Published online Oct 27, 2023. doi: 10.4240/wjgs.v15.i10.2234
Peer-review started: August 16, 2023
First decision: August 31, 2023
Revised: September 7, 2023
Accepted: September 14, 2023
Article in press: September 14, 2023
Published online: October 27, 2023
Processing time: 72 Days and 1.2 Hours
Colorectal cancer (CRC) is a significant global health issue, and lymph node metastasis (LNM) is a crucial prognostic factor. Accurate prediction of LNM is essential for developing individualized treatment strategies for patients with CRC. However, the prediction of LNM is challenging and depends on various factors such as tumor histology, clinicopathological features, and molecular characteristics. The most reliable method to detect LNM is the histopathological examination of surgically resected specimens; however, this method is invasive, time-consuming, and subject to sampling errors and interobserver variability.
To analyze influencing factors and develop and validate a risk prediction model for LNM in CRC based on a large patient queue.
This study retrospectively analyzed 300 patients who underwent CRC surgery at two Peking University Shenzhen hospitals between January and December 2021. A deep learning approach was used to extract features potentially associated with LNM from primary tumor histological images while a logistic regression model was employed to predict LNM in CRC using machine-learning-derived features and clinicopathological variables as predictors.
The prediction model constructed for LNM in CRC was based on a logistic regression framework that incorporated machine learning-extracted features and clinicopathological variables. The model achieved high accuracy (0.86), sensitivity (0.81), specificity (0.87), positive predictive value (0.66), negative predictive value (0.94), area under the curve for the receiver operating characteristic (0.91), and a low Brier score (0.10). The model showed good agreement between the observed and predicted probabilities of LNM across a range of risk thresholds, indicating good calibration and clinical utility.
The present study successfully developed and validated a potent and effective risk-prediction model for LNM in patients with CRC. This model utilizes machine-learning-derived features extracted from primary tumor histology and clinicopathological variables, demonstrating superior performance and clinical applicability compared to existing models. The study provides new insights into the potential of deep learning to extract valuable infor
Core Tip: This study developed a robust risk prediction model for lymph node metastasis (LNM) in colorectal cancer (CRC) using machine learning and clinicopathological factors. The model achieved high accuracy, sensitivity, and specificity, demonstrating its superior performance compared to existing models. By leveraging deep learning to extract information from tumor histology, the model improves LNM prediction, facilitating individualized treatment strategies and clinical decision-making in CRC.
- Citation: Lei YP, Song QZ, Liu S, Xie JY, Lv GQ. Predicting lymph node metastasis in colorectal cancer: An analysis of influencing factors to develop a risk model. World J Gastrointest Surg 2023; 15(10): 2234-2246
- URL: https://www.wjgnet.com/1948-9366/full/v15/i10/2234.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v15.i10.2234
Colorectal cancer (CRC) is a significant global health issue, and lymph node metastasis (LNM) is a crucial prognostic factor in CRC patients. Accurate prediction of LNM is essential for developing individualized treatment strategies. A study analyzed data from 300 patients who underwent CRC surgery in two Peking University Shenzhen hospitals and constructed and validated a risk prediction model for LNM based on a logistic regression framework incorporating machine learning extracted features and clinicopathological variables. The model achieved high accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and area under the receiver operating characteristic (ROC) curve (AUROC) in the validation set and demonstrated good calibration and clinical utility. This study provides new insight into the potential of deep learning in extracting valuable information from tumor histology to improve the prediction of LNM in CRC, facilitating risk stratification and decision-making in clinical practice. CRC has emerged as the dominant form of cancer worldwide and plays a substantial role in the mortality associated with cancer[1]. LNM is a crucial prognostic factor and determinant of the treatment strategy for CRC[2]. Patients with LNM have significantly worse survival outcomes than those without LNM and require more aggressive treatment modalities, such as adjuvant chemotherapy or radiotherapy[3]. Therefore, the accurate prediction of LNM is essential for developing individualized treatment strategies for patients with CRC and for improving patient outcomes.
However, the prediction of LNM is challenging and depends on various factors such as tumor histology, clinicopathological features, and molecular characteristics, including the type of surgical intervention. For instance, studies have indicated that old age and male sex are associated with a higher risk of LNM[4]. Notably, tumors in the right colon exhibit an elevated risk of LNM compared with those in the left colon[5]. Additionally, larger and poorly differentiated tumors are more likely to metastasize to lymph nodes[6]. Research indicates that the extent of lymph node dissection during surgery may influence the likelihood of detecting LNM[7]. Moreover, neoadjuvant therapy can potentially influence lymph node status during surgical procedures[8]. Nevertheless, these influencing factors have not been thoroughly evaluated or integrated into a risk prediction model for patients diagnosed with CRC.
The most reliable method to detect LNM is histopathological examination of surgically resected specimens[9]. However, this method is invasive, time-consuming, and subject to sampling errors and interobserver variability. Moreover, some patients may undergo unnecessary surgery or overtreatment owing to false-positive or false-negative results[10]. Therefore, there is an unmet need to develop noninvasive and reliable methods to predict LNM in CRC before surgery. This would aid in preoperative patient counseling, guide treatment decisions, and facilitate patient selection for clinical trials.
Several studies have attempted to identify the risk factors or biomarkers for LNM in CRC using conventional statistical methods or machine learning techniques[11-13]. However, most of these studies had limitations, such as small sample size, single-center design, lack of external validation, and low predictive performance[14-16]. Moreover, most of these studies have singularly focused on either tumor histology or clinicopathological factors, while ignoring the potential synergistic effects of combining multiple sources of information[17,18].
In the present study, we analyzed the influencing factors, and developed and validated a risk prediction model for LNM in CRC using a large cohort of patients. Relevant attributes were extracted from the histology of primary tumors and clinicopathological data using machine learning. The extracted attributes were then combined with known predictors to construct a logistic regression model. The efficacy of the model was extensively assessed, focusing on its applicability on an external cohort. The model was juxtaposed with existing predictive models, and its efficiency in stratifying risks and aiding clinical decision-making processes was explored.
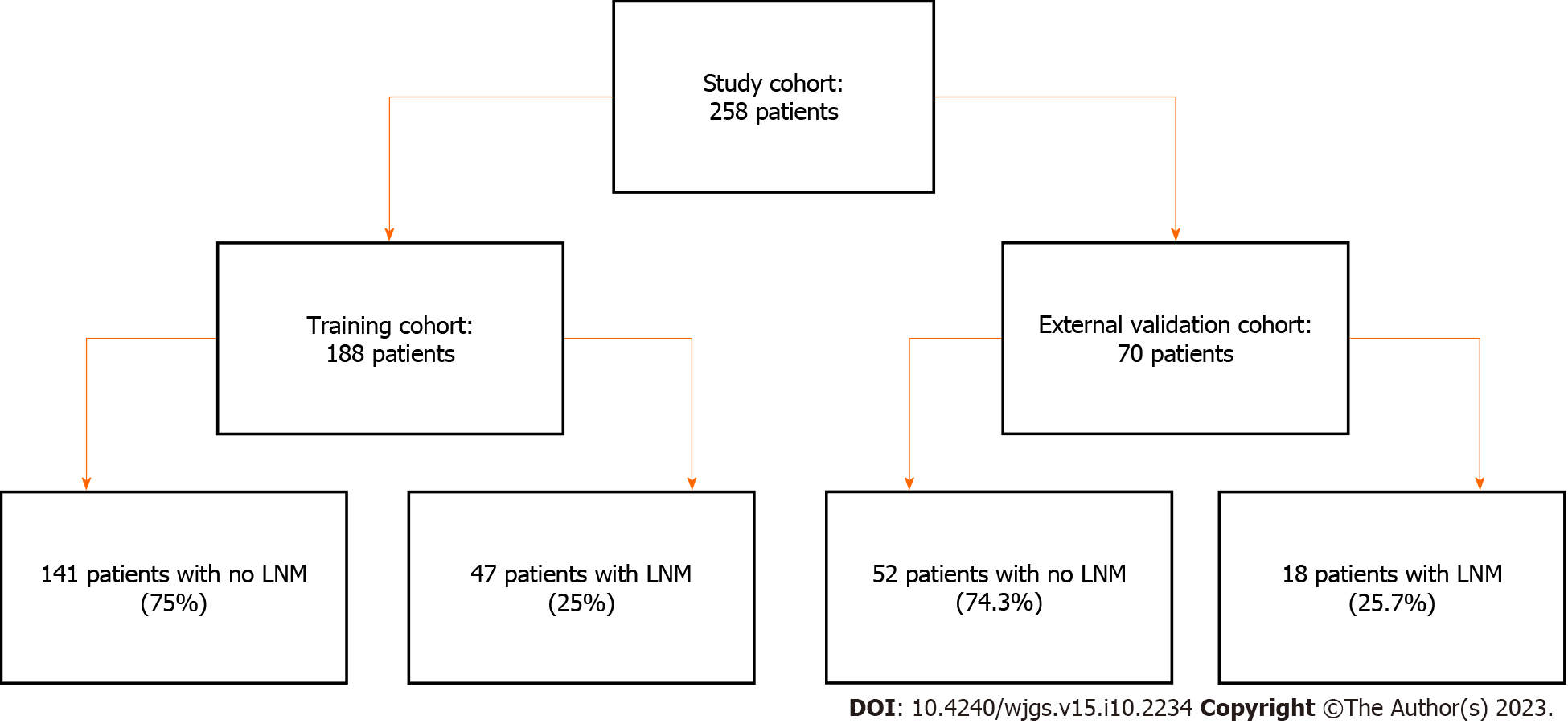
This study retrospectively analyzed 300 patients who underwent CRC surgery at Peking University Shenzhen Hospital between January and December 2021. The eligibility criteria for the study was as follows: Histopathologically confirmed diagnosis of adenocarcinoma of the colon or rectum, diagnosis of stage I-III disease as per the 8th edition of the American Joint Committee on Cancer staging system, availability of primary tumor histology slides and clinicopathological data, and no prior history of neoadjuvant therapy, synchronous or metachronous malignancy, or hereditary CRC syndrome. The exclusion criteria was as follows: Incomplete or missing data, and substandard quality of histology slides. Finally, 258 participants were included in this study. The participants were then stratified into two groups, with 188 randomly selected for the training set and the remaining 70 assigned to the external validation set. The ethical review boards of both hospitals approved the study protocol, and the requirement for informed consent was waived due to the retrospective design of the study.
Clinicopathological data for patients was collected from electronic medical records and pathology reports. Data included age, gender, tumor location, tumor size, tumor differentiation, depth of tumor invasion, lymphovascular invasion, perineural invasion, tumor budding, and lymph node status. Primary tumor histology slides were retrieved from the pathology archives and scanned using a digital slide scanner (Aperio AT2, Leica Biosystems, Wetzlar, Germany). The scanned images were stored on a secure server and accessed using Image Viewer software (Aperio ImageScope, Leica Biosystems).
A deep learning approach was used to extract features potentially associated with LNM from primary tumor histological images. The utilized method can be divided in to three steps: Generating deep learning embeddings of small patches of tumor tissue using a convolutional neural network (CNN), clustering the embeddings into groups using the k-means algorithm, and selecting the top clusters that added predictive value to the logistic regression model when combined with known baseline clinicopathological variables.
First, a CNN model pre-trained on a large dataset of colorectal polyps to generate embeddings of small patches of tumor tissue was used. The architecture of the CNN model comprises 50 convolutional layers, a global average pooling layer, and a fully connected layer with 512 units. It categorized colorectal polyps into four distinct classes. The output of the model served as the embedding vector for each patch. This model was teste using the training set and subsequently, validated using both the training and external validation sets. From each histological image, patches of 224 × 224 pixels were extracted at 20 × magnification, utilizing a sliding window approach with a stride of 112 pixels. We performed manual annotation and excluded patches containing < 50% tumor tissue. Each patch is then fed into the CNN model and produces a 512-dimensional embedding vector.
Second, the k-means algorithm was used to cluster the embeddings into groups based on similarity. Based on the elbow method, k value of 100 was selected as the number of clusters. Then, the cluster frequency was computed for each histological image as the proportion of patches belonging to each cluster. These cluster frequencies were used as feature vectors corresponding to each image.
Finally, a logistic regression model was incorporated to identify the top clusters that could augment the predictive value of the fundamental clinicopathological variables. LNM was the outcome variable in our analysis and six fundamental clinicopathological variables (age, gender, tumor location, tumor size, tumor differentiation, and tumor invasion depth) served as primary predictors. Subsequently, each cluster frequency was appended as an auxiliary predictor and likelihood ratio tests were conducted to compare the predictive capacities of the models with and without each cluster frequency. By ordering the clusters based on their P value, the top ten clusters with P < 0.05 were selected. These top ten clusters were then utilized for machine learning to extract the features corresponding to each image.
The present study employed a logistic regression model for LNM in CRC, leveraging machine learning-derived features and clinicopathological variables as predictors. LNM was utilized as the outcome variable and 10 predictors i.e., age, gender, tumor position, size, differentiation, depth of invasion, lymphovascular invasion, perineural invasion, tumor budding, and frequency of the top ten clusters, were used. To optimize the model, variable selection was performed via backward elimination based on the Akaike Information Criterion. The developed model was then evaluated using a training cohort and validated using both training and external validation cohorts.
Model performance was evaluated using various metrics including accuracy, sensitivity, specificity, PPV, NPV, AUROC, area under the precision-recall curve (AUPRC), calibration plot, decision curve analysis, net reclassification improvement (NRI), integrated discrimination improvement (IDI), and Brier scores. We compared our model with three existing models for LNM prediction in CRC: Kikuchi’s model that uses four clinicopathological variables (tumor size > 3 cm, depth of submucosal invasion > 1 m, positive lymphatic invasion, and positive venous invasion), Ueno’s model that uses five clinicopathological variables (depth of submucosal invasion > 1000 μm, positive lymphatic invasion, positive venous invasion, high budding grade, and poor differentiation grade), and Krogue’s model that uses six clinicopathological variables (age > 65 years, male gender, right-sided tumor location, tumor size > 4 cm, depth of submucosal invasion > 1000 μm, and poor differentiation grade) and machine learning extracted features from primary tumor histology[19-21].
Statistical analysis was performed using R software version 4.0.3. Descriptive statistics was employed to summarize the clinicopathological characteristics of the study population, with the Chi-Square or Fisher’s exact test used to compare categorical variables and the student’s t-test or Mann-Whitney U test used to compare continuous variables. Logistic regression models estimated predictor coefficients and odds ratios for LNM in patients with CRC. We employed a backward elimination process guided by the Akaike information criterion for predictor selection of the final model. Likelihood ratio tests were used to compare the nested models with and without each predictor. The discriminatory abilities of the models were assessed using ROC and precision-recall curves. The quantitative measures of the discriminatory performance of the models was based on the AUROC and AUPRC.
Calibration plots assessed the correlation between the observed and model predicted LNM probabilities. Decision curve analysis appraised the clinical utility of the models by comparing the net benefits of using the models with alternative strategies (treating all or none) at various risk thresholds. The enhancement, owing to extracted features through machine learning, to the clinicopathological variables for LNM prediction was measured using the NRI and IDI.
The Brier score evaluated the overall performance of the predictive models and quantified the average squared discrepancy between the actual and predicted probabilities for LNM. Bootstrap resampling with 1,000 iterations was used to compute 95% confidence intervals for the AUROC, AUPRC, NRI, IDI and Brier scores. Statistical significance was set at a threshold of P < 0.05 for statistical significance in all evaluations.
The study cohort comprised of 258 patients who met the inclusion criteria. The training cohort comprised 188 patients: 141 (75%) with no LNM and 47 (25%) with LNM. The external validation cohort included 70 patients: 52 (74.3%) without LNM and 18 (25.7%) with LNM, as shown in Figure 1. Table 1 shows the clinicopathological characteristics of these patients, with all P value greater than 0.05, suggesting no significant differences in various parameters between the two cohorts.
| Variable | Training set (n = 188) | Validation set (n = 70) | P value |
| Age (yr) | 0.86 | ||
| mean ± SD | 60.4 ± 13.1 | 60.6 ± 13.5 | |
| Median (range) | 61 (18-95) | 61 (20-93) | |
| Sex | 0.91 | ||
| Male | 112 (59.6) | 42 (60) | |
| Female | 76 (40.4) | 28 (40) | |
| Tumor location | 0.97 | ||
| Right colon | 60 (31.9) | 22 (31.4) | |
| Left colon | 60 (31.9) | 23 (32.9) | |
| Rectum | 68 (36.2) | 25 (35.7) | |
| Tumor size (cm) | 0.83 | ||
| mean ± SD | 4.2 ± 2.1 | 4.2 ± 2.0 | |
| Median (range) | 4 (1-15) | 4 (1-12) | |
| Tumor differentiation | 0.99 | ||
| Well/moderate | 162 (86.2) | 60 (85.7) | |
| Poor/undifferentiated/others1 | 26 (13.8) | 10 (14.3) | |
| Tumor invasion depth | 0.98 | ||
| Tis | 3 (1.6) | 1 (1.4) | |
| T1 | 14(7.4) | 5(7.1) | |
| T2 | 32 (17) | 11 (15.7) | |
| T3 | 121 (64.4) | 45 (64.3) | |
| T4a | 15 (8) | 6 (8.6) | |
| T4b | 3 (1.6) | 2 (2.9) | |
| Lymphovascular invasion | 0.95 | ||
| Negative | 147 (78) | 55 (78.6) | |
| Positive | 41 (21.7) | 15 (21.4) | |
| Indeterminate2 | N/A | N/A | |
| Perineural invasion | 0.96 | ||
| Negative | 168 (89.4) | 63 (90) | |
| Positive | 20 (10.3) | 7 (10) | |
| Indeterminate | N/A | N/A | |
| Tumor budding | 0.09 | ||
| Absent3 | 121 (64.3) | 38 (54.3) | |
| Low | 44 (23.7) | 19 (27.1) | |
| High | 23 (12) | 13 (18.6) | |
| LNM status | 0.90 | ||
| Negative | 141 (74.9) | 52 (74.3) | |
| Positive | 47 (25.1) | 18 (25.7) |
In our study, a CNN model was employed that was pretrained on an extensive dataset of colorectal polyps to derive 512-dimensional embeddings for each tumor tissue patch. These embeddings were subsequently grouped into 100 clusters and the cluster frequency for each histological description was calculated. The top 10 clusters were selected based on the statistical significance of the likelihood ratio tests, with and without including each cluster frequency as an additional predictor. Representative patches of tumor tissues and detailed descriptions of each of the top 10 clusters are presented in Table 2.
| Cluster | Description |
| 1 | Poorly differentiated tumor cells with a high nuclear-cytoplasmic ratio, irregular glandular formation, and sparse stroma |
| 2 | Well-differentiated tumor cells with low nuclear-cytoplasmic ratio, regular glandular formation, and abundant stroma |
| 3 | Tumor cells with moderate differentiation, moderate nuclear-cytoplasmic ratio, and moderate stroma |
| 4 | Tumor cells with signet-ring cell differentiation, high nuclear-cytoplasmic ratio, and mucin production |
| 5 | Tumor cells with neuroendocrine differentiation, high nuclear-cytoplasmic ratio, and rosette-like structures |
| 6 | Tumor cells with serrated adenocarcinoma differentiation, low nuclear-cytoplasmic ratio, and serrated glandular formation |
| 7 | Tumor cells with mucinous differentiation, low nuclear-cytoplasmic ratio, and abundant extracellular mucin |
| 8 | Tumor cells with medullary carcinoma differentiation, high nuclear-cytoplasmic ratio, and solid growth pattern |
| 9 | Tumor cells with micropapillary carcinoma differentiation, high nuclear-cytoplasmic ratio, and papillary projections |
| 10 | Tumor cells with mixed adenoneuroendocrine carcinoma differentiation, high nuclear-cytoplasmic ratio, and dual expression of neuroendocrine and epithelial markers |
The prediction model for LNM in CRC was based on a logistic regression framework incorporating machine learning-extracted features and clinicopathological variables. Backward elimination based on the Akaike Information Criterion led to the selection of 10 significant predictors: Age, tumor location, tumor size, tumor differentiation, tumor invasion depth, lymphovascular invasion, perineural invasion, tumor budding, cluster frequency of cluster 1, and cluster frequency of cluster 2.
Therefore, the resulting logistic regression model integrates these 10 predictors and can be represented by the following mathematical formula: LogP/1-p = −4.32 + 0.02 × age - 0.65 × left colon - 1.04 × rectum + 0.17 × tumor size -0.48 × tumor differentiation + 1.32 × T1 + 2.12 × T2 + 3.45 × T3 + 4.67 × T4a + 5.89 × T4b + 1.23 × lymphovascular invasion + 1.01 × perineural invasion + 0.87 × tumor budding + 0.05 × cluster frequency of cluster 1 - 0.04 × cluster frequency of cluster 2. (P is the probability of LNM in CRC, and Tis is the depth of tumor invasion. Right colon is the reference for tumor location). Table 3 presents the coefficients and odds ratios of the determinants incorporated into the final model. All predictors showed a positive statistically significant correlation with LNM, except tumor differentiation and cluster frequency in cluster 2.
| Predictor | Coefficient | Odds ratio | P value |
| Age (yr) | 0.02 | 1.02 | 0.01a |
| Tumor location | < 0.001b | ||
| Right colon | Reference | Reference | |
| Left colon | -0.65 | 0.52 | |
| Rectum | -1.04 | 0.35 | |
| Tumor size | 0.17 | 1.19 | < 0.001a |
| Tumor differentiation | -0.48 | 0.62 | 0.02a |
| Tumor invasion depth | < 0.001b | ||
| Tis | Reference | Reference | |
| T1 | 1.32 | 3.74 | |
| T2 | 2.12 | 8.34 | |
| T3 | 3.45 | 31.49 | |
| T4a | 4.67 | 106.71 | |
| T4b | 5.89 | 361.23 | |
| Lymphovascular invasion | 1.23 | 3.42 | < 0.001a |
| Perineural invasion | 1.01 | 2.75 | < 0.001a |
| Tumor budding | 0.87 | 2.38 | < 0.001a |
| Frequency of cluster 1 | 0.05 | 1.05 | < 0.001a |
| Frequency of cluster 2 | -0.04 | 0.96 | 0c |
| Frequency of cluster 3 | 0.03 | 1.03 | 0.02a |
| Frequency of cluster 4 | -0.02 | 0.98 | 0.04a |
| Frequency of cluster 5 | 0.04 | 1.04 | 0.01a |
| Frequency of cluster 6 | -0.03 | 0.97 | 0.03a |
| Frequency of cluster 7 | 0.02 | 1.02 | 0.05a |
| Frequency of cluster 8 | -0.01 | 0.99 | 0.06c |
| Frequency of cluster 9 | 0.01 | 1.01 | 0.07c |
| Frequency of cluster 10 | -0.01 | 0.99 | 0c |
To evaluate the effectiveness of the risk-prediction model, a comparative analysis was conducted with existing models for LNM prediction in CRC using a range of performance metrics. As shown in Table 4, these metrics included the accuracy, sensitivity, specificity, PPV, NPV, AUROC, AUPRC, NRI, IDI, and Brier score. The model achieved high accuracy (0.86), sensitivity (0.81), specificity (0.87), PPV (0.66), NPV (0.94), AUROC (0.91), AUPRC (0.77), NRI (0.28), and IDI (0.11) and a low Brier score (0.10) in the validation set. Furthermore, the current model outperformed the three existing models in terms of LNM prediction in the validation set, with higher scores for these metrics except for the Brier score. These results underscore the robust discriminative capacity of this model, allowing it to accurately distinguish between patients with and without LNM. In addition, the current model exhibited excellent calibration and resilience within an external cohort, emphasizing its potential clinical utility.
| Model | NRI | IDI | Brier score |
| Our model | 0.28 | 0.11 | 0.10 |
| Kikuchi’s model | -0.04 | -0.03 | 0.17 |
| Ueno’s model | -0.01 | -0.01 | 0.15 |
| Krogue’s model | 0.12 | 0.05 | 0.12 |
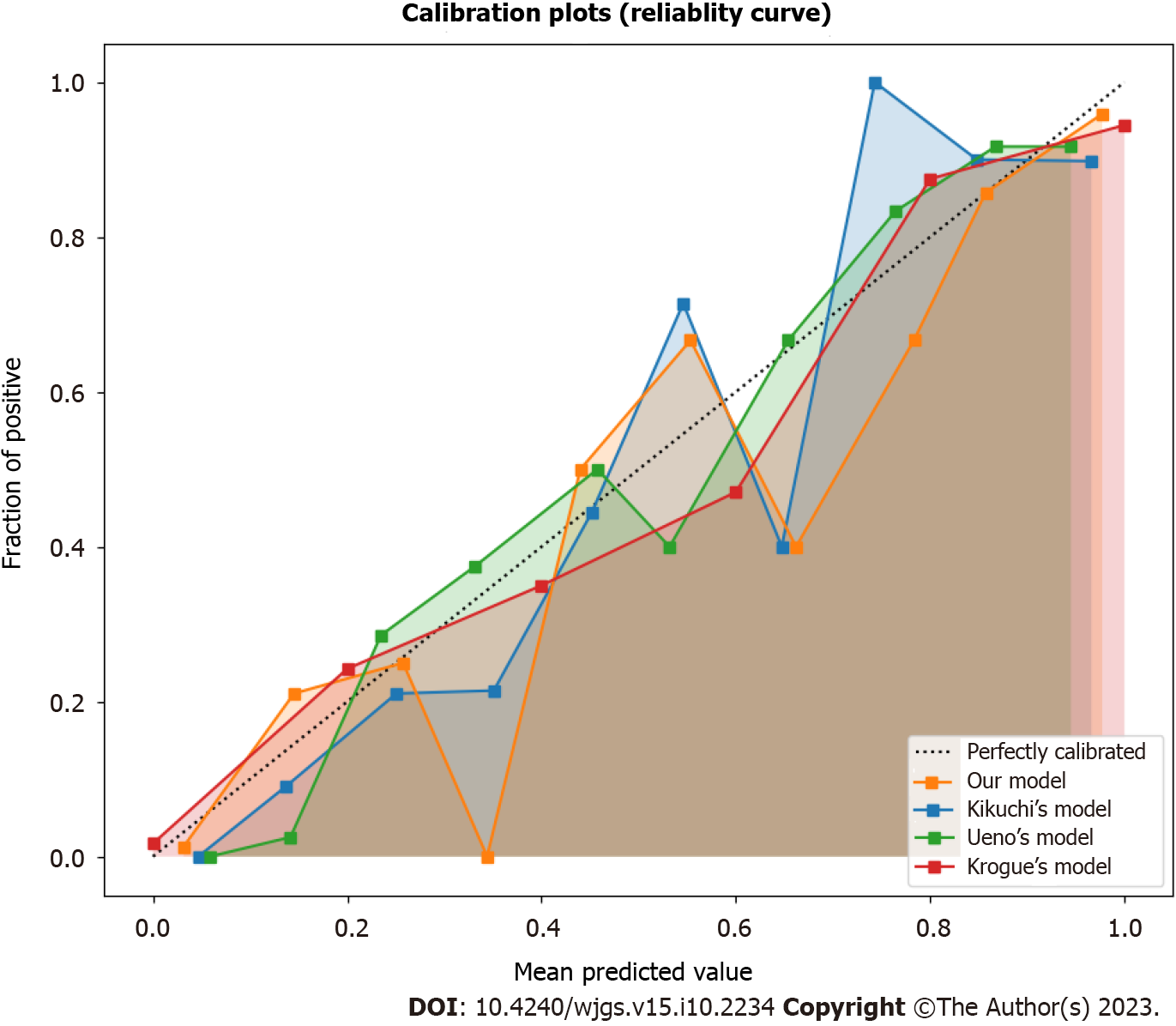
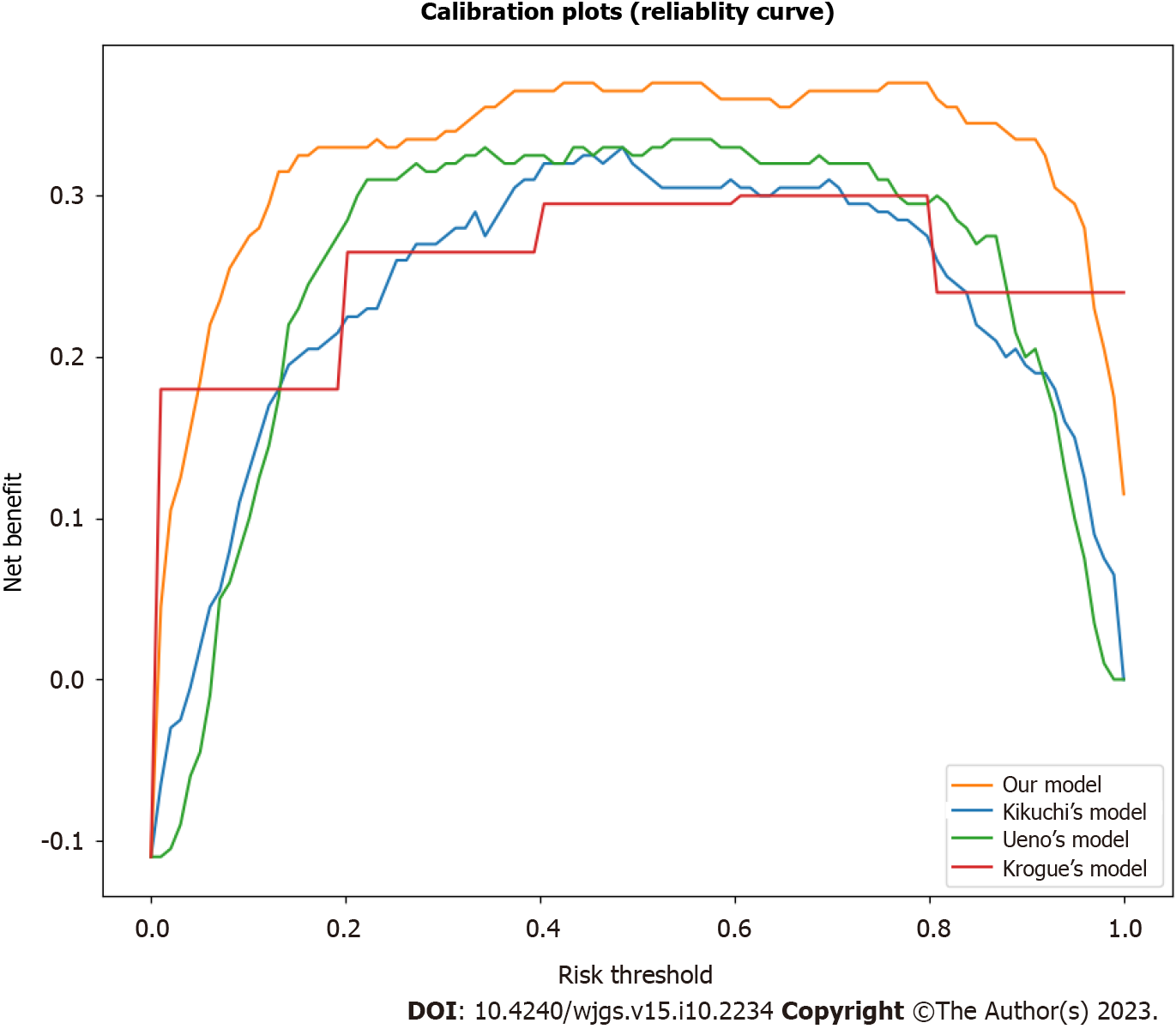
The calibration and decision curve analysis of the predicted probabilities of LNM between our risk prediction model and existing models in the validation set was assessed, as shown in Figures 2 and 3. The current model demonstrated good agreement between the observed and predicted probabilities of LNM across a range of risk thresholds, with a higher net benefit across a wide range of risk thresholds, indicating good calibration and clinical utility. However, the existing models showed some deviation from the ideal line with a lower net benefit, especially at low and high-risk thresholds, indicating poor calibration and clinical utility.
The risk prediction model devised in this study facilitated the classification of patients into various risk strata based on the calculated probability of LNM: Low (< 10%), intermediate (10%-30%), and high-risk (> 30%). As shown in Table 5, of the 70 CRC patients, 27 (38.6%) were classified as low-risk, 26 (37.1%) as intermediate-risk, and 17 (24.3%) as high-risk. Among the 18 cases of LNM, 1 (5.6%) was classified as low-risk, 6 (33.3%) as intermediate-risk, and 11 (61.1%) as high-risk. Appropriate therapeutic strategies should be judiciously selected in accordance with the assessment of LNM risk in patients with CRC.
| Risk group | Predicted probability of LNM | Number of patients | Number of LNMs |
| Low risk | < 10 | 27 (38.6) | 1 (5.6) |
| Intermediate risk | 10-30 | 26 (37.1) | 6 (33.3) |
| High risk | > 30 | 17 (24.3) | 11 (61.1) |
In the present study, we constructed and validated a risk-prediction model for LNM in CRC based on a large cohort of patients. For this purpose, machine learning techniques were used to extract features from primary tumor histology and clinicopathological data and combine them with established predictors to build a logistic regression model. The features extracted from machine learning provided additional predictive value for LNM in CRC, thereby improving the accuracy and discriminating capacity of the logistic regression model. Additionally, the model demonstrated good calibration and robustness in the validation set. Furthermore, the model can effectively stratify patients into different risk groups and guide the selection of appropriate treatment options.
To avoid the toxicity of over-therapeutic procedures in patients classified as low risk by the current model, curative treatment measures, such as endoscopic resection or local excision that eliminate the need for lymphadenectomy or adjuvant therapy, are recommended. For patients presenting with intermediate-risk T1 CRC, the recommendation is surgical resection accompanied by lymphadenectomy as curative treatment, with the decision for adjuvant therapy contingent on other prognostic factors. In contrast, patients with high-risk T1 CRC are recommended to be administered neoadjuvant therapy followed by surgical resection with lymphadenectomy for significantly minimizing the risk of recurrence.
Previous studies have shown that tumor histology contains rich information that reflects the biological behavior and molecular characteristics of tumor cells[22-24]. However, conventional histopathological evaluations are subjective, qualitative, and limited by human perception. Deep learning is a powerful tool that automatically learns complex and high-dimensional patterns from images and generates quantitative and objective features[25,26]. The current study used a pre-trained CNN model to generate the quantitative features of tumor histology. The top cluster frequencies were identified and added to the logistic regression model to enhance their predictive value when combined with baseline clinicopathological variables. These selected histological features may reflect the aggressiveness and invasiveness of the tumor cells and their interaction with the microenvironment, which may influence their ability to metastasize to the lymph nodes.
In this study, we opted for logistic regression as the optimum modeling method, driven by its simplicity, interpre
The present study provides valuable insights into the determinants of LNM in patients with CRC and presents an innovative risk prediction model; however, it has certain limitations. First, the retrospective design of the study may have contributed to selection bias. Second, we could not include potentially relevant risk factors, such as genetic and lifestyle influences, because of data unavailability. Third, model validation was performed exclusively within an independent cohort from the same institution, underscoring the need for future studies to validate the model across diverse populations and within a prospective framework. Finally, the limited size of the validation cohort could potentially affect the reliability of the validation findings. Despite these constraints, our model exhibited high efficacy and adeptness in distinguishing between patients with and without LNM, in addition to displaying sound calibration and robustness within an external cohort. This study forms a strong foundation for future efforts to develop and refine predictive models for LNM in patients with CRC.
In conclusion, a potent and effective risk-prediction model for LNM in patients with CRC was successfully developed and validated. The model utilized machine learning-derived features extracted from primary tumor histology and clinicopathological variables and demonstrated superior performance and clinical applicability compared to existing models. The study provided new insights into the potential of deep learning to extract valuable information from tumor histology, which can improve the prediction of LNM in CRC and facilitate risk stratification and decision-making in clinical practice. Further investigations are crucial to affirm the utility of the present model within larger and more heterogeneous cohorts and to probe the biological and molecular mechanisms underlying the features extracted using machine learning.
Colorectal cancer (CRC) is a significant global health issue, and accurate prediction of lymph node metastasis (LNM) is crucial for individualized treatment strategies. However, predicting LNM is challenging due to various factors and limitations in the histopathological examination method. This study aimed to develop a risk prediction model for LNM in CRC by incorporating machine learning and clinicopathological factors. The model demonstrated high accuracy, sensitivity, and specificity, providing valuable insights into the potential of deep learning in improving LNM prediction and guiding clinical decision-making for CRC patients.
Accurate prediction of LNM in CRC is crucial for improving patient outcomes and developing personalized treatment strategies. However, existing methods are invasive, time-consuming, and prone to errors. This study aimed to address these limitations by developing a risk prediction model using machine learning and clinicopathological factors. The motivation was to provide a more accurate and efficient approach for predicting LNM in CRC, enabling clinicians to make informed decisions regarding treatment and facilitating improved patient care.
The main objectives of this study were to analyze the factors influencing LNM in CRC, and to develop and validate a risk prediction model for LNM based on a large patient cohort. The study aimed to utilize machine learning techniques and clinicopathological factors to construct an accurate prediction model that outperforms existing models. The goal was to improve the prediction of LNM in CRC, enabling personalized treatment strategies and enhancing clinical decision-making. Additionally, the study sought to explore the potential of deep learning in extracting valuable information from tumor histology for improved risk stratification.
In this study, a retrospective analysis was conducted on 300 patients who underwent CRC surgery at two Peking University Shenzhen hospitals between January and December 2021. The main approach involved the development of a risk prediction model for LNM in CRC. A deep learning method was utilized to extract features from primary tumor histological images that could be associated with LNM. Additionally, a logistic regression model was used, incorporating these machine-learning-derived features along with clinicopathological variables, as predictors for LNM in CRC. The performance of the prediction model was evaluated based on accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), receiver operating characteristic (ROC), and Brier score to assess calibration and clinical utility.
The developed risk prediction model for LNM in CRC achieved excellent performance. The model demonstrated high accuracy (0.86), sensitivity (0.81), specificity (0.87), PPV (0.66), NPV (0.94), and area under the curve for the ROC (0.91). Additionally, it exhibited a low Brier score of 0.10. The observed and predicted probabilities of LNM showed strong agreement across various risk thresholds, indicating reliable calibration and clinical utility. These findings highlight the effectiveness and clinical applicability of the model, which utilizes machine-learning-derived features from primary tumor histology and clinicopathological variables.
The study successfully developed and validated a powerful risk prediction model for LNM in CRC. The model, incorporating machine-learning-derived features from primary tumor histology and clinicopathological variables, displayed superior performance and clinical applicability compared to existing models. By leveraging deep learning techniques, valuable information was extracted from tumor histology, leading to improved LNM prediction. This development has significant implications for individualized treatment strategies and clinical decision-making in CRC, enabling better risk stratification. The findings highlight the potential of machine learning and deep learning in enhancing LNM prediction and improving patient outcomes in CRC management.
The successful development and validation of a potent risk prediction model for LNM in CRC opens up promising research avenues. Further exploration can focus on refining the model by incorporating additional molecular characteristics and genetic data to enhance its predictive accuracy. Additionally, prospective studies can be conducted to validate the model’s performance in larger and diverse patient populations. Furthermore, the integration of real-time image analysis techniques and artificial intelligence algorithms can streamline the prediction process, enabling faster and more accurate LNM assessment. These advancements have the potential to revolutionize clinical practice and optimize treatment strategies for CRC patients.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Iruarrizaga-Lejarreta M, Spain; Landberg G, Sweden S-Editor: Qu XL L-Editor: A P-Editor: Yuan YY
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55686] [Article Influence: 7955.1] [Reference Citation Analysis (132)] |
| 2. | Benson AB, Venook AP, Al-Hawary MM, Cederquist L, Chen YJ, Ciombor KK, Cohen S, Cooper HS, Deming D, Engstrom PF, Garrido-Laguna I, Grem JL, Grothey A, Hochster HS, Hoffe S, Hunt S, Kamel A, Kirilcuk N, Krishnamurthi S, Messersmith WA, Meyerhardt J, Miller ED, Mulcahy MF, Murphy JD, Nurkin S, Saltz L, Sharma S, Shibata D, Skibber JM, Sofocleous CT, Stoffel EM, Stotsky-Himelfarb E, Willett CG, Wuthrick E, Gregory KM, Freedman-Cass DA. NCCN Guidelines Insights: Colon Cancer, Version 2.2018. J Natl Compr Canc Netw. 2018;16:359-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 635] [Cited by in RCA: 681] [Article Influence: 97.3] [Reference Citation Analysis (1)] |
| 3. | Qiu MZ, Cai MY, Zhang DS, Wang ZQ, Wang DS, Li YH, Xu RH. Clinicopathological characteristics and prognostic analysis of Lauren classification in gastric adenocarcinoma in China. J Transl Med. 2013;11:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 128] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 4. | Yahagi M, Okabayashi K, Hasegawa H, Tsuruta M, Kitagawa Y. The Worse Prognosis of Right-Sided Compared with Left-Sided Colon Cancers: a Systematic Review and Meta-analysis. J Gastrointest Surg. 2016;20:648-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 217] [Article Influence: 24.1] [Reference Citation Analysis (1)] |
| 5. | Hari DM, Leung AM, Lee JH, Sim MS, Vuong B, Chiu CG, Bilchik AJ. AJCC Cancer Staging Manual 7th edition criteria for colon cancer: do the complex modifications improve prognostic assessment? J Am Coll Surg. 2013;217:181-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 217] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 6. | Hornychova H, Melichar B, Tomsova M, Mergancova J, Urminska H, Ryska A. Tumor-infiltrating lymphocytes predict response to neoadjuvant chemotherapy in patients with breast carcinoma. Cancer Invest. 2008;26:1024-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 83] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 7. | van der Pas MH, Haglind E, Cuesta MA, Fürst A, Lacy AM, Hop WC, Bonjer HJ; COlorectal cancer Laparoscopic or Open Resection II (COLOR II) Study Group. Laparoscopic versus open surgery for rectal cancer (COLOR II): short-term outcomes of a randomised, phase 3 trial. Lancet Oncol. 2013;14:210-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1030] [Cited by in RCA: 1206] [Article Influence: 100.5] [Reference Citation Analysis (0)] |
| 8. | Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5537] [Cited by in RCA: 6446] [Article Influence: 429.7] [Reference Citation Analysis (0)] |
| 9. | Duffy MJ, Lamerz R, Haglund C, Nicolini A, Kalousová M, Holubec L, Sturgeon C. Tumor markers in colorectal cancer, gastric cancer and gastrointestinal stromal cancers: European group on tumor markers 2014 guidelines update. Int J Cancer. 2014;134:2513-2522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 248] [Article Influence: 22.5] [Reference Citation Analysis (1)] |
| 10. | Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1362] [Cited by in RCA: 1282] [Article Influence: 116.5] [Reference Citation Analysis (0)] |
| 11. | Gründner J, Prokosch HU, Stürzl M, Croner R, Christoph J, Toddenroth D. Predicting Clinical Outcomes in Colorectal Cancer Using Machine Learning. Stud Health Technol Inform. 2018;247:101-105. [PubMed] |
| 12. | Bychkov D, Linder N, Turkki R, Nordling S, Kovanen PE, Verrill C, Walliander M, Lundin M, Haglund C, Lundin J. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep. 2018;8:3395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 427] [Cited by in RCA: 363] [Article Influence: 51.9] [Reference Citation Analysis (0)] |
| 13. | Smith JJ, Chow OS, Gollub MJ, Nash GM, Temple LK, Weiser MR, Guillem JG, Paty PB, Avila K, Garcia-Aguilar J; Rectal Cancer Consortium. Organ Preservation in Rectal Adenocarcinoma: a phase II randomized controlled trial evaluating 3-year disease-free survival in patients with locally advanced rectal cancer treated with chemoradiation plus induction or consolidation chemotherapy, and total mesorectal excision or nonoperative management. BMC Cancer. 2015;15:767. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 290] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 14. | Liebig C, Ayala G, Wilks JA, Berger DH, Albo D. Perineural invasion in cancer: a review of the literature. Cancer. 2009;115:3379-3391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 856] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 15. | Backes Y, Elias SG, Groen JN, Schwartz MP, Wolfhagen FHJ, Geesing JMJ, Ter Borg F, van Bergeijk J, Spanier BWM, de Vos Tot Nederveen Cappel WH, Kessels K, Seldenrijk CA, Raicu MG, Drillenburg P, Milne AN, Kerkhof M, Seerden TCJ, Siersema PD, Vleggaar FP, Offerhaus GJA, Lacle MM, Moons LMG; Dutch T1 CRC Working Group. Histologic Factors Associated With Need for Surgery in Patients With Pedunculated T1 Colorectal Carcinomas. Gastroenterology. 2018;154:1647-1659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 16. | Watanabe T, Muro K, Ajioka Y, Hashiguchi Y, Ito Y, Saito Y, Hamaguchi T, Ishida H, Ishiguro M, Ishihara S, Kanemitsu Y, Kawano H, Kinugasa Y, Kokudo N, Murofushi K, Nakajima T, Oka S, Sakai Y, Tsuji A, Uehara K, Ueno H, Yamazaki K, Yoshida M, Yoshino T, Boku N, Fujimori T, Itabashi M, Koinuma N, Morita T, Nishimura G, Sakata Y, Shimada Y, Takahashi K, Tanaka S, Tsuruta O, Yamaguchi T, Yamaguchi N, Tanaka T, Kotake K, Sugihara K; Japanese Society for Cancer of the Colon and Rectum. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2016 for the treatment of colorectal cancer. Int J Clin Oncol. 2018;23:1-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 576] [Cited by in RCA: 603] [Article Influence: 86.1] [Reference Citation Analysis (0)] |
| 17. | Gunderson LL, Jessup JM, Sargent DJ, Greene FL, Stewart AK. Revised TN categorization for colon cancer based on national survival outcomes data. J Clin Oncol. 2010;28:264-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 437] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 18. | Kikuchi R, Takano M, Takagi K, Fujimoto N, Nozaki R, Fujiyoshi T, Uchida Y. Management of early invasive colorectal cancer. Risk of recurrence and clinical guidelines. Dis Colon Rectum. 1995;38:1286-1295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 440] [Article Influence: 14.7] [Reference Citation Analysis (1)] |
| 19. | Ueno H, Mochizuki H, Hashiguchi Y, Shimazaki H, Aida S, Hase K, Matsukuma S, Kanai T, Kurihara H, Ozawa K, Yoshimura K, Bekku S. Risk factors for an adverse outcome in early invasive colorectal carcinoma. Gastroenterology. 2004;127:385-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 501] [Cited by in RCA: 527] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 20. | Krogue JD, Azizi S, Tan F, Flament-Auvigne I, Brown T, Plass M, Reihs R, Müller H, Zatloukal K, Richeson P, Corrado GS, Peng LH, Mermel CH, Liu Y, Chen PC, Gombar S, Montine T, Shen J, Steiner DF, Wulczyn E. Predicting lymph node metastasis from primary tumor histology and clinicopathologic factors in colorectal cancer using deep learning. Commun Med (Lond). 2023;3:59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 21. | Huang YQ, Liang CH, He L, Tian J, Liang CS, Chen X, Ma ZL, Liu ZY. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol. 2016;34:2157-2164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 896] [Cited by in RCA: 1318] [Article Influence: 146.4] [Reference Citation Analysis (0)] |
| 22. | Overwater A, Kessels K, Elias SG, Backes Y, Spanier BWM, Seerden TCJ, Pullens HJM, de Vos Tot Nederveen Cappel WH, van den Blink A, Offerhaus GJA, van Bergeijk J, Kerkhof M, Geesing JMJ, Groen JN, van Lelyveld N, Ter Borg F, Wolfhagen F, Siersema PD, Lacle MM, Moons LMG; Dutch T1 CRC Working Group. Endoscopic resection of high-risk T1 colorectal carcinoma prior to surgical resection has no adverse effect on long-term outcomes. Gut. 2018;67:284-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 23. | Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, van der Laak JAWM; the CAMELYON16 Consortium, Hermsen M, Manson QF, Balkenhol M, Geessink O, Stathonikos N, van Dijk MC, Bult P, Beca F, Beck AH, Wang D, Khosla A, Gargeya R, Irshad H, Zhong A, Dou Q, Li Q, Chen H, Lin HJ, Heng PA, Haß C, Bruni E, Wong Q, Halici U, Öner MÜ, Cetin-Atalay R, Berseth M, Khvatkov V, Vylegzhanin A, Kraus O, Shaban M, Rajpoot N, Awan R, Sirinukunwattana K, Qaiser T, Tsang YW, Tellez D, Annuscheit J, Hufnagl P, Valkonen M, Kartasalo K, Latonen L, Ruusuvuori P, Liimatainen K, Albarqouni S, Mungal B, George A, Demirci S, Navab N, Watanabe S, Seno S, Takenaka Y, Matsuda H, Ahmady Phoulady H, Kovalev V, Kalinovsky A, Liauchuk V, Bueno G, Fernandez-Carrobles MM, Serrano I, Deniz O, Racoceanu D, Venâncio R. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA. 2017;318:2199-2210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1871] [Cited by in RCA: 1545] [Article Influence: 193.1] [Reference Citation Analysis (0)] |
| 24. | Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, Grabsch HI, Yoshikawa T, Brenner H, Chang-Claude J, Hoffmeister M, Trautwein C, Luedde T. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25:1054-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 992] [Cited by in RCA: 748] [Article Influence: 124.7] [Reference Citation Analysis (0)] |
| 25. | Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, Pencina MJ, Kattan MW. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21:128-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3195] [Cited by in RCA: 3319] [Article Influence: 221.3] [Reference Citation Analysis (1)] |
| 26. | Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1604] [Cited by in RCA: 2273] [Article Influence: 227.3] [Reference Citation Analysis (0)] |
| 27. | Roxburgh CS, McMillan DC, Richards CH, Atwan M, Anderson JH, Harvey T, Horgan PG, Foulis AK. The clinical utility of the combination of T stage and venous invasion to predict survival in patients undergoing surgery for colorectal cancer. Ann Surg. 2014;259:1156-1165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 28. | Kim CH, Huh JW, Kim HR, Kim YJ. Prognostic comparison between number and distribution of lymph node metastases in patients with right-sided colon cancer. Ann Surg Oncol. 2014;21:1361-1368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 29. | Phipps AI, Shi Q, Newcomb PA, Nelson GD, Sargent DJ, Alberts SR, Limburg PJ. Associations between cigarette smoking status and colon cancer prognosis among participants in North Central Cancer Treatment Group Phase III Trial N0147. J Clin Oncol. 2013;31:2016-2023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 30. | Binefa G, Rodríguez-Moranta F, Teule A, Medina-Hayas M. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol. 2014;20:6786-6808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 245] [Cited by in RCA: 256] [Article Influence: 23.3] [Reference Citation Analysis (3)] |