Published online Apr 15, 2014. doi: 10.4239/wjd.v5.i2.128
Revised: December 28, 2013
Accepted: January 17, 2014
Published online: April 15, 2014
Processing time: 149 Days and 16.3 Hours
Type 2 diabetes mellitus (T2DM) is a complex disease in which both genetic and environmental factors interact in determining impaired β-cell insulin secretion and peripheral insulin resistance. Insulin resistance in muscle, liver and fat is a prominent feature of most patients with T2DM and obesity, resulting in a reduced response of these tissues to insulin. Considerable evidence has been accumulated to indicate that heredity is a major determinant of insulin resistance and T2DM. It is believed that, among individuals destined to develop T2DM, hyperinsulinemia is the mechanism by which the pancreatic β-cell initially compensates for deteriorating peripheral insulin sensitivity, thus ensuring normal glucose tolerance. Most of these people will develop T2DM when β-cells fail to compensate. Despite the progress achieved in this field in recent years, the genetic causes of insulin resistance and T2DM remain elusive. Candidate gene association, linkage and genome-wide association studies have highlighted the role of genetic factors in the development of T2DM. Using these strategies, a large number of variants have been identified in many of these genes, most of which may influence both hepatic and peripheral insulin resistance, adipogenesis and β-cell mass and function. Recently, a new gene has been identified by our research group, the HMGA1 gene, whose loss of function can greatly raise the risk of developing T2DM in humans and mice. Functional genetic variants of the HMGA1 gene have been associated with insulin resistance syndromes among white Europeans, Chinese individuals and Americans of Hispanic ancestry. These findings may represent new ways to improve or even prevent T2DM.
Core tip: Despite the progress in clinical and laboratory investigations, the fundamental cause of type 2 diabetes mellitus (T2DM) remains uncertain. Candidate gene, linkage and genome-wide association studies have highlighted the role of genetics in the development of T2DM. Using these strategies, a large number of variants have been identified in many genes, most of which may influence an individual’s risk of developing T2DM. In this review, we compile information on genetic factors that influence the risk of T2DM. In addition, we discuss the results from recent studies on the role of HMGA1 on the issue, which might be important for future breakthroughs in this field.
- Citation: Brunetti A, Chiefari E, Foti D. Recent advances in the molecular genetics of type 2 diabetes mellitus. World J Diabetes 2014; 5(2): 128-140
- URL: https://www.wjgnet.com/1948-9358/full/v5/i2/128.htm
- DOI: https://dx.doi.org/10.4239/wjd.v5.i2.128
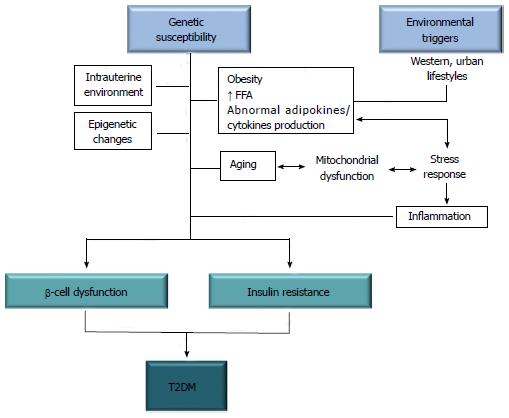
Type 2 diabetes mellitus (T2DM) is a chronic endocrine and metabolic disease that is often associated with being overweight or frank obesity. It affects millions of people worldwide, with a rapidly increasing incidence and prevalence[1,2]. The latest estimate from the International Diabetes Federation (http://www.idf.org) is equivalent to a global prevalence rate of 8.4% of the adult population, while worldwide diabetes cases hit a new record at 382 million in 2013. Among the determinants of this steadily increasing trend is the combination of genetic and environmental factors responsible for either a positive energy balance resulting in body fat accumulation and weight gain and/or a reduced energy expenditure from a reduction in physical activity and a sedentary lifestyle. Despite extensive attempts at clinical management of T2DM, many diabetic patients will develop a wide variety of long-term complications, including retinopathy, nephropathy and cardiovascular diseases that are among the most frequent causes of morbidity and mortality in affected people, whose effective prevention and treatment require enormous efforts and funding[3]. Typically, T2DM is presented as a common, heterogeneous, complex disease in which both predisposing genetic factors and precipitating environmental factors interact together and cause hyperglycemia, which constitutes the primary hallmark of T2DM[4,5]. Although still poorly understood, the role of genetics in T2DM is well documented. This is supported by a series of evidence, including the strong familial aggregation of the disease, in which the risk of developing T2DM is 40% for those who have an affected parent (higher if the mother rather than the father) and 70% if both parents are diabetics[6]. The highest risk in first-degree relatives, compared to the general population, persists even after removal from the family of origin, for example, as a result of adoption. Furthermore, in identical monozygotic twins (with identical genetic makeup), the concordance rate for the disease approaches 100%, much higher than that seen in non-identical (dizygotic) twins or among siblings[7]. Genetic predisposition in T2DM is also supported by the observation that differences in disease prevalence rates exist among populations, even after migration of entire ethnic groups to another country, thus independent from the environmental influences[8].
On the other hand, the role of environmental factors in influencing susceptibility to T2DM is equally well known. Among these factors are increased caloric intake and a sedentary lifestyle, two conditions common in populations with a higher standard of living and a more westernized lifestyle, responsible for most of the excess weight and obesity in the modern adult’s life[9]. The spread of the western way of life in developing countries also explains the epidemic explosion of the disease[1,2], whereas the existing epidemiological data show that the spatial and temporal distribution of T2DM in the geographical areas examined is comparable to the trend of being overweight and obesity[10]. The excess weight causes insulin resistance, which represents the initial step in the natural history of T2DM. Initially, in individuals destined to become diabetic, pancreatic β-cells compensate for the insulin resistance by secreting increased levels of insulin, thus ensuring post-prandial euglycemia[11]. Hyperglycemia in insulin resistant subjects develops later when the β-cells fail to compensate. Thus, from a pathophysiological standpoint, T2DM is characterized by a combination of peripheral insulin resistance and inadequate insulin secretion by the pancreatic β-cells. As supported by numerous studies in the literature[12,13], both defects are the result of a complex interaction between genetic and environmental factors (Figure 1), including chemical agents (calcium and zinc ions) and polluting organic substances that are suspected to play a role in amyloid fiber formation in pancreatic β-cells, thus contributing to the pathology of T2DM[14-17]. The involvement in the pathogenesis of T2DM of multiple genes that interact with each other in an epistatic manner may explain why, despite the enormous efforts made to date, the identification of genetic determinants responsible for an increased susceptibility to T2DM still remains unsolved[18,19].
The present review aims to give an overview of the recent findings in this context. We also discuss the results from some recent studies which might be important for future breakthroughs in this field.
Over the past few years, various international research centers have been involved in the study and identification of genes predisposing to T2DM using various methods of investigation. Linkage analysis was used to identify potential genes associated with the disease, starting from the analysis of families and then studying a small number of individuals genetically related to each other. Genotyping for genetic markers in family members with and without T2DM has allowed the identification of DNA regions containing loci associated with disease risk. Thanks to this method, the association of T2DM with the calpain-10 (CAPN10) gene[20] was initially identified and later its association with the transcription factor 7-like 2 (TCF7L2) gene[21], whose genetic variants in affected individuals increase the risk of diabetes approximately 1.5 times[19].
Another approach used was to search for genetic variants within functional candidate genes encoding for protein(s) with important implications for glucose homeostasis and positional candidate genes that have a genetic association on the basis of a previous linkage study. This experimental strategy is applied to population studies rather than studies of families. Association studies of functional candidate genes represent one of the most powerful approaches as the pathogenetic mechanism of any genetic abnormality would be easily explained. The limit of this strategy, however, is constituted by the fact that it allows focused attention on a single gene at a time. Although many studies have reported associations of functional and positional candidate genes with T2DM, only some of these showed a significant and reproducible association with the disease (Table 1).
| Gene | Chr | Odds ratio | RAF | Study | Function and probable mechanism | Ref |
| ADAMTS9 | 3 | 1.09-1.05 | 0.68-0.81 | MA | Metalloproteinase/Insulin action | [22–24] |
| ADCY5 | 3 | 1.12 | 0.78 | MA | Adenylyl cyclases/Insulin action | [25] |
| ANK1 | 8 | 1.09 | 0.76 | MA, CC | Cell stability/β-cell function | [26–28] |
| ANKRD55 | 5 | 1.08 | 0.7 | MA, CC | Insulin action | [26,27] |
| ANKS1A | 6 | 1.11 | 0.91 | GWAS | Pathway regulator/Unknown | [29] |
| ARAP1 | 11 | 1.08-1.14 | 0.81-0.88 | GWAS, MA | Actin cytoskeleton modulator/β-cell function | [22,24] |
| BCAR1 | 16 | 1.12 | 0.89 | MA, CC | Docking protein/β-cell function | [26,27] |
| BCL2 | 18 | 1.09 | 0.64 | GWAS | Cell death regulator/Unknown | [24] |
| BCL11A | 2 | 1.08-1.09 | 0.46 | MA | Zinc finger/β-cell function | [22] |
| CAMK1DCDC123 | 10 | 1.07-1.11 | 0.18 | LA, MA | Protein kinase/β-cell function | [22–24] |
| Mitotic protein/β-cell function | ||||||
| CAPN10 | 2 | 1.09-1.18 | 0.73-0.96 | MA | Calpain cysteine protease/Insulin action | [30–33] |
| CDKAL1 | 6 | 1.10-1.20 | 0.27-0.31 | GWAS, MA | β-cell function | [24,34–36] |
| CDKN2ACDKN2B | 9 | 1.19-1.20 | 0.82-0.83 | GWAS | Cyclin-dependent kinase inhibitor/β-cell function | [24,34,35] |
| CENTD2 | 11 | 1.08-1.13 | 0.81-0.88 | GWAS | β-cell function | [22,24] |
| CHCHD9TLE4 | 9 | 1.11-1.20 | 0.93 | MA | Unknown | [22] |
| CILP2 | 19 | 1.13 | 0.08 | MA, CC | Unknown | [26,27] |
| DGKB | 7 | 1.04-1.06 | 0.47-0.54 | MA | Diacylglycerol kinase/Insulin action | [24,25] |
| DUSP9 | X | 1.09-1.27 | 0.12-0.77 | MA | Phosphatase | [22,24] |
| FOLH1 | 11 | 1.10 | 0.09 | GWAS | Transmembrane glycoprotein/Unknown | [24] |
| FTO | 16 | 1.06-1.27 | 0.38-0.41 | GWAS, MA | Metabolic regulator/Insulin action | [24,37] |
| GATAD2A | 19 | 1.12 | 0.08 | GWAS | Transcriptional repressor/Unknown | [24] |
| GCK | 7 | 1.07 | 0.20 | MA | Glucokinase/Insulin action | [25] |
| GCKR | 2 | 1.06-1.09 | 0.59-0.62 | MA | Glucokinase regulator/Insulin action | [24,25] |
| GIPR | 19 | 1.10 | 0.27 | GWAS | G-protein coupled receptor/Unknown | [24] |
| GRB14 | 2 | 1.07 | 0.60 | MA, GCS | Adapter protein/Insulin action | [26,27] |
| HFE | 6 | 1.12 | 0.29 | MA | Membrane protein/Unknown | [38] |
| HHEX | 10 | 1.12-1.13 | 0.53-0.60 | AL, MA | Transcriptional repressor/ | [22,24,34,39] |
| IDE | Intracellular insulin degradation/ | |||||
| KIF11 | Motor protein | |||||
| HMG20A | 15 | 1.08 | 0.68 | MA, GCS | Chromatin-associated protein/Unknown | [26,27] |
| HMGA1 | 6 | 1.34-15.8 | 0.10 | GCS | Transcriptional regulator/Insulin action | [40-42] |
| HMGA2 | 12 | 1.10-1.20 | 0.09-0.10 | MA | Transcriptional regulator | [22,24] |
| HNF1A | 12 | 1.07-1.14 | 0.77-0.85 | MA | Pancreatic and liver transcriptional activator | [22,24] |
| HNF1B | 17 | 1.08-1.17 | 0.47-0.51 | GCS, MA | Transcription factor/β-cell function | [22,24] |
| IGF2BP2 | 3 | 1.14 | 0.29-0.32 | GWAS, MA | Binding protein/β-cell function | [22,24,34,35] |
| IRS1 | 2 | 1.09-1.12 | 0.64-0.67 | GCS, MA | Insulin signaling element/Insulin action | [22,24,43] |
| JAZF1 | 7 | 1.10 | 0.52 | MA | Zinc finger/β-cell function | [22,23] |
| KCNJ11 | 11 | 1.09-1.14 | 0.37-0.47 | GCS, MA | Potassium channel/β-cell function | [22,24,34,44] |
| KCNQ1 | 11 | 1.08-1.23 | 0.44 | GWAS | Potassium channel/β-cell function | [22,45,46] |
| KLF14 | 7 | 1.07-1.10 | 0.55 | MA | Transcription factor/Insulin action | [22] |
| KLHDC5 | 12 | 1.10 | 0.80 | MA, CC | Mitotic progression and cytokinesis/Unknown | [26,27] |
| LAMA1 | 18 | 1.13 | 0.38 | GWAS | Cellular migration mediator/Unknown | [29] |
| MC4R | 18 | 1.08 | 0.27 | MA, CC | G-protein–coupled receptor/Unknown | [26,27] |
| MTNR1B | 11 | 1.05-1.08 | 0.28-0.30 | GWAS, MA | Melatonin receptor/β-cell function | [24,47-49] |
| NOTCH2 | 1 | 1.06-1.13 | 0.10-0.11 | MA | Membrane receptor | [22-24] |
| PPARG | 3 | 1.11-1.17 | 0.85-0.88 | GCS, MA | Nuclear receptor/Insulin action | [22,24,34,50] |
| PRC1 | 15 | 1.07-1.10 | 0.22 | MA | Cytokinesis regulator | [22] |
| PROX1 | 1 | 1.07 | 0.50 | MA | Homeobox transcription factor/Insulin action | [25] |
| PTPRD | 9 | 1.57 | 0.10 | GWAS | Protein tyrosine phosphatase | [51] |
| RBMS1 | 2 | 1.11-1.08 | 0.79-0.83 | MA | DNA modulator/Insulin action | [24,52] |
| SLC2A2 | 3 | 1.06 | 0.74 | GWAS | Glucose sensor/β-cell function | [24] |
| SLC30A8 | 8 | 1.11-1.18 | 0.65-0.70 | GWAS, MA | Zinc efflux transporter/β-cell function | [22,24,25,34,53] |
| SREBF1 | 17 | 1.07 | 0.38 | GWAS | Lipid transcriptional regulator/Unknown | [24] |
| SRR | 17 | 1.28 | 0.69 | GWAS | Serine racemase | [51] |
| TCF7L2 | 10 | 1.31-1.71 | 0.26-0.30 | LA, MA,GWAS | Participates in the Wnt signaling pathway/β-cell function | [21,22,24,34] |
| THADA | 2 | 1.15 | 0.90 | MA | Thyroid adenoma-associated protein/β-cell function | [22-24] |
| TH/INS | 11 | 1.14 | 0.39 | GWAS | Catecholamine synthesis/Unknown | [24] |
| TLE1 | 9 | 1.07 | 0.57 | MA, CC | Transcriptional corepressor/Unknown | [26,27] |
| TP53INP1 | 8 | 1.06-1.11 | 0.48 | MA | Proapoptotic protein/Unknown | [22] |
| TSPAN8 | 12 | 1.06-1.09 | 0.27-0.71 | MA | Cell surface glycoprotein/β-cell function | [22-24] |
| LGR5 | G-protein coupled receptor/β-cell function | |||||
| WFS1 | 4 | 1.10-1.13 | 0.60-0.73 | GCS | Transmembrane protein/β-cell function | [22,24,54,55] |
| ZBED3 | 5 | 1.08-1.16 | 0.26 | MA | Zinc finger/b-cell function | [22] |
| ZFAND6 | 15 | 1.01-1.11 | 0.60-0.72 | MA | Zinc finger/β-cell function | [22,24] |
| ZMIZ1 | 10 | 1.08 | 0.52 | MA, CC | Transcriptional regulator/Unknown | [26,27] |
| Haplogroup B | mtDNA | 1.52 | 0.25 | GCS | [56] | |
| OriB | mtDNA | 1.10 | 0.30 | MA | [57] |
From 2007 onwards, the list of candidate genes has grown considerably, largely due to genome-wide association studies (GWAS), a technique commonly used to find links between genes and diseases across a substantial population. This strategy uses a database of over a million known genetic variants, which represent the majority of all common variants (minor allele frequency > 5%-10%), thus offering the possibility of simultaneously analyzing thousands of variations in a large number of patients and to perform meta-analysis of data from multiple studies. This methodology has helped to identify dozens of new associations between T2DM and genes with known or unknown functions (Table 1)[22-57], confirming some of the results from previous studies. However, despite the great potential of this approach, it is estimated that genetic variants identified through GWAS explain only 10% heritability for T2DM[58,59]. These relatively modest results can be explained taking into account some important limits of this strategy, such as the involvement of novel genetic variants not yet covered in the GWAS database, or the presence of variants with a frequency lower than the minimum threshold value. This means that the genes identified by GWAS so far are just the tip of the iceberg and that T2DM, far from being a condition limited to a few genetically and phenotypically prevalent forms, actually encompasses a heterogeneous group of genetically distinct disorders[18].
However, in many genetic studies carried out to date, the functional mechanism(s) by which the associated gene may increase susceptibility to T2DM is often poorly understood. In this respect, the intrinsic limitations of both the linkage analysis and GWAS are amplified by the fact that, in most cases, the genetic variants identified are located in non-coding regions of the DNA, whereby it becomes even more difficult to trace the role and influence of the associated gene in the development of the disease. In cases in which it was possible to ascertain the precise pathogenic mechanism, for example, through the study of association with the circulating levels of insulin or through the direct analysis of the gene’s protein product, it has been seen that most of the genes identified are involved in pancreatic β-cell mass and/or function, thus with implications in insulin secretion defects (Table 1). This observation suggests that most of the risk associated with T2DM in the general population relates to genetic defects in β-cells, while peripheral insulin resistance predominantly suffers from the environmental component[18,19,60].
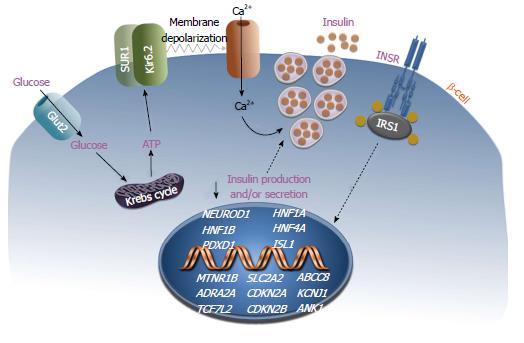
Figure 2 depicts some of the genes whose alteration confers an elevated risk of T2DM. Using the analysis of functional or positional candidate genes, several variants have been identified, including polymorphisms of the gene insulin receptor substrate-1 (IRS-1)[22,24,43]. The Gly972Arg variant of IRS-1 determines a defect in the binding of the p85 subunit of the phosphatidylinositol 3-kinase (PI3K) which in pancreatic β-cells causes a marked decrease in insulin secretion in response to glucose and sulfonylureas[61]. Other polymorphisms implicated in T2DM have been identified in the ABCC8 (also known as SUR1) and KCNJ11 genes, whose protein products take place in the formation of the Adenosine triphosphate (ATP)-sensitive potassium channel/sulfonylurea receptor of the pancreatic β-cell. The therapeutic response to sulfonylureas is compromised in patients with mutations in these genes. Other genes whose mutations were initially considered responsible for the less common forms of diabetes mellitus have subsequently been associated with an increased risk of T2DM[19]. Among these are the hepatocyte nuclear factor-1 homeobox A ( AHNF1A) gene, whose mutations are responsible for the most common monogenic form of MODY (MODY3), a form of maturity onset diabetes of the young (also known as HNF1A-MODY), and the gene hepatocyte nuclear factor-1 homeobox B (HNF1B), which determines a less frequent but more severe monogenic form of diabetes, the MODY5. Both of these genes encode nuclear transcription factors involved in the development and function of pancreatic islets.
As already mentioned, the association between TCF7L2 gene polymorphisms and susceptibility to T2DM was highlighted initially by linkage studies and confirmed thereafter by GWAS. However, only recently has the role played by the transcription factor TCF7L2 in the β-cell insulin secretion become evident[62]. Another gene that has recently been associated with T2DM is the melatonin receptor 1B (MTNR1B) gene which encodes for the receptor of the pineal hormone melatonin, MTNR1B, that is involved in the regulation and facilitation of sleep. Genetic variants of the MTNR1B gene, associated with gain-of-function of the MTNR1B receptor protein and a reduction in insulin secretion, have been reported in diabetic patients with abnormalities in melatonin secretion and circadian rhythm disorders of the sleep-wake cycle[63]. Another example of genetic abnormality associated with β-cell dysfunction and the risk of T2DM involves the ADRA2A gene that encodes for the alpha 2A-adrenergic receptor, which mediates the adrenergic suppression of insulin secretion[60]. Diabetic patients with polymorphisms of the ADRA2A gene may have overexpression of the alpha 2A receptor, resulting in insulin secretion deficiency. In pancreatic islets obtained from diabetic patients carrying this variant, pharmacological treatment with alpha (2A)-AR antagonists rescued insulin secretion[64].
Recently, large scale GWAS meta-analyses and imputation-based GWAS studies have demonstrated that the ankyrin 1 gene, a gene encoding for a protein of the ankyrin family, is associated with T2DM in different ethnicities[26-28]. Ankyrin 1 is typically expressed in the erythrocytes and functions as an adaptor molecule between membrane and skeleton proteins. Interestingly, mutations of this gene are known to determine hereditary spherocytosis. How this protein can be implicated in T2DM is not yet understood; however, ankyrin 1 is also expressed in β-cells, where a cognate protein, ankyrin B, plays a role in regulating ATP sensitivity by interacting with the sulphonylurea receptor isoform SUR1.
Another recent study has identified new loci and variants in a large-scale gene-centric meta-analysis that included the SLC2A2 (solute carrier 2A2) gene[24]. This gene encodes the glucose transporter Glut2, which is expressed in pancreatic β-cells, liver and kidney, and functions as a glucose sensor to maintain glucose homeostasis. These findings support a previously postulated role of Glut2 in T2DM[65]. Also, variants of genes involved in the cell cycle, like the CDKN2A and CDKN2B (cyclin-dependent kinase inhibitor 2A and 2B) genes, have been associated with T2DM. Although not proved in humans, data from animal models support the idea that these genetic variants may affect β-cell mass later in life[66].
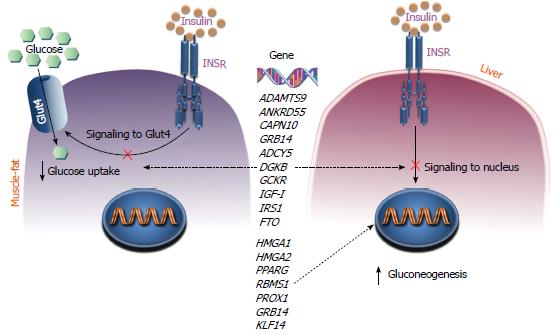
The first step in the mechanism of action of insulin is the interaction of the hormone with its specific receptor, the insulin receptor (INSR), on the cell surface of insulin responsive cells and tissues (Figure 3). The functional activation of INSR is a key moment in the pathophysiology of insulin action, followed by the selective activation of specific intracellular signaling pathways which are necessary for proper hormonal signal transduction. Although defects in INSR have been reported in a large number of patients with T2DM, mutations in the INSR gene have been found only in a small percentage (3%-4%) of these patients in whom genetic defects leading to receptor protein abnormalities were identified as cause of disease. However, certain patients with apparently normal INSR genes have reduced expression of both the INSR protein and INSR mRNA levels[13,18,19]. In these patients, it is possible that there are mutations in genes encoding trans-acting factors which regulate the level of INSR gene expression[40].
The mechanisms by which gene variants may impair insulin action in insulin target tissues are schematized in Figure 3. Among the genes involved in insulin resistance are those encoding for the glucokinase regulatory protein, GKRP, and the insulin-like growth factor-I, IGF-I. Genetic variants of these genes that predispose a person to develop insulin resistance have been recently identified by GWAS[25]. In addition, T2DM risk alleles at three loci (at FTO, KLF14 and PPARG) have been associated with higher fasting insulin (which is consistent with a primary defect on insulin action) and reduced insulin sensitivity[22]. In particular, variations in the fat mass and obesity-associated (FTO) gene appear to influence predisposition to T2DM through a positive effect on body mass index and obesity. Instead, the Krüppel-like factor 14 (KLF14) gene is considered a super gene with the ability to control other genes linked to body fat. The risk alleles at KLF14, along with those at peroxisome proliferator-activated receptor gamma (PPARG), appear to have a primary effect on insulin action which, unlike the alleles at FTO, is not driven by obesity[22].
A recently uncovered gene implicated in T2DM is the growth factor receptor-bound 14 (GRB14) gene[26,27], which codes for the Grb14 adaptor protein. Grb14 contains a C-terminal SH2 domain implicated in the interaction with a number of tyrosine kinase receptors and signaling proteins, and a domain called BPS (between pleckstrin homology), also required for binding to the INSR. This protein has been shown to specifically attenuate insulin action by inhibiting the catalytic activity of the INSR in insulin target tissues[67]. Many other recently identified diabetes-associated genes play still unknown roles in the pathophysiology of T2DM. Among them, the sterol regulatory element-binding transcription factor 1 (SREBF1) gene, which is involved in the transcriptional regulation of lipid homeostasis[24], and the high mobility group 20A (HMG20A) gene, which encodes a chromatin-associated protein and has previously been associated with a greater incidence of diabetes in obese subjects[26,27].
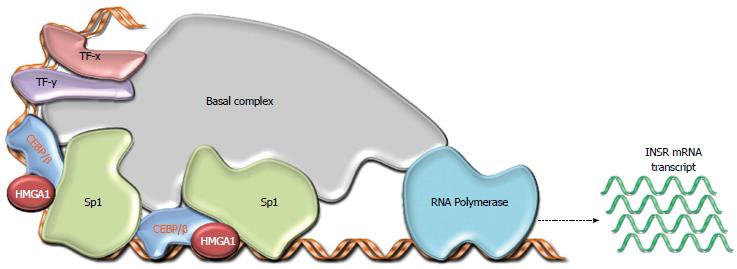
Among the group of genes recently associated with insulin resistance and T2DM is the HMGA1 gene, which encodes the architectural transcription factor, High Mobility Group A1 (HMGA1), a nonhistone basic protein that binds to AT-rich sequences of DNA via AT hooks, facilitating the assembly and stability of a multicomponent enhancer complex, the ‘‘enhanceosome’’, which drives gene transcription[68]. We previously found that HMGA1 is a key regulator of INSR gene expression[69-71] (Figure 4). Consistent with these findings, we identified two patients with insulin-resistant T2DM who had defects in HMGA1 expression and concomitant decreased INSR mRNA and protein in muscle, fat and circulating monocytes[72]. These individuals had normal INSR genes but had a novel genetic variant (c.*369del) in the 3’ noncoding region of the HMGA1 mRNA that contributed to the reduction of mRNA half-life and subsequent decline in HMGA1 expression. Epstein-Barr virus (EBV)-transformed lymphoblasts from these patients demonstrated defects in HMGA1 and INSR expression, indicating that the defects observed in vivo were not due to the altered metabolic state of the patients. In addition, the in vitro restoration of HMGA1 RNA and protein expression in these cells normalized INSR gene expression and restored both cell-surface INSR protein expression and insulin binding capacity[72]. The pathogenetic role of HMGA1 in T2DM was confirmed in genetically modified mice, in which the loss of HMGA1 expression (induced by disrupting the HMGA1 gene) considerably decreased INSR expression in the major target tissues of insulin action[72], thus supporting the concept that functional HMGA1 gene variants decrease INSR expression in human and mice.
In the context of these investigations, we later showed that four functional variants of the HMGA1 gene, leading to reduced INSR expression, were associated with insulin resistance and T2DM[40]. The most frequent functional HMGA1 variant, c.136-14_136-13insC (also designated rs146052672), was detected in 7%-8% of patients with diabetes in individuals of white European ancestry[40]. Analysis of cultured EBV-transformed lymphoblasts from patients with T2DM and the rs146052672 variant revealed that these cells had lower levels of HMGA1 and INSR protein than cells from either patients with wild-type T2DM or controls. Once again, in transformed lymphoblasts from the patients with the HMGA1 rs146052672 variant, restoration of HMGA1 protein expression by complementary DNA transfection (in the sense but not antisense direction) restored INSR protein expression and insulin binding to these cells[40]. Although not replicated in a heterogeneous French population[73], the HMGA1 rs146052672 variant was significantly associated with T2DM among Chinese[41] and Hispanic-American[38] individuals. Further evidence, implicating the HMGA1 locus as one conferring a high cross-race risk for the development of insulin resistant diseases, has been provided recently by showing that the HMGA1 rs146052672 variant significantly associates with the metabolic syndrome in Italian and Turkish individuals and predisposes these (and other) populations to the unfavorable anthropometric and metabolic traits of the metabolic syndrome[74,42].
Overall, these data are consistent with the impression that the association of HMGA1 gene variants with T2DM is accomplished through a pathogenetic mechanism related to peripheral insulin resistance. However, additional studies in vitro and in vivo, in normal and mutant mice, indicate that HMGA1, in addition to its role on INSR gene and protein expression, acts as a novel downstream target of the INSR signaling pathway[75], thus representing a critical nuclear mediator of insulin action and function. In this regard, evidence has been provided indicating that HMGA1 plays an essential role in the transcriptional regulation of a variety of insulin-target genes, such as the IGFBP-1 gene, as well as the gluconeogenic genes PEPCK and G6Pase[76], contributing to the transcriptional regulation of glucose homeostasis.
Significant advances have been made in recent years in relation to the pathogenesis of T2DM. This has significantly improved our knowledge of one of the most serious health threats in the world, allowing identification of genes and pathways involved in the development and progression of the disease. It has recently become possible to acquire molecular and genetic level information from an individual (i.e., DNA genotyping, gene expression, epigenomic profile, etc.). However, while such information is becoming increasingly available, how the identified genes and pathways impact on T2DM still remain largely unknown, due to the multifactorial nature of the disease. Understanding the pathogenesis of T2DM is necessary to enable the identification of prognostic and predictive biomarkers, as well as new therapeutic targets, which in turn should lead to improved outcomes in affected patients. Thus, once new therapeutic targets of interest are identified, it is necessary to develop molecules that can rescue function to disease-associated genes or pathways and conduct studies that provide new strategies for the treatment of T2DM.
T2DM is a heterogeneous disease with a strong genetic component and familial inheritance. Considerable effort has been made in the last decades to identify genes that may explain all the diabetic phenotypes. Currently, however, genetic studies on T2DM can explain only a small percentage of its heritability. Until now, the HMGA1 gene displays the strongest association with T2DM and its most frequent variant, rs146052672, confers the highest risk for human T2DM. Hence, from a strategic point of view, this finding suggests directing future research towards the identification of rare genetic variants with a stronger association, rather than common variants with a relatively small effect on the disease. It is evident that if a genetic variant confers a high susceptibility to T2DM it may become a useful biomarker to search for. For example, the genetic variants identified in the HMGA1 gene may represent a predictive marker for early detection of T2DM, especially in those individuals with a family history of the disease. Moreover, variants in the human HMGA1 gene may induce a different clinical course of disease compared to diabetic patients without the variant and may predict response to therapy, allowing identification of a priori patients who could most benefit from a specific pharmacological treatment[77]. Another important point in support of genetic studies in T2DM is the fact that they may integrate and improve our knowledge about the molecular mechanisms underpinning the pathophysiology of this disease.
P- Reviewers: Hssan M, Panchu P, Rosa C S- Editor: Qi Y L- Editor: Roemmele A E- Editor: Wu HL
| 1. | Danaei G, Finucane MM, Lu Y, Singh GM, Cowan MJ, Paciorek CJ, Lin JK, Farzadfar F, Khang YH, Stevens GA. National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2·7 million participants. Lancet. 2011;378:31-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2693] [Cited by in RCA: 2454] [Article Influence: 175.3] [Reference Citation Analysis (0)] |
| 2. | Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047-1053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9344] [Cited by in RCA: 8965] [Article Influence: 426.9] [Reference Citation Analysis (1)] |
| 3. | Krolewski AS, Warram JH, Freire MB. Epidemiology of late diabetic complications. A basis for the development and evaluation of preventive programs. Endocrinol Metab Clin North Am. 1996;25:217-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 74] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 4. | Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: pathogenesis and treatment. Lancet. 2008;371:2153-2156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 83] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 5. | Unger RH. Reinventing type 2 diabetes: pathogenesis, treatment, and prevention. JAMA. 2008;299:1185-1187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 6. | Groop L, Forsblom C, Lehtovirta M, Tuomi T, Karanko S, Nissén M, Ehrnström BO, Forsén B, Isomaa B, Snickars B. Metabolic consequences of a family history of NIDDM (the Botnia study): evidence for sex-specific parental effects. Diabetes. 1996;45:1585-1593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 197] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 7. | Weijnen CF, Rich SS, Meigs JB, Krolewski AS, Warram JH. Risk of diabetes in siblings of index cases with Type 2 diabetes: implications for genetic studies. Diabet Med. 2002;19:41-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 72] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 8. | Flegal KM, Ezzati TM, Harris MI, Haynes SG, Juarez RZ, Knowler WC, Perez-Stable EJ, Stern MP. Prevalence of diabetes in Mexican Americans, Cubans, and Puerto Ricans from the Hispanic Health and Nutrition Examination Survey, 1982-1984. Diabetes Care. 1991;14:628-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 193] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 9. | Neel JV. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet. 1962;14:353-362. [PubMed] |
| 10. | Hossain P, Kawar B, El Nahas M. Obesity and diabetes in the developing world--a growing challenge. N Engl J Med. 2007;356:213-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1382] [Cited by in RCA: 1354] [Article Influence: 75.2] [Reference Citation Analysis (0)] |
| 11. | Reaven GM. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes. 1988;37:1595-1607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2634] [Cited by in RCA: 2285] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 12. | Staiger H, Machicao F, Fritsche A, Häring HU. Pathomechanisms of type 2 diabetes genes. Endocr Rev. 2009;30:557-585. [PubMed] |
| 13. | Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, Berglund G, Altshuler D, Nilsson P, Groop L. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220-2232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 669] [Cited by in RCA: 626] [Article Influence: 36.8] [Reference Citation Analysis (1)] |
| 14. | Hebda JA, Miranker AD. The interplay of catalysis and toxicity by amyloid intermediates on lipid bilayers: insights from type II diabetes. Annu Rev Biophys. 2009;38:125-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 196] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 15. | Sciacca MF, Milardi D, Messina GM, Marletta G, Brender JR, Ramamoorthy A, La Rosa C. Cations as switches of amyloid-mediated membrane disruption mechanisms: calcium and IAPP. Biophys J. 2013;104:173-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 16. | Brender JR, Krishnamoorthy J, Messina GM, Deb A, Vivekanandan S, La Rosa C, Penner-Hahn JE, Ramamoorthy A. Zinc stabilization of prefibrillar oligomers of human islet amyloid polypeptide. Chem Commun (Camb). 2013;49:3339-3341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 17. | Audouze K, Brunak S, Grandjean P. A computational approach to chemical etiologies of diabetes. Sci Rep. 2013;3:2712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 18. | Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008;8:186-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 205] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 19. | Ahlqvist E, Ahluwalia TS, Groop L. Genetics of type 2 diabetes. Clin Chem. 2011;57:241-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 20. | Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PE. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163-175. [PubMed] |
| 21. | Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320-323. [PubMed] |
| 22. | Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, McCulloch LJ, Ferreira T, Grallert H, Amin N, Wu G, Willer CJ, Raychaudhuri S, McCarroll SA, Langenberg C, Hofmann OM, Dupuis J, Qi L, Segrè AV, van Hoek M, Navarro P, Ardlie K, Balkau B, Benediktsson R, Bennett AJ, Blagieva R, Boerwinkle E, Bonnycastle LL, Bengtsson Boström K, Bravenboer B, Bumpstead S, Burtt NP, Charpentier G, Chines PS, Cornelis M, Couper DJ, Crawford G, Doney AS, Elliott KS, Elliott AL, Erdos MR, Fox CS, Franklin CS, Ganser M, Gieger C, Grarup N, Green T, Griffin S, Groves CJ, Guiducci C, Hadjadj S, Hassanali N, Herder C, Isomaa B, Jackson AU, Johnson PR, Jørgensen T, Kao WH, Klopp N, Kong A, Kraft P, Kuusisto J, Lauritzen T, Li M, Lieverse A, Lindgren CM, Lyssenko V, Marre M, Meitinger T, Midthjell K, Morken MA, Narisu N, Nilsson P, Owen KR, Payne F, Perry JR, Petersen AK, Platou C, Proença C, Prokopenko I, Rathmann W, Rayner NW, Robertson NR, Rocheleau G, Roden M, Sampson MJ, Saxena R, Shields BM, Shrader P, Sigurdsson G, Sparsø T, Strassburger K, Stringham HM, Sun Q, Swift AJ, Thorand B, Tichet J, Tuomi T, van Dam RM, van Haeften TW, van Herpt T, van Vliet-Ostaptchouk JV, Walters GB, Weedon MN, Wijmenga C, Witteman J, Bergman RN, Cauchi S, Collins FS, Gloyn AL, Gyllensten U, Hansen T, Hide WA, Hitman GA, Hofman A, Hunter DJ, Hveem K, Laakso M, Mohlke KL, Morris AD, Palmer CN, Pramstaller PP, Rudan I, Sijbrands E, Stein LD, Tuomilehto J, Uitterlinden A, Walker M, Wareham NJ, Watanabe RM, Abecasis GR, Boehm BO, Campbell H, Daly MJ, Hattersley AT, Hu FB, Meigs JB, Pankow JS, Pedersen O, Wichmann HE, Barroso I, Florez JC, Frayling TM, Groop L, Sladek R, Thorsteinsdottir U, Wilson JF, Illig T, Froguel P, van Duijn CM, Stefansson K, Altshuler D, Boehnke M, McCarthy MI; MAGIC investigators; GIANT Consortium. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1472] [Cited by in RCA: 1385] [Article Influence: 92.3] [Reference Citation Analysis (0)] |
| 23. | Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Boström KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jørgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjögren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ; Wellcome Trust Case Control Consortium, Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1460] [Cited by in RCA: 1395] [Article Influence: 82.1] [Reference Citation Analysis (0)] |
| 24. | Saxena R, Elbers CC, Guo Y, Peter I, Gaunt TR, Mega JL, Lanktree MB, Tare A, Castillo BA, Li YR, Johnson T, Bruinenberg M, Gilbert-Diamond D, Rajagopalan R, Voight BF, Balasubramanyam A, Barnard J, Bauer F, Baumert J, Bhangale T, Böhm BO, Braund PS, Burton PR, Chandrupatla HR, Clarke R, Cooper-DeHoff RM, Crook ED, Davey-Smith G, Day IN, de Boer A, de Groot MC, Drenos F, Ferguson J, Fox CS, Furlong CE, Gibson Q, Gieger C, Gilhuijs-Pederson LA, Glessner JT, Goel A, Gong Y, Grant SF, Grobbee DE, Hastie C, Humphries SE, Kim CE, Kivimaki M, Kleber M, Meisinger C, Kumari M, Langaee TY, Lawlor DA, Li M, Lobmeyer MT, Maitland-van der Zee AH, Meijs MF, Molony CM, Morrow DA, Murugesan G, Musani SK, Nelson CP, Newhouse SJ, O’Connell JR, Padmanabhan S, Palmen J, Patel SR, Pepine CJ, Pettinger M, Price TS, Rafelt S, Ranchalis J, Rasheed A, Rosenthal E, Ruczinski I, Shah S, Shen H, Silbernagel G, Smith EN, Spijkerman AW, Stanton A, Steffes MW, Thorand B, Trip M, van der Harst P, van der A DL, van Iperen EP, van Setten J, van Vliet-Ostaptchouk JV, Verweij N, Wolffenbuttel BH, Young T, Zafarmand MH, Zmuda JM; Look AHEAD Research Group; DIAGRAM consortium, Boehnke M, Altshuler D, McCarthy M, Kao WH, Pankow JS, Cappola TP, Sever P, Poulter N, Caulfield M, Dominiczak A, Shields DC, Bhatt DL, Zhang L, Curtis SP, Danesh J, Casas JP, van der Schouw YT, Onland-Moret NC, Doevendans PA, Dorn GW 2nd, Farrall M, FitzGerald GA, Hamsten A, Hegele R, Hingorani AD, Hofker MH, Huggins GS, Illig T, Jarvik GP, Johnson JA, Klungel OH, Knowler WC, Koenig W, März W, Meigs JB, Melander O, Munroe PB, Mitchell BD, Bielinski SJ, Rader DJ, Reilly MP, Rich SS, Rotter JI, Saleheen D, Samani NJ, Schadt EE, Shuldiner AR, Silverstein R, Kottke-Marchant K, Talmud PJ, Watkins H, Asselbergs FW, de Bakker PI, McCaffery J, Wijmenga C, Sabatine MS, Wilson JG, Reiner A, Bowden DW, Hakonarson H, Siscovick DS, Keating BJ. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet. 2012;90:410-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 200] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 25. | Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Mägi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparsø T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proença C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Böttcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jørgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martínez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orrù M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvänen AC, Tanaka T, Thorand B, Tichet J, Tönjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Ríos M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators, Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1722] [Cited by in RCA: 1721] [Article Influence: 114.7] [Reference Citation Analysis (0)] |
| 26. | Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Müller-Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJ, Vedantam S, Chen H, Florez JC, Fox C, Liu CT, Rybin D, Couper DJ, Kao WH, Li M, Cornelis MC, Kraft P, Sun Q, van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JR, Platou CG, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stančáková A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, Crenshaw AT, Doney AS, Dorkhan M, Edkins S, Emilsson V, Eury E, Forsen T, Gertow K, Gigante B, Grant GB, Groves CJ, Guiducci C, Herder C, Hreidarsson AB, Hui J, James A, Jonsson A, Rathmann W, Klopp N, Kravic J, Krjutškov K, Langford C, Leander K, Lindholm E, Lobbens S, Männistö S, Mirza G, Mühleisen TW, Musk B, Parkin M, Rallidis L, Saramies J, Sennblad B, Shah S, Sigurðsson G, Silveira A, Steinbach G, Thorand B, Trakalo J, Veglia F, Wennauer R, Winckler W, Zabaneh D, Campbell H, van Duijn C, Uitterlinden AG, Hofman A, Sijbrands E, Abecasis GR, Owen KR, Zeggini E, Trip MD, Forouhi NG, Syvänen AC, Eriksson JG, Peltonen L, Nöthen MM, Balkau B, Palmer CN, Lyssenko V, Tuomi T, Isomaa B, Hunter DJ, Qi L; Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium, Shuldiner AR, Roden M, Barroso I, Wilsgaard T, Beilby J, Hovingh K, Price JF, Wilson JF, Rauramaa R, Lakka TA, Lind L, Dedoussis G, Njølstad I, Pedersen NL, Khaw KT, Wareham NJ, Keinanen-Kiukaanniemi SM, Saaristo TE, Korpi-Hyövälti E, Saltevo J, Laakso M, Kuusisto J, Metspalu A, Collins FS, Mohlke KL, Bergman RN, Tuomilehto J, Boehm BO, Gieger C, Hveem K, Cauchi S, Froguel P, Baldassarre D, Tremoli E, Humphries SE, Saleheen D, Danesh J, Ingelsson E, Ripatti S, Salomaa V, Erbel R, Jöckel KH, Moebus S, Peters A, Illig T, de Faire U, Hamsten A, Morris AD, Donnelly PJ, Frayling TM, Hattersley AT, Boerwinkle E, Melander O, Kathiresan S, Nilsson PM, Deloukas P, Thorsteinsdottir U, Groop LC, Stefansson K, Hu F, Pankow JS, Dupuis J, Meigs JB, Altshuler D, Boehnke M, McCarthy MI; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981-990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1706] [Cited by in RCA: 1477] [Article Influence: 113.6] [Reference Citation Analysis (0)] |
| 27. | Harder MN, Ribel-Madsen R, Justesen JM, Sparsø T, Andersson EA, Grarup N, Jørgensen T, Linneberg A, Hansen T, Pedersen O. Type 2 diabetes risk alleles near BCAR1 and in ANK1 associate with decreased β-cell function whereas risk alleles near ANKRD55 and GRB14 associate with decreased insulin sensitivity in the Danish Inter99 cohort. J Clin Endocrinol Metab. 2013;98:E801-E806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 28. | Imamura M, Maeda S, Yamauchi T, Hara K, Yasuda K, Morizono T, Takahashi A, Horikoshi M, Nakamura M, Fujita H. A single-nucleotide polymorphism in ANK1 is associated with susceptibility to type 2 diabetes in Japanese populations. Hum Mol Genet. 2012;21:3042-3049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 29. | Perry JR, Voight BF, Yengo L, Amin N, Dupuis J, Ganser M, Grallert H, Navarro P, Li M, Qi L. Stratifying type 2 diabetes cases by BMI identifies genetic risk variants in LAMA1 and enrichment for risk variants in lean compared to obese cases. PLoS Genet. 2012;8:e1002741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 168] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 30. | Weedon MN, Schwarz PE, Horikawa Y, Iwasaki N, Illig T, Holle R, Rathmann W, Selisko T, Schulze J, Owen KR. Meta-analysis and a large association study confirm a role for calpain-10 variation in type 2 diabetes susceptibility. Am J Hum Genet. 2003;73:1208-1212. [PubMed] |
| 31. | Jensen DP, Urhammer SA, Eiberg H, Borch-Johnsen K, Jørgensen T, Hansen T, Pedersen O. Variation in CAPN10 in relation to type 2 diabetes, obesity and quantitative metabolic traits: studies in 6018 whites. Mol Genet Metab. 2006;89:360-367. [PubMed] |
| 32. | Tsuchiya T, Schwarz PE, Bosque-Plata LD, Geoffrey Hayes M, Dina C, Froguel P, Wayne Towers G, Fischer S, Temelkova-Kurktschiev T, Rietzsch H. Association of the calpain-10 gene with type 2 diabetes in Europeans: results of pooled and meta-analyses. Mol Genet Metab. 2006;89:174-184. [PubMed] |
| 33. | Song Y, Niu T, Manson JE, Kwiatkowski DJ, Liu S. Are variants in the CAPN10 gene related to risk of type 2 diabetes? A quantitative assessment of population and family-based association studies. Am J Hum Genet. 2004;74:208-222. [PubMed] |
| 34. | Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331-1336. [PubMed] |
| 35. | Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341-1345. [PubMed] |
| 36. | Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770-775. [PubMed] |
| 37. | Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336-1341. [PubMed] |
| 38. | Rong Y, Bao W, Rong S, Fang M, Wang D, Yao P, Hu FB, Liu L. Hemochromatosis gene (HFE) polymorphisms and risk of type 2 diabetes mellitus: a meta-analysis. Am J Epidemiol. 2012;176:461-472. [PubMed] |
| 39. | Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881-885. [PubMed] |
| 40. | Chiefari E, Tanyolaç S, Paonessa F, Pullinger CR, Capula C, Iiritano S, Mazza T, Forlin M, Fusco A, Durlach V. Functional variants of the HMGA1 gene and type 2 diabetes mellitus. JAMA. 2011;305:903-912. [PubMed] |
| 41. | Liu L, Ding H, Wang HR, Xu YJ, Cui GL, Wang PH, Yuan G, Yu XF, Wang DW. Polymorphism of HMGA1 is associated with increased risk of type 2 diabetes among Chinese individuals. Diabetologia. 2012;55:1685-1688. [PubMed] |
| 42. | Kröger H, Donner I, Skiello G. Influence of a new virostatic compound on the induction of enzymes in rat liver. Arzneimittelforschung. 1975;25:1426-1429. [PubMed] |
| 43. | Almind K, Bjørbaek C, Vestergaard H, Hansen T, Echwald S, Pedersen O. Aminoacid polymorphisms of insulin receptor substrate-1 in non-insulin-dependent diabetes mellitus. Lancet. 1993;342:828-832. [PubMed] |
| 44. | Hani EH, Boutin P, Durand E, Inoue H, Permutt MA, Velho G, Froguel P. Missense mutations in the pancreatic islet beta cell inwardly rectifying K+ channel gene (KIR6.2/BIR): a meta-analysis suggests a role in the polygenic basis of Type II diabetes mellitus in Caucasians. Diabetologia. 1998;41:1511-1515. [PubMed] |
| 45. | Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K, Jørgensen T. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40:1098-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 531] [Cited by in RCA: 531] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 46. | Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40:1092-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 559] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 47. | Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spégel P, Bugliani M, Saxena R, Fex M, Pulizzi N. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41:82-88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 608] [Cited by in RCA: 547] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 48. | Bouatia-Naji N, Bonnefond A, Cavalcanti-Proença C, Sparsø T, Holmkvist J, Marchand M, Delplanque J, Lobbens S, Rocheleau G, Durand E. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41:89-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 498] [Cited by in RCA: 445] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 49. | Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 618] [Cited by in RCA: 570] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 50. | Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Mykkänen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20:284-287. [PubMed] |
| 51. | Tsai FJ, Yang CF, Chen CC, Chuang LM, Lu CH, Chang CT, Wang TY, Chen RH, Shiu CF, Liu YM. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet. 2010;6:e1000847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 267] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 52. | Qi L, Cornelis MC, Kraft P, Stanya KJ, Linda Kao WH, Pankow JS, Dupuis J, Florez JC, Fox CS, Paré G. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum Mol Genet. 2010;19:2706-2715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 148] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 53. | Kong A, Steinthorsdottir V, Masson G, Thorleifsson G, Sulem P, Besenbacher S, Jonasdottir A, Sigurdsson A, Kristinsson KT, Jonasdottir A. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 454] [Cited by in RCA: 423] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 54. | Minton JA, Hattersley AT, Owen K, McCarthy MI, Walker M, Latif F, Barrett T, Frayling TM. Association studies of genetic variation in the WFS1 gene and type 2 diabetes in U.K. populations. Diabetes. 2002;51:1287-1290. [PubMed] |
| 55. | Sandhu MS, Weedon MN, Fawcett KA, Wasson J, Debenham SL, Daly A, Lango H, Frayling TM, Neumann RJ, Sherva R. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet. 2007;39:951-953. [PubMed] |
| 56. | Liou CW, Chen JB, Tiao MM, Weng SW, Huang TL, Chuang JH, Chen SD, Chuang YC, Lee WC, Lin TK. Mitochondrial DNA coding and control region variants as genetic risk factors for type 2 diabetes. Diabetes. 2012;61:2642-2651. [PubMed] |
| 57. | Ye Z, Gillson C, Sims M, Khaw KT, Plotka M, Poulton J, Langenberg C, Wareham NJ. The association of the mitochondrial DNA OriB variant (16184-16193 polycytosine tract) with type 2 diabetes in Europid populations. Diabetologia. 2013;56:1907-1913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 58. | McCarthy MI, Zeggini E. Genome-wide association studies in type 2 diabetes. Curr Diab Rep. 2009;9:164-171. [PubMed] |
| 59. | Imamura M, Maeda S. Genetics of type 2 diabetes: the GWAS era and future perspectives [Review]. Endocr J. 2011;58:723-739. [PubMed] |
| 60. | Ingelsson E, Langenberg C, Hivert MF, Prokopenko I, Lyssenko V, Dupuis J, Mägi R, Sharp S, Jackson AU, Assimes TL. Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes. 2010;59:1266-1275. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 224] [Cited by in RCA: 203] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 61. | Sesti G, Marini MA, Cardellini M, Sciacqua A, Frontoni S, Andreozzi F, Irace C, Lauro D, Gnasso A, Federici M. The Arg972 variant in insulin receptor substrate-1 is associated with an increased risk of secondary failure to sulfonylurea in patients with type 2 diabetes. Diabetes Care. 2004;27:1394-1398. [PubMed] |
| 62. | Villareal DT, Robertson H, Bell GI, Patterson BW, Tran H, Wice B, Polonsky KS. TCF7L2 variant rs7903146 affects the risk of type 2 diabetes by modulating incretin action. Diabetes. 2010;59:479-485. [PubMed] |
| 63. | Mulder H, Nagorny CL, Lyssenko V, Groop L. Melatonin receptors in pancreatic islets: good morning to a novel type 2 diabetes gene. Diabetologia. 2009;52:1240-1249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 107] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 64. | Rosengren AH, Jokubka R, Tojjar D, Granhall C, Hansson O, Li DQ, Nagaraj V, Reinbothe TM, Tuncel J, Eliasson L. Overexpression of alpha2A-adrenergic receptors contributes to type 2 diabetes. Science. 2010;327:217-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 205] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 65. | Permutt MA, Koranyi L, Keller K, Lacy PE, Scharp DW, Mueckler M. Cloning and functional expression of a human pancreatic islet glucose-transporter cDNA. Proc Natl Acad Sci USA. 1989;86:8688-8692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 54] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 66. | McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med. 2010;363:2339-2350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 690] [Cited by in RCA: 625] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 67. | Béréziat V, Kasus-Jacobi A, Perdereau D, Cariou B, Girard J, Burnol AF. Inhibition of insulin receptor catalytic activity by the molecular adapter Grb14. J Biol Chem. 2002;277:4845-4852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 87] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 68. | Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 547] [Cited by in RCA: 577] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 69. | Brunetti A, Brunetti L, Foti D, Accili D, Goldfine ID. Human diabetes associated with defects in nuclear regulatory proteins for the insulin receptor gene. J Clin Invest. 1996;97:258-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 70. | Brunetti A, Manfioletti G, Chiefari E, Goldfine ID, Foti D. Transcriptional regulation of human insulin receptor gene by the high-mobility group protein HMGI(Y). FASEB J. 2001;15:492-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 77] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 71. | Foti D, Iuliano R, Chiefari E, Brunetti A. A nucleoprotein complex containing Sp1, C/EBP beta, and HMGI-Y controls human insulin receptor gene transcription. Mol Cell Biol. 2003;23:2720-2732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 107] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 72. | Foti D, Chiefari E, Fedele M, Iuliano R, Brunetti L, Paonessa F, Manfioletti G, Barbetti F, Brunetti A, Croce CM. Lack of the architectural factor HMGA1 causes insulin resistance and diabetes in humans and mice. Nat Med. 2005;11:765-773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 173] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 73. | Marquez M, Huyvaert M, Perry JR, Pearson RD, Falchi M, Morris AP, Vivequin S, Lobbens S, Yengo L, Gaget S. Low-frequency variants in HMGA1 are not associated with type 2 diabetes risk. Diabetes. 2012;61:524-530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 74. | Chiefari E, Tanyolaç S, Iiritano S, Sciacqua A, Capula C, Arcidiacono B, Nocera A, Possidente K, Baudi F, Ventura V. A polymorphism of HMGA1 is associated with increased risk of metabolic syndrome and related components. Sci Rep. 2013;3:1491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 75. | Chiefari E, Nevolo MT, Arcidiacono B, Maurizio E, Nocera A, Iiritano S, Sgarra R, Possidente K, Palmieri C, Paonessa F. HMGA1 is a novel downstream nuclear target of the insulin receptor signaling pathway. Sci Rep. 2012;2:251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 76. | Iiritano S, Chiefari E, Ventura V, Arcidiacono B, Possidente K, Nocera A, Nevolo MT, Fedele M, Greco A, Greco M. The HMGA1-IGF-I/IGFBP system: a novel pathway for modulating glucose uptake. Mol Endocrinol. 2012;26:1578-1589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 77. | Smith RJ, Nathan DM, Arslanian SA, Groop L, Rizza RA, Rotter JI. Individualizing therapies in type 2 diabetes mellitus based on patient characteristics: what we know and what we need to know. J Clin Endocrinol Metab. 2010;95:1566-1574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 76] [Article Influence: 5.1] [Reference Citation Analysis (0)] |