)] |
| 19. | Elhoseny M, Shankar K. Optimal bilateral filter and convolutional neural network based denoising method of medical image measurements. Measurement. 2019;143:125-135. [DOI] [Full Text] |
| 20. | Michael PF, Yoon HJ. Survey of image denoising methods for medical image classification. Proceedings of SPIE 11314, Medical Imaging; 2006 March 13; San Diego, California, USA: SPIE, 2006: 383-394. [DOI] [Full Text] |
| 21. | Huynh HT, Karademir I, Oto A, Suzuki K. Computerized liver volumetry on MRI by using 3D geodesic active contour segmentation. AJR Am J Roentgenol. 2014;202:152-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 22. | Suchman LA. Plans and situated actions: the problem of human-machine communication. Cambridge: Cambridge University Press, 1987: 1-220. |
| 23. | Le DC, Chinnasarn K, Chansangrat J, Keeratibharat N, Horkaew P. Semi-automatic liver segmentation based on probabilistic models and anatomical constraints. Sci Rep. 2021;11:6106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Moghbel M, Mashohor S, Mahmud R, Saripan MI. Review of liver segmentation and computer assisted detection/diagnosis methods in computed tomography. Artif Intell Rev. 2018;50:497-537. [RCA] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 25. | Ansari MY, Abdalla A, Ansari MY, Ansari MI, Malluhi B, Mohanty S, Mishra S, Singh SS, Abinahed J, Al-Ansari A, Balakrishnan S, Dakua SP. Practical utility of liver segmentation methods in clinical surgeries and interventions. BMC Med Imaging. 2022;22:97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 26. | Heimann T, van Ginneken B, Styner MA, Arzhaeva Y, Aurich V, Bauer C, Beck A, Becker C, Beichel R, Bekes G, Bello F, Binnig G, Bischof H, Bornik A, Cashman PM, Chi Y, Cordova A, Dawant BM, Fidrich M, Furst JD, Furukawa D, Grenacher L, Hornegger J, Kainmüller D, Kitney RI, Kobatake H, Lamecker H, Lange T, Lee J, Lennon B, Li R, Li S, Meinzer HP, Nemeth G, Raicu DS, Rau AM, van Rikxoort EM, Rousson M, Rusko L, Saddi KA, Schmidt G, Seghers D, Shimizu A, Slagmolen P, Sorantin E, Soza G, Susomboon R, Waite JM, Wimmer A, Wolf I. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans Med Imaging. 2009;28:1251-1265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 734] [Cited by in RCA: 532] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 27. | Soler L, Hostettler A, Agnus V, Charnoz A, Fasquel J, Moreau J, Osswald A, Bou-hadjar M, Marescaux J. 3D image reconstruction for comparison of algorithm database: A patient specific anatomical and medical image database. IRCAD, Strasbourg, France, Tech. Rep (2010).. |
| 28. | Le DC, Chansangrat J, Keeratibharat N, Horkaew P. Symmetric Reconstruction of Functional Liver Segments and Cross-Individual Correspondence of Hepatectomy. Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 29. | Kumar SS, Moni RS, Rajeesh J. Automatic liver and lesion segmentation: a primary step in diagnosis of liver diseases. SIViP. 2013;7:163-172. [DOI] [Full Text] |
| 30. | Chen X, Udupa JK, Bagci U, Zhuge Y, Yao J. Medical image segmentation by combining graph cuts and oriented active appearance models. IEEE Trans Image Process. 2012;21:2035-2046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 111] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 31. | Huang L, Weng M, Shuai H, Huang Y, Sun J, Gao F. Automatic Liver Segmentation from CT Images Using Single-Block Linear Detection. Biomed Res Int. 2016;2016:9420148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 32. | Wu W, Zhou Z, Wu S, Zhang Y. Automatic Liver Segmentation on Volumetric CT Images Using Supervoxel-Based Graph Cuts. Comput Math Methods Med. 2016;2016:9093721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Mohamed RG, Seada NA, Hamdy S, Mostafa MG. An Adaptive Method for Fully Automatic Liver Segmentation in Medical MRI-Images. Int J Comput Appl. 2017;179:12-18. [DOI] [Full Text] |
| 34. | Zheng R, Wang Q, Lv S, Li C, Wang C, Chen W, Wang H. Automatic Liver Tumor Segmentation on Dynamic Contrast Enhanced MRI Using 4D Information: Deep Learning Model Based on 3D Convolution and Convolutional LSTM. IEEE Trans Med Imaging. 2022;41:2965-2976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 35. | Kainmüller D, Lange T, Lamecker H. Shape constrained automatic segmentation of the liver based on a heuristic intensity model. Proceedings of MICCAI Workshop 3D Segmentation in the Clinic; 2007 Oct 29; Brisbane, Australia. Berlin: Springer, 2007: 109-116. |
| 36. | Erdt M, Steger S, Kirschner M, Wesarg S. Fast automatic liver segmentation combining learned shape priors with observed shape deviation. Proceedings of IEEE 23rd Int. Symp. on Computer-Based Medical Systems (CBMS); 2010 Oct 12; Bentley, WA, Australia: IEEE, 2010: 249-254. [DOI] [Full Text] |
| 37. | Li G, Chen X, Shi F, Zhu W, Tian J, Xiang D. Automatic Liver Segmentation Based on Shape Constraints and Deformable Graph Cut in CT Images. IEEE Trans Image Process. 2015;24:5315-5329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 38. | Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation From CT Volumes. IEEE Trans Med Imaging. 2018;37:2663-2674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1078] [Cited by in RCA: 873] [Article Influence: 124.7] [Reference Citation Analysis (0)] |
| 39. | He R, Xu S, Liu Y, Li Q, Zhao N, Yuan Y, Zhang H. Three-Dimensional Liver Image Segmentation Using Generative Adversarial Networks Based on Feature Restoration. Front Med (Lausanne). 2021;8:794969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 40. | Lee SG, Kim E, Bae JS, Kim JH, Yoon S. Robust End-to-End Focal Liver Lesion Detection Using Unregistered Multiphase Computed Tomography Images. IEEE Trans Emerg Top Comput Intell. 2023;7:319-329. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 41. | Yang X, Yu HC, Choi Y, Lee W, Wang B, Yang J, Hwang H, Kim JH, Song J, Cho BH, You H. A hybrid semi-automatic method for liver segmentation based on level-set methods using multiple seed points. Comput Methods Programs Biomed. 2014;113:69-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 42. | Maklad AS, Matsuhiro M, Suzuki H, Kawata Y, Niki N, Satake M, Moriyama N, Utsunomiya T, Shimada M. Blood vessel-based liver segmentation using the portal phase of an abdominal CT dataset. Med Phys. 2013;40:113501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 43. | Chartrand G, Cresson T, Chav R, Gotra A, Tang A, De Guise JA. Liver Segmentation on CT and MR Using Laplacian Mesh Optimization. IEEE Trans Biomed Eng. 2017;64:2110-2121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 44. | Chen Y, Wang Z, Zhao W, Yang X. Liver segmentation from CT images based on region growing method. Proceedings of. 3rd Int. Conference on Bioinformatics and Bio-medical Engineering; 2009 Jun 11; Beijing, China. IEEE, 2009: 1-4. [DOI] [Full Text] |
| 45. | Liao M, Zhao YQ, Wang W, Zeng YZ, Yang Q, Shih FY, Zou BJ. Efficient liver segmentation in CT images based on graph cuts and bottleneck detection. Phys Med. 2016;32:1383-1396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 46. | Lu F, Wu F, Hu P, Peng Z, Kong D. Automatic 3D liver location and segmentation via convolutional neural network and graph cut. Int J Comput Assist Radiol Surg. 2017;12:171-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 142] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 47. | Selver MA, Kocaoğlu A, Demir GK, Doğan H, Dicle O, Güzeliş C. Patient oriented and robust automatic liver segmentation for pre-evaluation of liver transplantation. Comput Biol Med. 2008;38:765-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 48. | Foruzan AH, Zoroofi RA, Hori M, Sato Y. A knowledge-based technique for liver segmentation in CT data. Comput Med Imaging Graph. 2009;33:567-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 49. | Antonidoss A, Kaliyamurthie KP. Segmentation from images using adaptive threshold. Middle-East J Scient Res. 2014;20:479-484. |
| 50. | Avşar TS, Arıca S. Automatic Segmentation of Computed Tomography Images of Liver Using Watershed and Thresholding Algorithms. Proceedings of European Medical and Biological Engineering Conference and Nordic-Baltic Conference on Biomedical Engineering and Medical Physics (EMBEC & NBC), 2017 June 11; Tampere, Finland. Singapore: Springer, 2017: 414–417. [DOI] [Full Text] |
| 51. | Beck A, Aurich V. HepaTux-A semiautomatic liver segmentation system. Proceedings of MICCAI Workshop 3D Segmentation in the Clinic; 2007 Oct 29; Brisbane, Australia. Berlin: Springer, 2007: 225-234. |
| 52. | Ruskó L, Bekes G, Németh G, Fidrich M. Fully Automatic Liver Segmentation for Contrast Enhanced CT Images. Proceedings of MICCAI Workshop 3D Segmentation in the Clinic; 2007 Oct 29; Brisbane, Australia. Berlin: Springer, 2007: 143-150. |
| 53. | Lu XQ, Wu JS, Ren XY, Zhang BH, Li YH. The study and application of the improved region growing algorithm for liver segmentation. Optik. 2014;125:2142-2147. [DOI] [Full Text] |
| 54. | Lim SJ, Jeong YY, Ho YS. Segmentation of the Liver Using the Deformable Contour Method on CT Images. Proceedings of Advances in Multimedia Information Processing - PCM 2005; 2005 Nov 11; Jeju Island, Korea. Berlin: Springer, 2005: 570-581. [RCA] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 55. | Chi Y, Cashman P, Bello F, Kitney RI. A discussion on the evaluation of a new automatic liver volume segmentation method for specified CT image datasets. Proceedings of MICCAI Workshop 3D Segmentation in the Clinic; 2007 Oct 29; Brisbane, Australia. Berlin: Springer, 2007. 167-175. |
| 56. | Ciecholewski M. Automatic Liver Segmentation from 2D CT Images Using an Approximate Contour Model. J Sign Process Syst. 2014;74:151-174. [DOI] [Full Text] |
| 57. | Li D, Liu L, Chen J, Li H, Yin Y. A multistep liver segmentation strategy by combining level set based method with texture analysis for CT images. Proceedings of International Conference on Orange Technologies; 2014 Nov 20; Xi'an, China. IEEE, 2014: 109-112. [DOI] [Full Text] |
| 58. | Hu P, Wu F, Peng J, Liang P, Kong D. Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol. 2016;61:8676-8698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 110] [Article Influence: 12.2] [Reference Citation Analysis (1)] |
| 59. | Le TS, Tran DL. A Robust Liver Segmentation in CT-images Using 3D Level-Set Developed with the Edge and the Region Information. Proceedings of International Conference on Intelligent Information Technology; 2018 Feb 26; Ha Noi, Viet Nam. New York: ACM, 2018: 1-8. [DOI] [Full Text] |
| 60. | Yi F, Moon I, Image segmentation: A survey of graph-cut methods. Proceedings of International Conference on Systems and Informatics (ICSAI 2012); 2012 May 19; Yantai, China. IEEE, 2012: 1936-1941. [DOI] [Full Text] |
| 61. | Beichel R, Bornik A, Bauer C, Sorantin E. Liver segmentation in contrast enhanced CT data using graph cuts and interactive 3D segmentation refinement methods. Med Phys. 2012;39:1361-1373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 62. | Huang Q, Ding H, Wang X, Wang G. Fully automatic liver segmentation in CT images using modified graph cuts and feature detection. Comput Biol Med. 2018;95:198-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 63. | Yang S, Hongjinda S, Hanna SS, Gallinger S, Wei AC, Kiss A, Law C. Utility of preoperative imaging in evaluating colorectal liver metastases declines over time. HPB (Oxford). 2010;12:605-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 64. | Hu H, Zheng Q, Huang Y, Huang XW, Lai ZC, Liu J, Xie X, Feng ST, Wang W, Lu M. A non-smooth tumor margin on preoperative imaging assesses microvascular invasion of hepatocellular carcinoma: A systematic review and meta-analysis. Sci Rep. 2017;7:15375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (1)] |
| 65. | Michels NA. Newer anatomy of the liver and its variant blood supply and collateral circulation. Am J Surg. 1966;112:337-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 579] [Cited by in RCA: 560] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 66. | Hiatt JR, Gabbay J, Busuttil RW. Surgical anatomy of the hepatic arteries in 1000 cases. Ann Surg. 1994;220:50-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 545] [Cited by in RCA: 501] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 67. | Choi TW, Chung JW, Kim HC, Lee M, Choi JW, Jae HJ, Hur S. Anatomic Variations of the Hepatic Artery in 5625 Patients. Radiol Cardiothorac Imaging. 2021;3:e210007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 68. | Catalano OA, Singh AH, Uppot RN, Hahn PF, Ferrone CR, Sahani DV. Vascular and biliary variants in the liver: implications for liver surgery. Radiographics. 2008;28:359-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 154] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 69. | Faria LL, Darce GF, Bordini AL, Herman P, Jeismann VB, de Oliveira IS, Ortega CD, Rocha MS. Liver Surgery: Important Considerations for Pre- and Postoperative Imaging. Radiographics. 2022;42:722-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 70. | Fang CH, You JH, Lau WY, Lai EC, Fan YF, Zhong SZ, Li KX, Chen ZX, Su ZH, Bao SS. Anatomical variations of hepatic veins: three-dimensional computed tomography scans of 200 subjects. World J Surg. 2012;36:120-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 71. | Sureka B, Sharma N, Khera PS, Garg PK, Yadav T. Hepatic vein variations in 500 patients: surgical and radiological significance. Br J Radiol. 2019;92:20190487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 72. | Cawich SO, Sinanan A, Deshpande RR, Gardner MT, Pearce NW, Naraynsingh V. Anatomic variations of the intra-hepatic biliary tree in the Caribbean: A systematic review. World J Gastrointest Endosc. 2021;13:170-183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 5] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 73. | Smith TJ, Korngold E, Orloff SL. Preoperative imaging in colorectal liver metastases: Current practices. Curr Surg Rep. 2014;2:39. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 74. | Van Cutsem E, Nordlinger B, Adam R, Köhne CH, Pozzo C, Poston G, Ychou M, Rougier P; European Colorectal Metastases Treatment Group. Towards a pan-European consensus on the treatment of patients with colorectal liver metastases. Eur J Cancer. 2006;42:2212-2221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 430] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 75. | Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244:254-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 826] [Cited by in RCA: 1011] [Article Influence: 53.2] [Reference Citation Analysis (0)] |
| 76. | Patel S, Cheek S, Osman H, Jeyarajah DR. MRI with gadoxetate disodium for colorectal liver metastasis: is it the new "imaging modality of choice"? J Gastrointest Surg. 2014;18:2130-2135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 77. | Frankel TL, Gian RK, Jarnagin WR. Preoperative imaging for hepatic resection of colorectal cancer metastasis. J Gastrointest Oncol. 2012;3:11-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 26] [Reference Citation Analysis (0)] |
| 78. | van Kessel CS, Buckens CF, van den Bosch MA, van Leeuwen MS, van Hillegersberg R, Verkooijen HM. Preoperative imaging of colorectal liver metastases after neoadjuvant chemotherapy: a meta-analysis. Ann Surg Oncol. 2012;19:2805-2813. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 79. | Lucchese AM, Kalil AN, Schwengber A, Suwa E, Rolim de Moura GG. Usefulness of intraoperative ultrasonography in liver resections due to colon cancer metastasis. Int J Surg. 2015;20:140-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 80. | Hoareau J, Venara A, Lebigot J, Hamel JF, Lermite E, Caroli-Bosc FX, Aube C. Intraoperative Contrast-Enhanced Ultrasound in Colorectal Liver Metastasis Surgery Improves the Identification and Characterization of Nodules. World J Surg. 2016;40:190-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 81. | Langella S, Ardito F, Russolillo N, Panettieri E, Perotti S, Mele C, Giuliante F, Ferrero A. Intraoperative Ultrasound Staging for Colorectal Liver Metastases in the Era of Liver-Specific Magnetic Resonance Imaging: Is It Still Worthwhile? J Oncol. 2019;2019:1369274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 82. | Huf S, Platz Batista da Silva N, Wiesinger I, Hornung M, Scherer MN, Lang S, Stroszczynski C, Fischer T, Jung EM. Analysis of Liver Tumors Using Preoperative and Intraoperative Contrast-Enhanced Ultrasound (CEUS/IOCEUS) by Radiologists in Comparison to Magnetic Resonance Imaging and Histopathology. Rofo. 2017;189:431-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 83. | Niekel MC, Bipat S, Stoker J. Diagnostic imaging of colorectal liver metastases with CT, MR imaging, FDG PET, and/or FDG PET/CT: a meta-analysis of prospective studies including patients who have not previously undergone treatment. Radiology. 2010;257:674-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 393] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 84. | Egger J, Busse H, Brandmaier P, Seider D, Gawlitza M, Strocka S, Voglreiter P, Dokter M, Hofmann M, Kainz B, Hann A, Chen X, Alhonnoro T, Pollari M, Schmalstieg D, Moche M. Interactive Volumetry Of Liver Ablation Zones. Sci Rep. 2015;5:15373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 85. | Wu PH, Bedoya M, White J, Brace CL. Feature-based automated segmentation of ablation zones by fuzzy c-mean clustering during low-dose computed tomography. Med Phys. 2021;48:703-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 86. | Bai Z, Jiang H, Li S, Yao YD. Liver Tumor Segmentation Based on Multi-Scale Candidate Generation and Fractal Residual Network. IEEE Access. 2019;7:82122-82133. [DOI] [Full Text] |
| 87. | Xi XF, Wang L, Sheng VS, Cui Z, Fu B, Hu F. Cascade U-ResNets for Simultaneous Liver and Lesion Segmentation. IEEE Access. 2020;8:68944-68952. [DOI] [Full Text] |
| 88. | Han Y, Li X, Wang B, Wang L. Boundary Loss-Based 2.5D Fully Convolutional Neural Networks Approach for Segmentation: A Case Study of the Liver and Tumor on Computed Tomography. Algorithms. 2021;14. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 89. | Meng L, Zhang Q, Bu S. Two-Stage Liver and Tumor Segmentation Algorithm Based on Convolutional Neural Network. Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 90. | Child CG, Turcotte JG. Surgery and portal hypertension. Major Probl Clin Surg. 1964;1:1-85. [PubMed] |
| 91. | Pugh RN, Murray-Lyon IM, Dawson JL, Pietroni MC, Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg. 1973;60:646-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5490] [Cited by in RCA: 5738] [Article Influence: 110.3] [Reference Citation Analysis (2)] |
| 93. | Minagawa M, Makuuchi M, Torzilli G, Takayama T, Kawasaki S, Kosuge T, Yamamoto J, Imamura H. Extension of the frontiers of surgical indications in the treatment of liver metastases from colorectal cancer: long-term results. Ann Surg. 2000;231:487-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 466] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 94. | Aloia TA, Vauthey JN, Loyer EM, Ribero D, Pawlik TM, Wei SH, Curley SA, Zorzi D, Abdalla EK. Solitary colorectal liver metastasis: resection determines outcome. Arch Surg. 2006;141:460-6; discussion 466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 326] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 95. | Shoup M, Gonen M, D'Angelica M, Jarnagin WR, DeMatteo RP, Schwartz LH, Tuorto S, Blumgart LH, Fong Y. Volumetric analysis predicts hepatic dysfunction in patients undergoing major liver resection. J Gastrointest Surg. 2003;7:325-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 340] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 96. | Abdalla EK, Barnett CC, Doherty D, Curley SA, Vauthey JN. Extended hepatectomy in patients with hepatobiliary malignancies with and without preoperative portal vein embolization. Arch Surg. 2002;137:675-80; discussion 680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 337] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 97. | Kishi Y, Abdalla EK, Chun YS, Zorzi D, Madoff DC, Wallace MJ, Curley SA, Vauthey JN. Three hundred and one consecutive extended right hepatectomies: evaluation of outcome based on systematic liver volumetry. Ann Surg. 2009;250:540-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 364] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 98. | Fan ST, Lo CM, Liu CL, Yong BH, Chan JK, Ng IO. Safety of donors in live donor liver transplantation using right lobe grafts. Arch Surg. 2000;135:336-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 336] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 99. | Clavien PA, Emond J, Vauthey JN, Belghiti J, Chari RS, Strasberg SM. Protection of the liver during hepatic surgery. J Gastrointest Surg. 2004;8:313-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 57] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 100. | Hshieh TT, Sundaram V. Liver transplantation for hepatocellular carcinoma: are international guidelines possible? Hepatobiliary Surg Nutr. 2013;2:113-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 101. | Vauthey JN, Abdalla EK, Doherty DA, Gertsch P, Fenstermacher MJ, Loyer EM, Lerut J, Materne R, Wang X, Encarnacion A, Herron D, Mathey C, Ferrari G, Charnsangavej C, Do KA, Denys A. Body surface area and body weight predict total liver volume in Western adults. Liver Transpl. 2002;8:233-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 443] [Cited by in RCA: 465] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 102. | Ribero D, Amisano M, Bertuzzo F, Langella S, Lo Tesoriere R, Ferrero A, Regge D, Capussotti L. Measured versus estimated total liver volume to preoperatively assess the adequacy of the future liver remnant: which method should we use? Ann Surg. 2013;258:801-6; discussion 806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 103. | Shimoda M, Hariyama M, Oshiro Y, Suzuki S. Development of new software enabling automatic identification of the optimal anatomical liver resectable region, incorporating preoperative liver function. Oncol Lett. 2019;18:6639-6647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 104. | Selle D, Preim B, Schenk A, Peitgen HO. Analysis of vasculature for liver surgical planning. IEEE Trans Med Imaging. 2002;21:1344-1357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 228] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 105. | Yang X, Yang JD, Hwang HP, Yu HC, Ahn S, Kim BW, You H. Segmentation of liver and vessels from CT images and classification of liver segments for preoperative liver surgical planning in living donor liver transplantation. Comput Methods Programs Biomed. 2018;158:41-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 106. | Huang SH, Wang BL, Cheng M, Wu WL, Huang XY, Ju Y. A fast method to segment the liver according to Couinaud's classification. Proceedings of Medical Imaging and Informatics, Gao X, Müller H, Loomes MJ, Comley R, Luo S, editors. Lecture Notes in Computer Science, Springer, Berlin, Germany. 2008; 4987: 270-276. [DOI] [Full Text] |
| 107. | Debarba HG, Zanchet DJ, Fracaro D, Maciel A, Kalil AN. Efficient liver surgery planning in 3D based on functional segment classification and volumetric information. Annu Int Conf IEEE Eng Med Biol Soc. 2010;2010:4797-4800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 108. | Chen Y, Yue X, Zhong C, Wang G. Functional Region Annotation of Liver CT Image Based on Vascular Tree. Biomed Res Int. 2016;2016:5428737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 109. | Zhang Q, Fan YF, Wan JF, Liu YX. An Efficient and Clinical-Oriented 3D Liver Segmentation Method. IEEE Access. 2017;5:18737-18744. [DOI] [Full Text] |
| 110. | Boltcheva D, Passat N, Agnus V, Col MA JD, Ronse C, Soler L, Automatic anatomical segmentation of the liver by separation planes. Proceedings of SPIE 6141, Medical Imaging; 2006 March 13; San Diego, California, USA: SPIE, 2006: 383-394. [DOI] [Full Text] |
| 111. | Oliveira D, Feitosa R, Correia M. Automatic Couinaud liver and veins segmentation from CT images. Proceedings of 1st Int. Conference on Bio-inspired Systems and Signal Processing (BIOSTEC 2008); 2008 Jan 28; Madeira, Portugal. SciTePress, 2008: 249-252. [DOI] [Full Text] |
| 112. | Ruskó L, Mátéka I, Kriston A. Virtual volume resection using multi-resolution triangular representation of B-spline surfaces. Comput Methods Programs Biomed. 2013;111:315-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Reference Citation Analysis (0)] |
| 113. | Butdee C, Pluempitiwiriyawej C, Tanpowpong N. 3D plane cuts and cubic Bézier curve for CT liver volume segmentation according to Couinaud's classification, Songk. J Sci Tech. 2017;39:793-801. [DOI] [Full Text] |
| 114. | Lebre MA, Vacavant A, Grand-Brochier M, Rositi H, Abergel A, Chabrot P, Magnin B. Automatic segmentation methods for liver and hepatic vessels from CT and MRI volumes, applied to the Couinaud scheme. Comput Biol Med. 2019;110:42-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 115. | Le DC, Chansangrat J, Keeratibharat N, Horkaew P. Functional Segmentation for Pre-operative Liver Resection Based on Hepatic Vascular Networks. IEEE Access. 2021;9:15485-15498. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 116. | Alirr OI, Abd Rahni AA. Automatic atlas-based liver segmental anatomy identification for hepatic surgical planning. Int J Comput Assist Radiol Surg. 2020;15:239-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 117. | Chansangrat J, Keeratibharat N. Portal vein embolization: rationale, techniques, outcomes and novel strategies. Hepat Oncol. 2021;8:HEP42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 118. | Ge PL, Du SD, Mao YL. Advances in preoperative assessment of liver function. Hepatobiliary Pancreat Dis Int. 2014;13:361-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 119. | Lim MC, Tan CH, Cai J, Zheng J, Kow AW. CT volumetry of the liver: where does it stand in clinical practice? Clin Radiol. 2014;69:887-895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 120. | Nanashima A, Yamaguchi H, Shibasaki S, Sawai T, Yamaguchi E, Yasutake T, Tsuji T, Jibiki M, Nakagoe T, Ayabe H. Measurement of serum hyaluronic acid level during the perioperative period of liver resection for evaluation of functional liver reserve. J Gastroenterol Hepatol. 2001;16:1158-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 121. | Lee SG, Hwang S. How I do it: assessment of hepatic functional reserve for indication of hepatic resection. J Hepatobiliary Pancreat Surg. 2005;12:38-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 66] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 122. | Yokoyama Y, Ebata T, Igami T, Sugawara G, Mizuno T, Yamaguchi J, Nagino M. The Predictive Value of Indocyanine Green Clearance in Future Liver Remnant for Posthepatectomy Liver Failure Following Hepatectomy with Extrahepatic Bile Duct Resection. World J Surg. 2016;40:1440-1447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 123. | Abdalla EK, Denys A, Chevalier P, Nemr RA, Vauthey JN. Total and segmental liver volume variations: implications for liver surgery. Surgery. 2004;135:404-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 186] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 124. | Dello SA, Stoot JH, van Stiphout RS, Bloemen JG, Wigmore SJ, Dejong CH, van Dam RM. Prospective volumetric assessment of the liver on a personal computer by nonradiologists prior to partial hepatectomy. World J Surg. 2011;35:386-392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 125. | Simpson AL, Geller DA, Hemming AW, Jarnagin WR, Clements LW, D'Angelica MI, Dumpuri P, Gönen M, Zendejas I, Miga MI, Stefansic JD. Liver planning software accurately predicts postoperative liver volume and measures early regeneration. J Am Coll Surg. 2014;219:199-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (2)] |
| 126. | Shindoh J, Truty MJ, Aloia TA, Curley SA, Zimmitti G, Huang SY, Mahvash A, Gupta S, Wallace MJ, Vauthey JN. Kinetic growth rate after portal vein embolization predicts posthepatectomy outcomes: toward zero liver-related mortality in patients with colorectal liver metastases and small future liver remnant. J Am Coll Surg. 2013;216:201-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 236] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 127. | Shi M, Guo RP, Lin XJ, Zhang YQ, Chen MS, Zhang CQ, Lau WY, Li JQ. Partial hepatectomy with wide versus narrow resection margin for solitary hepatocellular carcinoma: a prospective randomized trial. Ann Surg. 2007;245:36-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 404] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 128. | Fang C, Zhang P, Qi X. Digital and intelligent liver surgery in the new era: Prospects and dilemmas. EBioMedicine. 2019;41:693-701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 129. | Schneider C, Allam M, Stoyanov D, Hawkes DJ, Gurusamy K, Davidson BR. Performance of image guided navigation in laparoscopic liver surgery - A systematic review. Surg Oncol. 2021;38:101637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 130. | Baiocchi GL, Diana M, Boni L. Indocyanine green-based fluorescence imaging in visceral and hepatobiliary and pancreatic surgery: State of the art and future directions. World J Gastroenterol. 2018;24:2921-2930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 98] [Cited by in RCA: 93] [Article Influence: 13.3] [Reference Citation Analysis (2)] |
| 131. | Potharazu AV, Gangemi A. Indocyanine green (ICG) fluorescence in robotic hepatobiliary surgery: A systematic review. Int J Med Robot. 2023;19:e2485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 132. | Pollmann L, Juratli M, Roushansarai N, Pascher A, Hölzen JP. Quantification of Indocyanine Green Fluorescence Imaging in General, Visceral and Transplant Surgery. J Clin Med. 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 133. | Šteňo A, Buvala J, Babková V, Kiss A, Toma D, Lysak A. Current Limitations of Intraoperative Ultrasound in Brain Tumor Surgery. Front Oncol. 2021;11:659048. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 134. | Reitinger B, Bornik A, Beichel R, Schmalstieg D. Liver surgery planning using virtual reality. IEEE Comput Graph Appl. 2006;26:36-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 52] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 135. | Khor WS, Baker B, Amin K, Chan A, Patel K, Wong J. Augmented and virtual reality in surgery-the digital surgical environment: applications, limitations and legal pitfalls. Ann Transl Med. 2016;4:454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 160] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 136. | Longo UG, De Salvatore S, Candela V, Zollo G, Calabrese G, Fioravanti S, Giannone L, Marchetti A, De Marinis MG, Denaro V. Augmented Reality, Virtual Reality and Artificial Intelligence in Orthopedic Surgery: A Systematic Review. Appl Sci. 2021;11:3253. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 137. | Bourdel N, Collins T, Pizarro D, Bartoli A, Da Ines D, Perreira B, Canis M. Augmented reality in gynecologic surgery: evaluation of potential benefits for myomectomy in an experimental uterine model. Surg Endosc. 2017;31:456-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 138. | Choi PT, Lam KC, Lui LM. FLASH: Fast Landmark Aligned Spherical Harmonic Parameterization for Genus-0 Closed Brain Surfaces. SIAM J Imaging Sci. 2015;8:67-94. [DOI] [Full Text] |
| 139. | Bidgood WD Jr, Horii SC. Introduction to the ACR-NEMA DICOM standard. Radiographics. 1992;12:345-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 96] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 140. | Graham RN, Perriss RW, Scarsbrook AF. DICOM demystified: a review of digital file formats and their use in radiological practice. Clin Radiol. 2005;60:1133-1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 71] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 141. | Bidgood WD Jr, Horii SC, Prior FW, Van Syckle DE. Understanding and using DICOM, the data interchange standard for biomedical imaging. J Am Med Inform Assoc. 1997;4:199-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 188] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 142. | NIfTI. Neuroimaging Informatics Technology Initiative (NIfTI). 18 Dec 2013. [cited 25 Jun 2023]. Available from: https://nifti.nimh.nih.gov/. |
| 143. | FDA. Software as a Medical Device (SaMD). 12 April 2018. [cited 26 Jun 2023]. Available from: https://www.fda.gov/medical-devices/digital-health-center-excellence/software-medical-device-samd. |
| 144. | IMDRF. Software as a Medical Device: Possible Framework for Risk Categorization and Corresponding Considerations. 18 Sep 2014. [cited 26 Jun 2023]. Available from: https://www.imdrf.org/documents/software-medical-device-possible-framework-risk-categorization-and-corresponding-considerations. |
| 145. | CDRH. General Principles of Software Validation; Final Guidance for Industry and FDA Staff, Food and Drug Administration, U.S. Department Of Health and Human Services. 11 Jan 2002. [cited 26 Jun 2023]. Available from: https://www.fda.gov/media/73141/download. |