Published online Oct 27, 2023. doi: 10.4240/wjgs.v15.i10.2154
Peer-review started: April 28, 2023
First decision: July 23, 2023
Revised: August 4, 2023
Accepted: August 17, 2023
Article in press: August 17, 2023
Published online: October 27, 2023
Processing time: 182 Days and 1.1 Hours
Our previous study found that the telomerase-associated protein 1 (TEP1, rs938886 and rs1713449) and homo sapiens RecQ like helicase 5 (RECQL5, rs820196) single nucleotide polymorphisms (SNPs) were associated with changes in heart rate (HR) ≥ 30% during peritoneal lavage with distilled water after gastrectomy. This study established a single tube method for detecting these three SNPs using two-dimensional (2D) polymerase chain reaction (PCR), and investigated whether SNP-SNP and SNP-environment interactions increase the risk of high HR variability (HRV).
To investigate whether genotypes, genetic patterns, SNP-SNP and SNP-envi
2D PCR was used to establish a single-tube method to detect TEP1 rs938886 and rs1713449 and RECQL5 rs820196, and the results were compared with those of sanger sequencing. After adjusting for confounders such as age, sex, smoking, hypertension, and thyroid dysfunction, a nonconditional logistic regression model was used to assess the associations between the genotypes and the genetic patterns (codominant, dominant, overdominant, recessive, and additive) of the three SNPs and a risk ≥ 15% or ≥ 30% of a sudden drop in HR during post
The coincidence rate between the 2D PCR and sequencing was 100%. When the HRV cutoff value was 15%, the patients with the RECQL5 (rs820196) TC genotype had a higher risk of high HRV than those who had the TT genotype (odds ratio = 1.97; 95%CI: 1.05-3.70; P = 0.045). Under the codominant and overdominant models, the TC genotype of RECQL5 (rs820196) was associated with a higher risk of HR decrease relative to the TT and TT + CC genotypes (P = 0.031 and 0.016, respectively). When the HRV cutoff value was 30%, patients carrying the GC-TC genotypes of rs938886 and rs820196 showed a higher HRV risk when compared with the GG–TT genotype carriers (P = 0.01). In the three-factor model of rs938886, rs820196, and rs1713449, patients carrying the GC-TC-CT genotype had a higher risk of HRV compared with the wild-type GG-TT-CC carriers (P = 0.01). For rs820196, nonsmokers with the TC genotype had a higher HRV risk compared with nonsmokers carrying the TT genotype (P = 0.04). When the HRV cutoff value was 15%, patients carrying the TT-TT and the TC-CT genotypes of rs820196 and rs1713449 showed a higher HRV risk when compared with TT-CC genotype carriers (P = 0.04 and 0.01, respe
The polymorphism of the RECQL5 and TEP1 genes were associated with HRV during peritoneal lavage with distilled water after gastrectomy.
Core Tip: Our previous study found that peritoneal lavage with distilled water may cause a sudden decrease in heart rate (HR) in some patients during clinical gastrectomy, which was related to telomerase-associated protein 1 and homo sapiens RecQ like helicase 5 gene polymorphisms. Here, instead of Sanger sequencing, we developed a single-tube method using two-dimensional polymerase chain reaction to genotyping single nucleotide polymorphisms (SNPs) quickly and economically. We also investigated whether genotypes, genetic patterns and the interaction effects of SNP-SNP and SNP-environment were associated with a risk of high HR variability. This study helps clinicians to better assess the perioperative risk of patients undergoing gastrectomy.
- Citation: Yao S, Yuan Y, Zhang J, Yu Y, Luo GH. Gene polymorphisms associated with sudden decreases in heart rate during extensive peritoneal lavage with distilled water after gastrectomy. World J Gastrointest Surg 2023; 15(10): 2154-2170
- URL: https://www.wjgnet.com/1948-9366/full/v15/i10/2154.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v15.i10.2154
Gastric cancer currently ranks fifth in incidence and fourth in mortality worldwide[1]. Peritoneal metastasis is the most frequent pattern of postoperative recurrence in patients with gastric cancer. In order to reduce abdominal metastasis and planting caused by surgery, distilled water or normal saline are normally used to wash the peritoneum after radical distal gastrectomy. The Chinese SEIPLUS trial reported that, for extensive intraoperative peritoneal lavage (EIPL), a new prophylactic strategy for the prevention of the peritoneal metastasis of locally advanced gastric cancer, the short-term postoperative complication rate following surgery alone (17%) was significantly higher than that following combined surgery and multiple warm physiological saline lavages (11.1%)[2]. However, they found that addition of EIPL with saline did not improve the 3-year survival rate of advanced gastric cancer patients compared with surgery alone[3]. A latest randomized study evaluated the efficacy and long-term outcome of advanced gastric cancer patients with EIPL[4]. It was found that EIPL can reduce the possibility of perioperative complications including ileus and abdominal abscess. The overall survival curve and recurrence-free survival curve were better in the EIPL group. A multicenter study found that EIPL with saline after surgery did not provide a survival benefit compared with surgery alone and should not be recommended for patients undergoing curative gastrectomy for gastric cancer[5]. Distilled water can induce osmotic lysis in cancer cells. Takemoto et al[6] demonstrated the cytocidal effects of distilled-water-induced hypotonic shock on colorectal cancer cells, which supported the use of peritoneal lavage with distilled water to remove and kill colorectal cells during surgery. Recently, a randomized trial was performed to assess the survival impact of extensive peritoneal lavage using distilled water or saline at high volumes after pancreatic resection for pancreatic ductal adenocarcinoma, which suggested that lavage with distilled water or saline could become standard practice during surgery for pancreatic cancer if it proves to be beneficial to the long-term prognosis of the patient[7].
Our previous study found that peritoneal lavage with distilled water may have caused a sudden decrease in heart rate in some patients during clinical gastrectomy. To investigate whether gene polymorphisms are associated with high heart rate variability (HRV), we genotyped 194 patients who underwent distal gastrectomy and identified three single nucleotide polymorphisms (SNPs) (TEP1 rs938886 and rs1713449 and RECQL5 rs820196) associated with a risk of high HRV using whole-exome sequencing[8]. In this study, two-dimensional (2D) polymerase chain reaction (PCR) was used to establish a single-tube method to detect these three SNPs in 192 patients (two were excluded due to incomplete clinical data). Single-gene analysis may not be appropriate for the further study of complex traits because the main effect of a single locus may be too limited for observation. Therefore, SNP-SNP and SNP-environment interactions among the three variants from the two selected genes were tested using a generalized multifactor dimensionality reduction (GMDR) approach.
A total of 192 patients (137 males and 55 females) scheduled to undergo distal gastrectomy were enrolled. Patients were diagnosed according to clinical and pathological data, and those with a history of other cancers or arrhythmia were excluded. Participant demographic data (age, sex, hypertension, diabetes, thyroid dysfunction, and smoking status) were collected using a standard clinical information questionnaire. Evaluation of the genomic differences of these gastric cancer patients was conducted in a previous study[8]. Patients were divided into two groups according to the changes in HR (using 30% and 15% as cutoffs). Change in HR = (ultimate HR before lavage-HR after lavage)/HR before lavage × 100%. This study received ethical approval from the institutional review board of the Third Affiliated Hospital of Soochow University and was conducted in accordance with the Helsinki Declaration on human medical research.
Standard techniques were adopted for the collection of venous blood samples from the participants. Whole blood was stored at -80 ℃ for subsequent SNP analysis. Genomic DNA was extracted using the TIANamp Blood DNA Kit (Tiangen Biotech, Beijing, China). According to the principles of 2D PCR and amplification-refractory mutation system PCR, the mutation sites of the TEP1 (rs1713449), TEP1 (rs938886), and RECQL5 (rs820196) genes were designed at the 3' ends of the specific forward primers. The respective specific reverse primers were designed according to the DNA sequence using the PrimerPremier5.0 software, wherein the primer for TEP1 (rs1713449) was designed according to the reverse complementary strand. Three fluorescein amidite tags and three hexachloro-fluorescein tags were used to label the wild-type and mutant forward primers (respectively) of TEP1 (rs1713449), TEP1 (rs938886), and RECQL5 (rs820196). The sequences of the primers and probes are listed in Supplementary Table 1, which were synthesized by Sangon Biotech (Shanghai, China). The optimized 2D multiplex PCR system included 2 μL genomic DNA, 2.5 μL 10 × Immobuffer (Bioline, London, United Kingdom), 0.75 μL 25 mmol/L MgCl2 (Bioline), 0.7 μL 10 mmol/L dNTPs (Takara Bio, Shiga, Japan), 0.5 μL 5 U/μL IMMOLASE DNA polymerase (Bioline), 0.4 μL each probe (10 μM), 0.1 μL each forward primer (10 μM; 0.2 μL for TEP1 rs938886), 0.6 μL reverse primer (10 μM), and deionized water, to make a total volume of 25 μL. The PCRs were carried out with the following thermal cycling conditions: Initial denaturation of DNA at 95 ℃ for 10 min; amplification for five cycles at 95 ℃ for 20 s and 60 ℃ for 15 s; and 35 cycles at 95 ℃ for 20 s, 72 ℃ for 1 s, and 60 ℃ for 15 s. The fluorescence acquisition commenced with heating at 95 ℃ for 15 s and 30 ℃ for 4 min; the temperature was gradually increased from 30 ℃ to 70 ℃ with a ramp rate of 0.1 ℃/s, and the fluorescence signal was acquired continuously. The final step was cooling at 40 ℃ for 30 s. The results of 2D PCR were compared with sanger sequencing.
Statistical analyses were performed using SAS version 9.4 (Cary, NC, United States), and the nominal P value ≤ 0.05 was considered the significance threshold. Normally and non-normally distributed continuous variables were compared using student’s t test and Mann–Whitney U test, respectively, and the variables were expressed as mean ± SD. The genotype and allelic frequency distributions of polymorphisms between two groups were compared using the χ2 test and the Hardy–Weinberg equilibrium. Associations between polymorphisms and HR change were assessed by calculating odds ratios (ORs) with 95%CIs using logistic regression analysis adjusted for age, sex, hypertension, thyroid dysfunction, and smoking status. The pairwise linkage disequilibrium (LD) and frequency of haplotypes were calculated with Haploview 4.2. GMDR was used to analyze the SNP-SNP and SNP-environment interactions. P < 0.05 was considered statistically significant.
We recruited 192 patients with gastric cancer. The basic characteristics are presented in Tables 1 and 2. The HRV cutoff values used were 30% and 15%. There were no significant differences in the baseline demography, smoking status, hypertension, diabetes, and thyroid dysfunction between the two groups (P > 0.05).
| Variable | HRV ≥ 30% | HRV < 30% | χ2 | P value |
| Sex | 1.828 | 0.18 | ||
| Female | 9 (16.36) | 46 (83.64) | ||
| Male | 13 (9.49) | 124 (90.51) | ||
| Age (yr) | 0.098 | 0.75 | ||
| < 60 | 5 (14.29) | 30 (85.71) | ||
| ≥ 60 | 17 (10.83) | 140 (89.17) | ||
| Smoking status | 0.592 | 0.44 | ||
| No | 20 (12.58) | 139 (87.42) | ||
| Yes | 2 (6.06) | 31 (93.94) | ||
| Hypertension | 0.213 | 0.64 | ||
| No | 13 (10.66) | 109 (89.34) | ||
| Yes | 9 (12.86) | 61 (87.14) | ||
| Diabetes | 0.299 | 0.58 | ||
| No | 19 (10.80) | 157 (89.20) | ||
| Yes | 3 (18.75) | 13 (81.25) | ||
| Blood type | 0.964 | 0.81 | ||
| A | 9 (12.33) | 64 (87.67) | ||
| AB | 1 (5.00) | 19 (95.00) | ||
| B | 5 (11.36) | 39 (88.64) | ||
| O | 7 (12.73) | 49 (87.27) | ||
| Thyroid function | 0.005 | 0.95 | ||
| Normal | 20 (11.70) | 151 (88.30) | ||
| Abnormal | 2 (9.52) | 19 (90.48) |
| Variable | HRV ≥ 15% | HRV < 15% | χ2 | P value |
| Sex | 0.015 | 0.90 | ||
| Female | 21 (38.18) | 34 (61.82) | ||
| Male | 51 (37.23) | 86 (62.77) | ||
| Age (yr) | 1.232 | 0.27 | ||
| < 60 | 16 (45.71) | 19 (54.29) | ||
| ≥ 60 | 56 (35.67) | 101 (64.33) | ||
| Smoking status | 0.412 | 0.52 | ||
| No | 58 (36.48) | 101 (63.52) | ||
| Yes | 14 (42.42) | 19 (57.58) | ||
| Hypertension | 0.006 | 0.94 | ||
| No | 46 (37.70) | 76 (62.30) | ||
| Yes | 26 (37.14) | 44 (62.86) | ||
| Diabetes | 2.618 | 0.11 | ||
| No | 63 (35.80) | 113 (64.20) | ||
| Yes | 9 (56.25) | 7 (43.75) | ||
| Blood type | 1.803 | 0.61 | ||
| A | 27 (36.99) | 46 (63.01) | ||
| AB | 5 (25.00) | 15 (75.00) | ||
| B | 17 (38.64) | 27 (61.36) | ||
| O | 23 (41.82) | 32 (58.18) | ||
| Thyroid function | 0.289 | 0.59 | ||
| Normal | 63 (36.84) | 108 (63.16) | ||
| Abnormal | 9 (42.86) | 12 (57.14) |
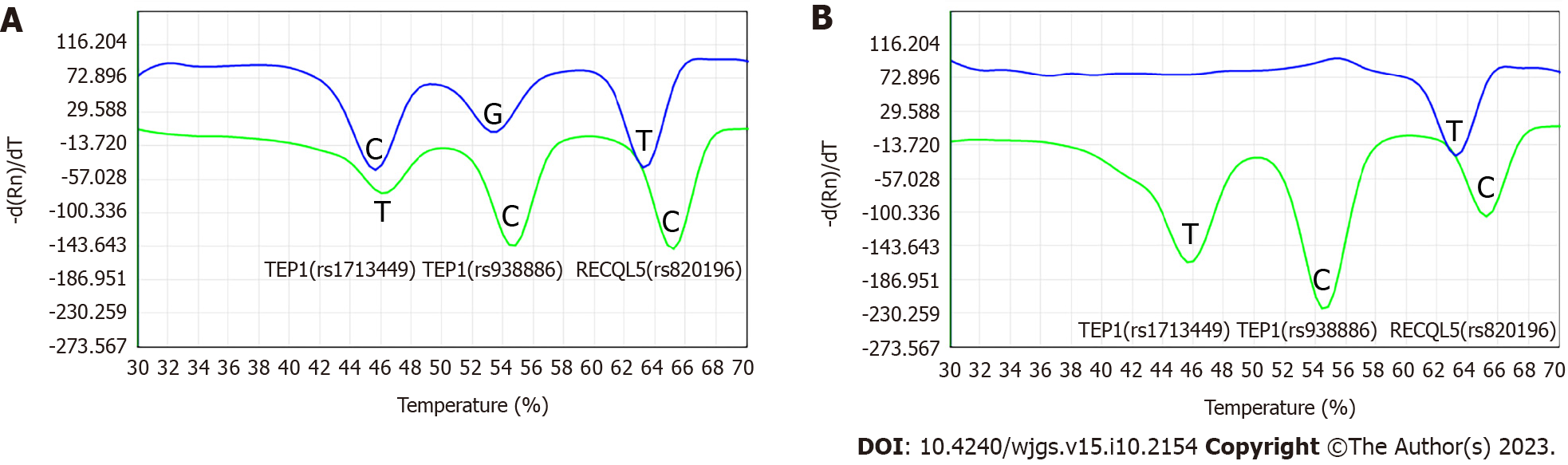
Figure 1 shows the melting curves of six alleles of the three genes detected by 2D PCR. Six melting valleys were clearly identified (Figure 1A) and six different alleles were intuitively typed. The genotypes of TEP1 (rs1713449), TEP1 (rs938886), and RECQL5 (rs820196) in 190 gastric cancer patients investigated by 2D PCR were completely consistent with those determined by sanger sequencing (the remaining two cases were not validated because of sample loss, Figure 1).
Genotypes and allele frequencies of TEP1 (rs938886), TEP1 (rs1713449) and RECQL5 (rs820196) in patients with a 30% or 15% decrease in HR as the cutoff points were compared. When the patients were grouped according to the 30% decrease in HR cutoff, there were no significant differences in the distribution of heterozygous and homozygous mutant and wild-type genotypes (Table 3, P > 0.05). However, when the patients were grouped using the 15% decrease in HR cutoff, the distribution of the RECQL5 (rs820196) genotype frequencies was significantly different between the two groups. For RECQL5 (rs820196), compared with individuals with the TT genotype, patients with the TC genotype had a significantly increased risk of adverse HR decline, with an OR of 1.97 (95%CI: 1.05-3.70), which implies that RECQL5 (rs820196) was associated with HRV in the allelic models (Table 4, P < 0.05).
| SNP | HRV ≥ 30% | HRV < 30% | χ2 | P value | OR (95%CI) |
| TEP1 (rs938886) | 1.30 | 0.52 | |||
| GG | 9 (10.00) | 81 (90.00) | 1.00 (Reference) | ||
| GC | 12 (14.12) | 73 (85.88) | 1.63 (0.63, 4.20) | ||
| CC | 1 (5.88) | 16 (94.12) | 0.72 (0.081, 6.39) | ||
| 0.016 | 0.90 | ||||
| G | 30 (11.32) | 235 (88.68) | 1.00 (Reference) | ||
| C | 14 (11.76) | 105 (88.24) | 1.16 (0.58, 2.31) | ||
| TEP1 (rs1713449) | 1.54 | 0.46 | |||
| CC | 9 (10.11) | 80 (89.89) | 1.00 (Reference) | ||
| CT | 12 (14.29) | 72 (85.71) | 1.63 (0.63, 4.21) | ||
| TT | 1 (5.26) | 18 (94.74) | 0.62 (0.071, 5.43) | ||
| 0.0001 | 0.99 | ||||
| C | 30 (11.45) | 232 (88.55) | 1.00 (Reference) | ||
| T | 14 (11.48) | 108 (88.52) | 1.11 (0.56, 2.22) | ||
| RECQL5 (rs820196) | 3.93 | 0.14 | |||
| TT | 7 (8.05) | 80 (91.95) | 1.00 (Reference) | ||
| TC | 14 (16.47) | 71 (83.53) | 2.39 (0.90, 6.33) | ||
| CC | 1 (5.00) | 19 (95.00) | 0.65 (0.073, 5.72) | ||
| 0.33 | 0.57 | ||||
| T | 28 (10.81) | 231 (89.19) | 1.00 (Reference) | ||
| C | 16 (12.80) | 109 (87.20) | 1.27 (0.66, 2.47) | ||
| SNP | HRV ≥ 15% | HRV < 15% | χ2 | P value | OR (95%CI) |
| TEP1 (rs938886) | 1.56 | 0.46 | |||
| GG | 30 (33.33) | 60 (66.67) | 1.00 (Reference) | ||
| GC | 34 (40.00) | 51 (60.00) | 1.34 (0.72, 2.52) | ||
| CC | 8 (47.06) | 9 (52.94) | 1.74 (0.59, 5.15) | ||
| 1.50 | 0.22 | ||||
| G | 94 (35.47) | 171 (64.53) | 1.00 (Reference) | ||
| C | 50 (42.02) | 69 (57.98) | 1.30 (0.83, 2.04) | ||
| TEP1 (rs1713449) | 2.02 | 0.36 | |||
| CC | 29 (32.58) | 60 (67.42) | 1.00 (Reference) | ||
| CT | 34 (40.48) | 50 (59.52) | 1.42 (0.75, 2.67) | ||
| TT | 9 (47.37) | 10 (52.63) | 1.85 (0.66, 5.21) | ||
| 2.00 | 0.16 | ||||
| C | 92 (35.11) | 170 (64.89) | 1.00 (Reference) | ||
| T | 52 (42.62) | 70 (57.38) | 1.36 (0.87, 2.13) | ||
| RECQL5 (rs820196) | 6.20 | 0.045 | |||
| TT | 27 (31.03) | 60 (68.97) | 1.00 (Reference) | ||
| TC | 40 (47.06) | 45 (52.94) | 1.97 (1.05, 3.70)a | ||
| CC | 5 (25.00) | 15 (75.00) | 0.69 (0.22, 2.13) | ||
| 0.49 | 0.48 | ||||
| T | 94 (36.29) | 165 (63.71) | 1.00 (Reference) | ||
| C | 50 (40.00) | 75 (60.00) | 1.15 (0.74, 1.78) |
We next applied five genetic models (codominant, dominant, overdominant, recessive, and additive) to further analyze the relationships between the three SNPs and HRV. No significant differences were found between the patients in the HRV ≥ 30% group and the control group for the studied SNPs in all the genetic models (P > 0.05, Table 5). However, when the patients were grouped according to the 15% HRV cutoff, RECQL5 (rs820196) was associated with a higher risk of HR change in the codominant and overdominant models. In the codominant model, the TC genotype was associated with a higher risk relative to the TT genotype (OR = 2.0; 95%CI: 1.06-3.76; P = 0.031). In the overdominant model, the TC genotype was associated with a higher risk relative to the TT and CC genotypes (OR = 2.10; 95%CI: 1.15-3.84; P = 0.016, Table 6).
| Inheritance model | TEP1 (rs938886) | TEP1 (rs1713449) | RECQL5 (rs820196) | |||
| OR (95%CI) | P value | OR (95%CI) | P value | OR (95%CI) | P value | |
| Codominant | ||||||
| Aa vs AA | 1.67 (0.64, 4.31) | 0.293 | 1.67 (0.65, 4.34) | 0.289 | 2.35 (0.88, 6.24) | 0.087 |
| aa vs AA | 0.62 (0.068, 5.70) | 0.675 | 0.54 (0.060, 4.84) | 0.582 | 0.58 (0.065, 5.21) | 0.628 |
| Dominant (Aa + aa vs AA) | 1.49 (0.59, 3.76) | 0.403 | 1.46 (0.58, 3.68) | 0.429 | 2.02 (0.78, 5.25) | 0.151 |
| Overdominant (Aa vs AA + aa) | 1.69 (0.68, 4.24) | 0.261 | 1.74 (0.69, 4.35) | 0.238 | 2.54 (0.99, 6.49) | 0.051 |
| Recessive (aa vs Aa + AA) | 0.56 (0.067, 4.64) | 0.590 | 0.48 (0.059, 3.92) | 0.494 | 0.41 (0.050, 3.27) | 0.396 |
| Additive (AA vs Aa vs aa) | 1.18 (0.57, 2.42) | 0.657 | 1.12 (0.55, 2.27) | 0.752 | 1.28 (0.65, 2.51) | 0.472 |
| Inheritance model | TEP1 (rs938886) | TEP1 (rs1713449) | RECQL5 (rs820196) | |||
| OR (95%CI) | P value | OR (95%CI) | P value | OR (95%CI) | P value | |
| Codominant | ||||||
| Aa vs AA | 1.36 (0.72, 2.55) | 0.341 | 1.43 (0.76, 2.69) | 0.271 | 2.00 (1.06, 3.76) | 0.031a |
| aa vs AA | 1.31 (0.75, 2.29) | 0.345 | 1.34 (0.79, 2.28) | 0.279 | 0.82 (0.46, 1.45) | 0.492 |
| Dominant (Aa + aa vs AA) | 1.40 (0.77, 2.56) | 0.273 | 1.49 (0.81, 2.72) | 0.196 | 1.64 (0.90, 2.98) | 0.109 |
| Overdominant (Aa vs AA + aa) | 1.23 (0.67, 2.23) | 0.506 | 1.27 (0.70, 2.31) | 0.441 | 2.10 (1.15, 3.84) | 0.016b |
| Recessive (aa vs Aa + AA) | 1.50 (0.53, 4.25) | 0.442 | 1.56 (0.58, 4.16) | 0.378 | 0.50 (0.17, 1.46) | 0.202 |
| Additive (AA vs Aa vs aa) | 1.33 (0.83, 2.12) | 0.233 | 1.38 (0.88, 2.18) | 0.166 | 1.15 (0.74, 1.80) | 0.537 |
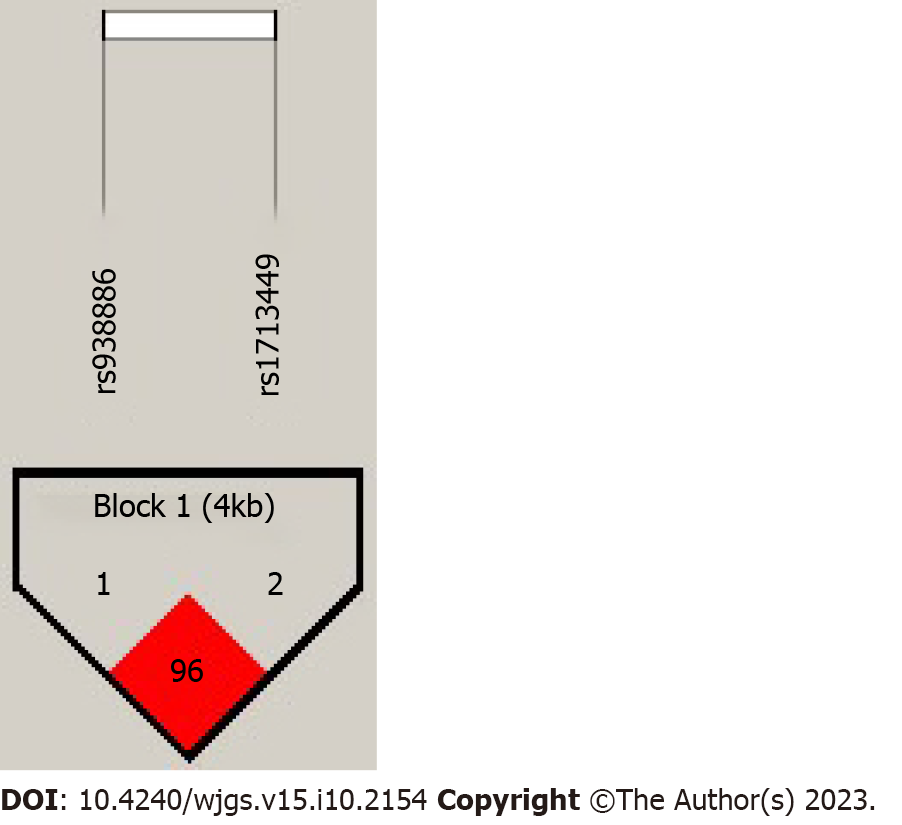
The results of the pairwise LD analysis of the two TEP1 SNPs are presented in Figure 2. We detected a haplotype block with a strong LD between the rs938886 and rs1713449 SNPs. The D value was 1, indicating that these two SNPs tend to be co-inherited. The haplotype analysis indicated that, regardless of whether the patients were grouped according to the 30% or 15% HRV cutoff, the rs938886-C/rs1713449-T and rs938886-G/rs1713449-T haplotypes were not associated with HRV risk (Tables 7 and 8, P > 0.05).
| TEP1 (rs938886) | TEP1 (rs1713449) | Frequency | HRV ≥ 30% | HRV < 30% | OR (95%CI) | P value |
| G | C | 0.682 | 30 (11.45) | 232 (88.55) | 1.00 (Reference) | - |
| C | T | 0.310 | 14 (11.76) | 105 (88.24) | 1.15 (0.58, 2.28) | 0.70 |
| G | T | 0.008 | 0 (00.00) | 3 (100.00) | - | - |
| TEP1 (rs938886) | TEP1 (rs1713449) | Frequency | HRV ≥ 15% | HRV < 15% | OR (95%CI) | P value |
| G | C | 0.682 | 92 (35.11) | 170 (64.89) | 1.00 (Reference) | - |
| C | T | 0.310 | 50 (42.02) | 69 (57.98) | 1.33 (0.85-2.08) | 0.22 |
| G | T | 0.008 | 2 (66.67) | 1 (33.33) | 4.13 (0.37, 46.59) | 0.25 |
The GMDR approach was used to evaluate the effect of SNP-SNP interactions among the three TEP1 and RECQL5 SNPs. The results obtained from the GMDR analysis of the one-to three-locus models in the patients with HRV ≥ 30% are summarized in Table 9. The one-locus model of RECQL5 rs820196 was the best model of SNP-SNP interaction, recording the highest cross-validation consistency (CVC) of 10/10 and a testing accuracy of 0.6127 (P value based on 1000 permutations, P = 0.032).
| Best model | Training balanced accuracy | Testing balanced accuracy | Cross-validation consistency | P value |
| rs820196 | 0.6116 | 0.6127 | 10/10 | 0.032a |
| rs938886, rs820196 | 0.6495 | 0.5782 | 10/10 | 0.151 |
| rs938886, rs820196, rs1713449 | 0.6495 | 0.5753 | 10/10 | 0.168 |
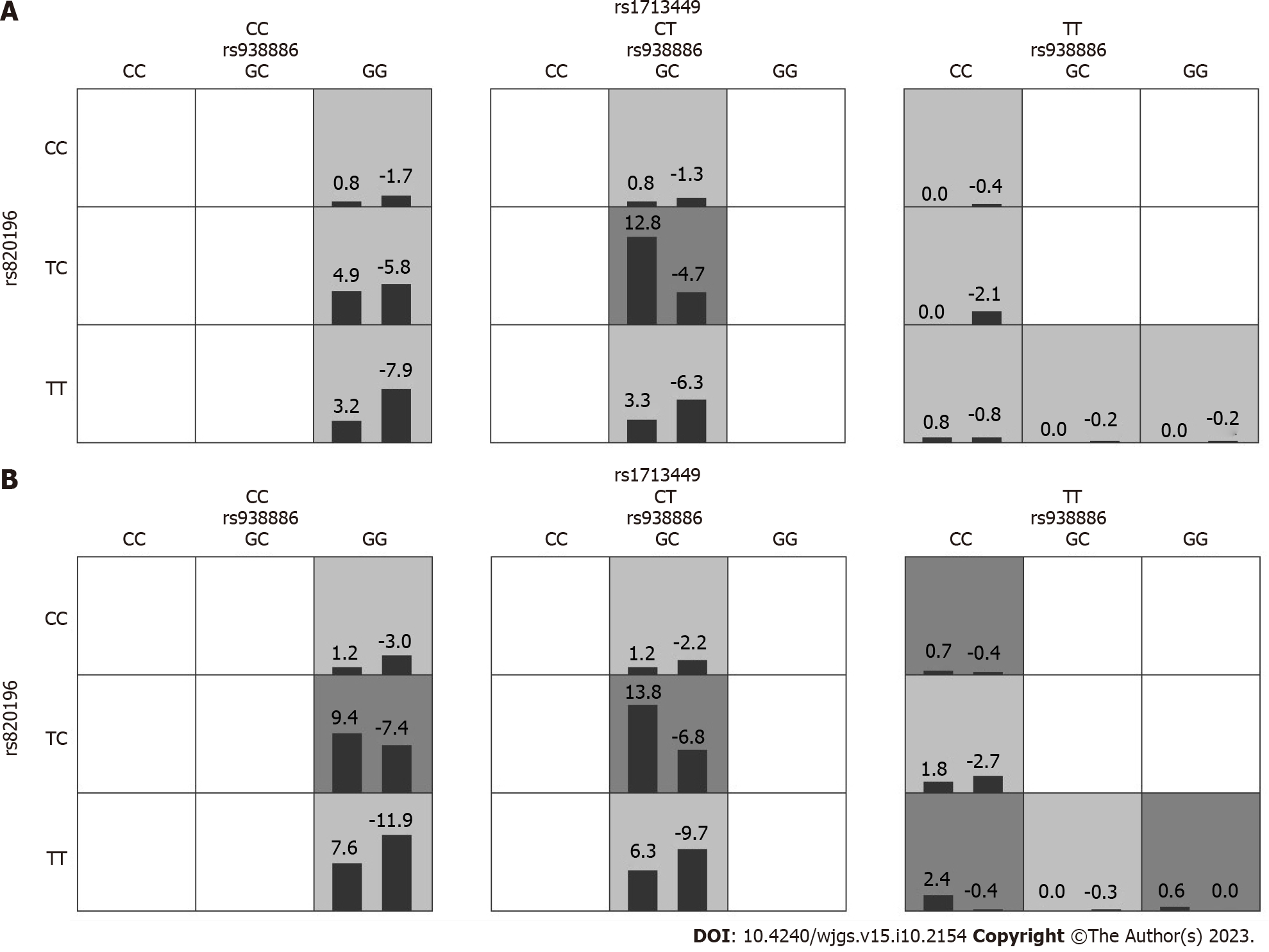
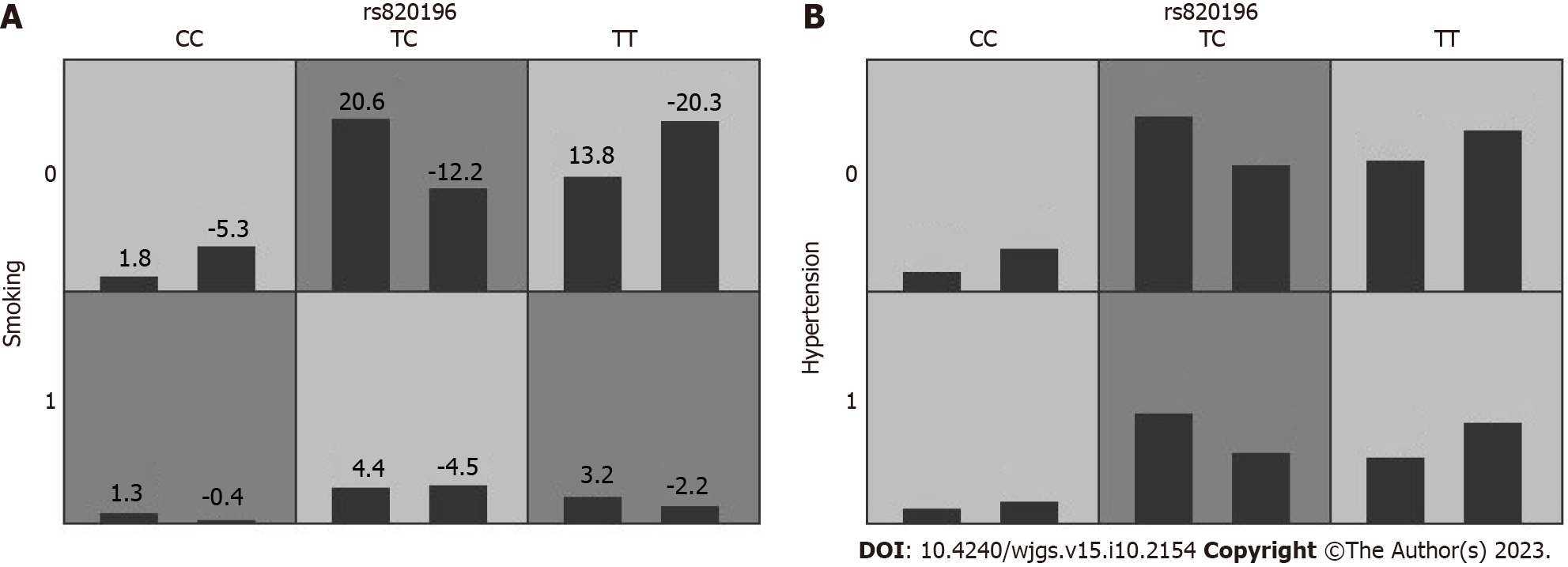
In order to obtain an OR with a 95%CI for the joint effects of SNP-SNP interactions on HRV, we also conducted interaction analyses of the GMDR models using logistic regression. After adjusting for the factors of age, gender, smoking, hypertension, and thyroid function by multivariate logistic regression analysis, the combination of the rs93886 and rs820196 SNPs (GC-TC) was correlated with a higher risk of decreased HR ≥ 30% (Figure 3A). One-to three-locus models in the patients with decreased HR ≥15% are showed in Figure 3B. Table 10 describes the results generated from the GMDR method for the two-way and three-way gene-gene interaction analyses using covariate adjustment. Compared with the wild-type, individuals with the GC-TC (rs93886 and rs820196, respectively) genotype and the GC-TC-CT (rs93886, rs820196, and rs1713449, respectively) genotype showed a significantly increased HRV risk (OR = 6.16 and 6.27 with P = 0.01).
| SNP genotypes | HRV ≥ 30% (n = 22) | HRV < 30% (n = 170) | OR (95%CI) | P value | ||
| rs938886 rs820196 | ||||||
| GG | TT | 3 (6.67) | 42 (93.33) | 1.00 (Reference) | - | |
| GG | TC | 5 (14.29) | 30 (85.71) | 2.91 (0.63, 13.55) | 0.17 | |
| GG | CC | 1 (10.00) | 9 (90.00) | 2.19 (0.19, 25.37) | 0.53 | |
| GC | TT | 3 (8.11) | 34 (91.89) | 1.65 (0.31, 8.74) | 0.56 | |
| GC | TC | 9 (22.50) | 31 (77.50) | 6.16 (1.53, 24.83) | 0.01a | |
| CC | TT | 1 (20.00) | 4 (80.00) | 5.84 (0.46, 74.57) | 0.17 | |
| Others | 0 (0.00) | 20 (100.00) | - | - | ||
| rs938886 rs820196 rs1713449 | ||||||
| GG | TT | CC | 3 (6.82) | 41 (93.18) | 1.00 (Reference) | - |
| GG | TC | CC | 5 (14.29) | 30 (85.71) | 2.97 (0.64, 13.79) | 0.17 |
| GG | CC | CC | 1 (10.00) | 9 (90.00) | 2.23 (0.19, 25.87) | 0.52 |
| GC | TT | CT | 3 (8.33) | 33 (91.67) | 1.73 (0.33, 9.18) | 0.52 |
| GC | TC | CT | 9 (22.50) | 31 (77.50) | 6.27 (1.56, 25.28) | 0.01b |
| CC | TT | TT | 1 (20.00) | 4 (80.00) | 5.94 (0.47, 75.98) | 0.17 |
| Others | 0 (0.00) | 22 (100.00) | - | - | ||
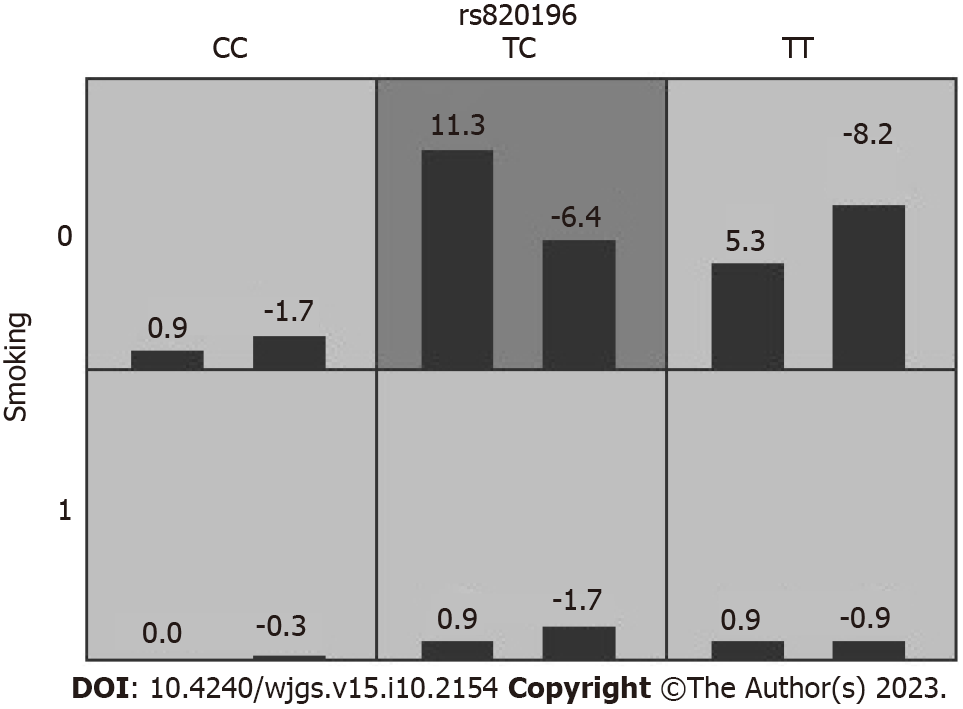
Table 11 describes the results generated with the GMDR method for the SNP-environment interaction analysis using covariate adjustment. The rs820196-smoking two-factor model was found to exhibit a statistically significant association with HRV ≥ 30% (P = 0.045) (Figure 4).
| Best model | Training balanced accuracy | Testing balanced accuracy | CVC | P value |
| SNP-smoking interaction1 | ||||
| Smoking | 0.5265 | 0.5135 | 10/10 | 0.263 |
| rs820196-smoking | 0.6281 | 0.6021 | 10/10 | 0.045a |
| rs938886-rs820196-smoking | 0.6703 | 0.5947 | 8/10 | 0.099 |
| SNP-hypertension interaction2 | ||||
| Hypertension | 0.5292 | 0.3308 | 10/10 | 0.800 |
| rs820196-hypertension | 0.6117 | 0.5648 | 10/10 | 0.190 |
| rs820196-rs1713449-hypertension | 0.6996 | 0.5063 | 8/10 | 0.426 |
| SNP-thyroid function interaction3 | ||||
| Thyroid function | 0.5123 | 0.4980 | 10/10 | 0.194 |
| rs820196-thyroid function | 0.6263 | 0.5895 | 10/10 | 0.104 |
| rs938886-rs820196-thyroid function | 0.6665 | 0.5727 | 8/10 | 0.153 |
After adjusting for age, gender, hypertension, and thyroid function, logistics regression showed that the nonsmokers with the rs820196 TC genotype had a significantly higher risk of HRV than the nonsmokers with the TT genotype (P = 0.04, Table 12).
| Smoking | rs820196 genotype | HRV ≥ 30% (n = 22) | HRV < 30% (n = 170) | OR (95%CI) | P value |
| No | TT | 6 (7.89) | 70 (92.11) | 1.00 (Reference) | - |
| Yes | TT | 1 (9.09) | 10 (90.91) | 1.47 (0.15, 14.27) | 0.74 |
| No | TC | 13 (19.70) | 53 (80.30) | 2.91 (1.03, 8.20) | 0.04a |
| Yes | TC | 1 (5.26) | 18 (94.74) | 0.80 (0.088, 7.36) | 0.85 |
| No | CC | 1 (5.88) | 16 (94.12) | 0.81 (0.089, 7.35) | 0.85 |
| Other | 0 (0.00) | 3 (100.00) | - | - | |
The results obtained from the GMDR analysis of the one-to three-locus models in the patients with HRV ≥ 15% are summarized in Table 13. The one-locus model of RECQL5 rs820196 was the best model of SNP-SNP interaction, recording the highest CVC of 10/10 and a testing accuracy of 0.5920 (P = 0.039).
| Best model | Training balanced accuracy | Testing balanced accuracy | CVC | P value |
| rs820196 | 0.5885 | 0.5920 | 10/10 | 0.039a |
| rs820196, rs1713449 | 0.6301 | 0.5798 | 9/10 | 0.052 |
| rs938886, rs820196, rs1713449 | 0.6335 | 0.5885 | 10/10 | 0.066 |
Table 14 summarizes the results obtained from the GMDR analysis for the two- to three-loci models of gene–gene interactions. Compared with the TT-CC (rs820196-rs1713449) genotype, the TT-TT and TC-CT genotypes were associated with a higher risk of HRV ≥ 15% (P = 0.04 and 0.01, respectively). Compared with the GG-CC-TT (rs938886-rs1713449-rs820196) genotype, the GC-CT-TC genotype showed a significantly increased HRV risk (P = 0.02).
| SNPs genotypes | HRV ≥ 15% (n = 72) | HRV < 15% (n = 120) | OR (95%CI) | P value | ||
| rs820196 rs1713449 | ||||||
| TT | CC | 12 (27.27) | 32 (72.73) | 1.00 (Reference) | - | |
| TT | CT | 10 (27.78) | 26 (72.22) | 1.03 (0.38, 2.79) | 0.95 | |
| TT | TT | 5 (71.43) | 2 (28.57) | 6.87(1.15, 41.09) | 0.04a | |
| TC | CC | 15 (42.86) | 20 (57.14) | 2.07 (0.79, 5.45) | 0.14 | |
| TC | CT | 22 (55.00) | 18 (45.00) | 3.26 (1.30, 8.20) | 0.01b | |
| TC | TT | 3 (30.00) | 7 (70.00) | 1.06 (0.22, 5.06) | 0.94 | |
| CC | CC | 2 (20.00) | 8 (80.00) | 0.56 (0.098, 3.21) | 0.52 | |
| CC | CT | 2 (25.00) | 6 (75.00) | 0.87 (0.15, 5.11) | 0.88 | |
| CC | TT | 1 (50.00) | 1 (50.00) | 2.90 (0.17, 50.84) | 0.47 | |
| rs938886 rs1713449 rs820196 | ||||||
| GG | CC | TT | 12 (27.27) | 32 (72.73) | 1.00 (Reference) | |
| GG | CC | TC | 15 (42.86) | 20 (57.14) | 1.87 (0.74, 4.77) | 0.19 |
| GG | CC | CC | 2 (20.00) | 8 (80.00) | 0.51 (0.09, 2.87) | 0.45 |
| GC | CT | TT | 10 (27.78) | 26 (72.22) | 0.93 (0.36, 2.44) | 0.89 |
| GC | CT | TC | 22 (55.00) | 18 (45.00) | 2.96 (1.21, 7.22) | 0.02c |
| GC | CT | CC | 2 (25.00) | 6 (75.00) | 0.79 (0.14, 4.57) | 0.79 |
| CC | TT | TT | 4 (80.00) | 1 (20.00) | 9.71 (0.97, 96.94) | 0.05 |
| CC | TT | TC | 3 (30.00) | 7 (70.00) | 0.98 (0.21, 4.63) | 0.98 |
| Others | 2 (50.0) | 2 (50.0) | - | - | ||
Table 15 and Figure 5 describe the results generated with the GMDR method for the SNP-environment interaction analysis using covariate adjustment. The rs820196-smoking and rs820196-hypertension two-factor models were found to exhibit a statistically significant association with HRV ≥ 15% (P < 0.05).
| Best model | Training balanced accuracy | Testing balanced accuracy | CVC | P value |
| SNP-smoking interaction1 | ||||
| Smoking | 0.5191 | 0.5171 | 10/10 | 0.349 |
| rs820196-smoking | 0.6148 | 0.5954 | 10/10 | 0.022a |
| rs820196-rs1713449-smoking | 0.6510 | 0.5764 | 9/10 | 0.065 |
| SNP-hypertension interaction2 | ||||
| Hypertension | 0.5156 | 0.3564 | 10/10 | 0.995 |
| rs820196-hypertension | 0.5885 | 0.5920 | 10/10 | 0.043b |
| rs820196-rs1713449-hypertension | 0.6434 | 0.5343 | 9/10 | 0.219 |
| SNP-thyroid function interaction3 | ||||
| Thyroid function | 0.5126 | 0.5113 | 10/10 | 0.381 |
| rs820196-thyroid function | 0.5888 | 0.5814 | 10/10 | 0.065 |
| rs820196-rs1713449-thyroid function | 0.6411 | 0.5706 | 9/10 | 0.075 |
After adjusting for age, gender, and thyroid function, logistics regression showed that the nonsmokers with the rs820196 TC genotype had a significantly higher risk of HRV ≥ 15% than the nonsmokers with the TT genotype (P = 0.01, Table 16).
| Environment | rs820196 genotype | HRV ≥ 15% (n = 72) | HRV < 15% (n = 120) | OR (95%CI) | P value |
| Smoking | |||||
| No | TT | 22 (28.95) | 54 (71.05) | 1.00 (Reference) | - |
| Yes | TT | 5 (45.45) | 6 (54.55) | 2.12 (0.57, 7.95) | 0.26 |
| No | TC | 33 (50.00) | 33 (50.00) | 2.53 (1.26, 5.08) | 0.01a |
| Yes | TC | 7 (36.84) | 12 (63.16) | 1.43 (0.48, 4.22) | 0.52 |
| No | CC | 3 (17.65) | 14 (82.35) | 0.48 (0.12, 1.89) | 0.30 |
| Yes | CC | 2 (66.67) | 1 (33.33) | 4.68 (0.39, 56.33) | 0.22 |
| Hypertension | |||||
| No | TT | 18 (32.73) | 37 (67.27) | 1.00 (Reference) | - |
| Yes | TT | 9 (28.13) | 23 (71.88) | 0.77 (0.28, 2.11) | 0.62 |
| No | TC | 25 (46.30) | 29 (53.70) | 1.75 (0.80, 3.82) | 0.16 |
| Yes | TC | 15 (48.39) | 16 (51.61) | 1.90 (0.75, 4.78) | 0.18 |
| No | CC | 3 (23.08) | 10 (76.92) | 0.58 (0.14, 2.42) | 0.45 |
| Yes | CC | 2 (28.57) | 5 (71.43) | 0.72 (0.12, 4.26) | 0.72 |
RECQL5 protein is involved in the regulation of transcription elongation, the DNA damage response and DNA replication[9,10]. Mutations in RECQ helicases are associated with the genetic disorders bloom syndrome, werner syndrome, and Rothmund-Thomson syndrome, which are characterized by chromosomal instability, premature aging, and a predisposition to cancer[11-13]. RECQL5-knockout mice are more likely to develop cancer, and human cells deficient in RECQL5 display chromosomal instability and elevated sister chromatid exchange events, similar to cells deficient in any of the other RECQ helicases[14]. According to a recent study, RECQL5 promotes metastasis and resistance to cisplatin in non-small cell lung cancer[15]. A large case-control study suggests that RECQL5 is a new moderate-risk breast cancer gene[16]. RECQL5 protein overexpression in breast cancer is strongly correlated with poor prognosis and survival, and with therapeutic resistance. A small molecule targeting RECQL5 was able to preferentially kill RECQL5-expressing breast cancers and led to the efficient sensitization of cisplatin-resistant breast cancers[17]. Philip et al[18] identified RECQL5 as a novel pharmacological target for expanding Poly (ADPRibose) Polymerase inhibitor based treatment horizon for homologous recombination-proficient cancers. Low RECQL5 expression indicates poor prognosis in gastric carcinoma and is an independent prognostic factor[19]. To date, no study has shown that RECQL5 is associated with arrhythmias, although a variant in chromodomain helicase DNA-binding protein 4 associated with childhood idiopathic epilepsy with sinus arrhythmia has been reported[20]. Given the cardinal role of the RECQL5 protein in genome stability[21], one might speculate that DNA disrepair caused by RECQL5 mutations is the probable cause of myocardial apoptosis and arrhythmia. Further investigations are required to determine the risk of sudden cardiogenic arrhythmia caused by RECQL5 mutations.
In this study, patients with gastric cancer were divided into two groups according to two different cutoffs for HRV: 30% and 15%. There were no significant differences in gender, age, smoking history, hypertension, diabetes, blood type, and thyroid function between the two groups regardless of the cutoff used, indicating that these factors had no effect on HRV. The distribution of the RECQL5 (rs820196) genotype frequency in patients with HRV ≥ 15% differed from that in the control group. The relationships between these SNPs and HRV risk were evaluated by the inheritance models. Under the co-dominant and overdominant models, the TC genotype was associated with a higher risk of HR decrease relative to the TT and TT + CC genotypes.
TEP1 is a component of the telomerase ribonucleoprotein complex, and is responsible for catalyzing the addition of new synthetic telomere sequences to chromosome ends[22]. Genetic variations in telomere-associated pathway genes might affect telomere length and chromosomal stability, and subsequently disease susceptibility. In this study, the LD analysis showed that TEP1 rs938886 and TEP1 rs1713449 had a strong linkage relationship. Unexpectedly, there were no differences among the three haplotypes (P > 0.05).
GMDR is a novel and powerful statistical tool for detecting and modeling epistasis. It is a non-parametric and model-free alternative to logistic regression for detecting and characterizing non-linear interactions among discrete genetic and environmental attributes[23]. It is mainly used to detect gene-gene and gene-environment interactions underlying complex traits in epidemiological and genetic research. In this study, after adjusting for a multitude of covariates, we found that the best one-factor model was rs820196, whether the HRV cutoff was 30% or 15%. Logistic regression analysis was performed for better risk assessment. When the HRV cutoff value was 30%, there was a significant gene–gene interaction between rs938886 and rs820196. The subjects carrying the GC-TC genotypes of rs938886 and rs820196 showed a higher HRV risk when compared with the GG-TT genotype carriers. In the three-factor model of rs938886, rs820196, and rs1713449, the patients carrying the GC-TC-CT genotype had a higher risk of HRV compared with the GG-TT-CC wild-type genotype. We also found a potential gene-environment interaction between rs820196 and smoking, such that the nonsmokers carrying the TC genotype of rs820196 had a higher HRV risk compared with the nonsmokers carrying the TT genotype. When the HRV cutoff was 15%, patients carrying the TT-TT and TC-CT genotypes of rs820196 and rs1713449 showed a higher HRV risk when compared with the TT-CC genotype carriers. Patients carrying the GC-CT-TC genotypes of rs938886, rs1713449, and rs820196 showed a higher HRV risk when compared with the GG-CC-TT genotype carriers. When the HRV cutoff was 15%, the best-fitting models for SNP-environment interactions were rs820196-smoking and rs820196-hypertension. Consistent with the results of the previous grouping, the nonsmokers carrying the TC genotype of rs820196 had a higher HRV risk compared with the nonsmokers carrying the TT genotype.
Although several positive associations were observed, some limitations of this study should be considered. First, the sample size was small, with only 192 patients being enrolled. Second, all the participants were recruited from the same hospital; hence, inherent selection bias was unavoidable. Third, was the prognosis of patients with high HRV poorer than that of those with low HRV? Did the patients with high HRV ever have arrhythmias in daily life? These questions need to be investigated. Given these limitations, further studies with larger sample sizes and more comprehensive clinical information will be required to confirm our findings.
This study on RECQL5 and TEP1 genetic polymorphisms may help uncover the underlying mechanisms of arrhythmia phenotypic variation. The 2D PCR used in this study helped to screen the three SNP loci more quickly and economically. In the future, when performing tumor resection and peritoneal lavage with distilled water, we suggest anesthesiologists assess the risk of sudden HR drop based on the genetic polymorphisms of RECQL5 (rs820196) and TEP1 (rs938886 and rs1713449), and medical history. If patients are at high risk and the baseline HR is < 40 beats/min, vasopressors such as norepinephrine, epinephrine, dopamine, and phenylephrine will be recommended before surgery. During the perioperative period, all patients are routinely monitored for arterial blood pressure by electrocardiography. An anesthetic machine is used to support breathing and monitor end-expiratory carbon dioxide partial pressure. Once the HR decreases by 30% or < 40 beats/min after lavage, vasopressors should be used immediately. If cardiac arrest occurs, cardiac compression should be performed immediately, so that the heartbeat pause time is strictly limited to 1 min. Extensive peritoneal lavage with warm distilled water is widely used in surgery for breast cancer, lung cancer and gastrointestinal cancer. The purpose of this study was to screen high-risk groups through the SNP detection of high-risk genes, and focus on improving safety during the perioperative period.
In conclusion, our results showed, for the first time, that polymorphisms of the RECQL5 and TEP1 genes were associated with sudden decreases in HR during abdominal lavage in patients with gastric cancer. Nonsmokers carrying the TC genotype of rs820196 and the GC-CT-TC genotype carriers of rs938886, rs1713449 and rs820196 were found to have a higher HRV risk.
Peritoneal lavage with distilled water to kill residual tumor cells is a routine procedure in gastrectomy, but this procedure often causes a sudden decrease in heart rate (HR) in some patients.
To investigate whether there are differences in genetic background between patients with discordant HR changes and help clinicians to better assess the perioperative risk of patients undergoing gastrectomy.
To investigate whether genotypes, genetic patterns, single nucleotide polymorphism (SNP)-SNP and SNP-environment interactions were associated with high heart rate variability (HRV).
A total of 192 patients who underwent distal gastrectomy were divided into two groups according to changes in HR (using 30% and 15% as cutoffs). Two-dimensional polymerase chain reaction was used to establish a single-tube method to detect telomerase-associated protein 1 (TEP1) rs938886 and rs1713449 and RecQ like helicase 5 (RECQL5) rs820196. Genotypes, genetic patterns and the interaction of SNP-SNP and SNP-environment were analyzed by non-conditional logistic regression model and generalized multifactor dimensionality reduction.
The polymorphism of the RECQL5 gene (rs820196) was associated with a sudden decrease in HR during abdominal lavage in patients with gastric cancer. Rs820196-smoking and rs820196-hypertension were associated with HRV ≥ 15%. Nonsmokers carrying the TC genotype of rs820196 and patients carrying the GC-CT-TC genotype of rs938886, rs1713449 and rs820196 had higher HRV risk.
The polymorphisms of RECQL5 (TC genotype of rs820196) and TEP1 (GC-CT genotype of rs938886 and rs1713449) genes were associated with HRV.
HRV risk assessment in patients who are about to undergo peritoneal lavage is helpful for perioperative safety. This cost-effective SNP screening method can be extended to other patients undergoing tumor resection (such as breast cancer, lung cancer and other gastrointestinal cancer) and multicenter studies.
The authors thank Professor Yueping Shen from Department of Epidemiology and Biostatistics, Medical College of Soochow University for providing data statistical analysis support.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fujino Y, Japan; Ueda H, Japan S-Editor: Qu XL L-Editor: A P-Editor: Wu RR
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64542] [Article Influence: 16135.5] [Reference Citation Analysis (176)] |
| 2. | Guo J, Xu A, Sun X, Zhao X, Xia Y, Rao H, Zhang Y, Zhang R, Chen L, Zhang T, Li G, Xu H, Xu D. Combined Surgery and Extensive Intraoperative Peritoneal Lavage vs Surgery Alone for Treatment of Locally Advanced Gastric Cancer: The SEIPLUS Randomized Clinical Trial. JAMA Surg. 2019;154:610-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 3. | Guo J, Xu A, Sun X, Zhao X, Xia Y, Rao H, Zhang Y, Zhang R, Chen L, Zhang T, Li G, Xu H, Xu D. Three-year outcomes of the randomized phase III SEIPLUS trial of extensive intraoperative peritoneal lavage for locally advanced gastric cancer. Nat Commun. 2021;12:6598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 4. | Song ED, Xia HB, Zhang LX, Ma J, Luo PQ, Yang LZ, Xiang BH, Zhou BC, Chen L, Sheng H, Fang Y, Han WX, Wei ZJ, Xu AM. Efficacy and outcome of extensive intraoperative peritoneal lavage plus surgery vs surgery alone with advanced gastric cancer patients. World J Gastrointest Surg. 2023;15:430-439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 5. | Yang HK, Ji J, Han SU, Terashima M, Li G, Kim HH, Law S, Shabbir A, Song KY, Hyung WJ, Kosai NR, Kono K, Misawa K, Yabusaki H, Kinoshita T, Lau PC, Kim YW, Rao JR, Ng E, Yamada T, Yoshida K, Park DJ, Tai BC, So JBY; EXPEL study group. Extensive peritoneal lavage with saline after curative gastrectomy for gastric cancer (EXPEL): a multicentre randomised controlled trial. Lancet Gastroenterol Hepatol. 2021;6:120-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 6. | Takemoto K, Shiozaki A, Ichikawa D, Komatsu S, Konishi H, Nako Y, Murayama Y, Kuriu Y, Nakanishi M, Fujiwara H, Okamoto K, Sakakura C, Nakahari T, Marunaka Y, Otuji E. Evaluation of the efficacy of peritoneal lavage with distilled water in colorectal cancer surgery: in vitro and in vivo study. J Gastroenterol. 2015;50:287-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 7. | Saif A, Teke M, Ryan C, Papai E, Nevler A, Hernandez JM, Lavu H. The WASH (Water or Saline at High Volumes) Trial: A Randomized Trial to Assess the Survival Impact of Extensive Peritoneal Lavage Using Distilled Water or Saline at High Volumes After Pancreatic Resection for Pancreatic Ductal Adenocarcinoma. Ann Surg Oncol. 2022;29:5372-5374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Yuan Y, Yao S, Luo GH, Zhang XY. Impact of metabolism-related mutations on the heart rate of gastric cancer patients after peritoneal lavage. World J Clin Cases. 2021;9:1318-1328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (3)] |
| 9. | Saponaro M, Kantidakis T, Mitter R, Kelly GP, Heron M, Williams H, Söding J, Stewart A, Svejstrup JQ. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 2014;157:1037-1049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 164] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 10. | Hamadeh Z, Lansdorp P. RECQL5 at the Intersection of Replication and Transcription. Front Cell Dev Biol. 2020;8:324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 11. | Bugreev DV, Mazina OM, Mazin AV. Bloom syndrome helicase stimulates RAD51 DNA strand exchange activity through a novel mechanism. J Biol Chem. 2009;284:26349-26359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 12. | Dhillon KK, Sidorova J, Saintigny Y, Poot M, Gollahon K, Rabinovitch PS, Monnat RJ Jr. Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell. 2007;6:53-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 13. | Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 498] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 14. | Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073-3084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 285] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 15. | Xia HW, Zhang ZQ, Yuan J, Niu QL. Human RECQL5 promotes metastasis and resistance to cisplatin in non-small cell lung cancer. Life Sci. 2021;265:118768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | Marchena-Perea EM, Salazar-Hidalgo ME, Gómez-Sanz A, Arranz-Ledo M, Barroso A, Fernández V, Tejera-Pérez H, Pita G, Núñez-Torres R, Pombo L, Morales-Chamorro R, Cano-Cano JM, Soriano MDC, Garre P, Durán M, Currás-Freixes M, de la Hoya M, Osorio A. A Large Case-Control Study Performed in Spanish Population Suggests That RECQL5 Is the Only RECQ Helicase Involved in Breast Cancer Susceptibility. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 17. | Chakraborty S, Dutta K, Gupta P, Das A, Ghosh SK, Patro BS. Targeting RECQL5 Functions, by a Small Molecule, Selectively Kills Breast Cancer in Vitro and in Vivo. J Med Chem. 2021;64:1524-1544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 18. | Philip KT, Dutta K, Chakraborty S, Patro BS. Functional inhibition of RECQL5 helicase elicits non-homologous end joining response and sensitivity of breast cancers to PARP inhibitor. Int J Biochem Cell Biol. 2023;161:106443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Reference Citation Analysis (0)] |
| 19. | Lin Y, Chen H, Wang X, Xiang J, Wang H, Peng J. Mining the role of RECQL5 in gastric cancer and seeking potential regulatory network by bioinformatics analysis. Exp Mol Pathol. 2020;115:104477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 20. | Liu XR, Ye TT, Zhang WJ, Guo X, Wang J, Huang SP, Xie LS, Song XW, Deng WW, Li BM, He N, Wu QY, Zhuang MZ, Xu M, Shi YW, Su T, Yi YH, Liao WP; China Epilepsy Gene 1. 0 Project. CHD4 variants are associated with childhood idiopathic epilepsy with sinus arrhythmia. CNS Neurosci Ther. 2021;27:1146-1156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 21. | Lu H, Davis AJ. Human RecQ Helicases in DNA Double-Strand Break Repair. Front Cell Dev Biol. 2021;9:640755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 22. | Poderycki MJ, Rome LH, Harrington L, Kickhoefer VA. The p80 homology region of TEP1 is sufficient for its association with the telomerase and vault RNAs, and the vault particle. Nucleic Acids Res. 2005;33:893-902. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 24] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 23. | Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet. 2007;80:1125-1137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 454] [Article Influence: 25.2] [Reference Citation Analysis (0)] |