Published online Jul 15, 2025. doi: 10.4239/wjd.v16.i7.106218
Revised: April 8, 2025
Accepted: May 27, 2025
Published online: July 15, 2025
Processing time: 146 Days and 7.2 Hours
Diabetes mellitus (DM) comprises distinct subtypes-including type 1 DM, type 2 DM, and gestational DM - all characterized by chronic hyperglycemia and sub
Core Tip: Metabolomics offers key insights into the distinct metabolic pathways of each diabetes mellitus subtype. By integrating multiple omics layers-genomic, transcriptomic, proteomic, and microbiomic - researchers can refine disease classification, identify novel biomarkers, and develop personalized interventions, substantially enhancing the efficacy of diabetes management.
- Citation: Song CM, Lin TH, Huang HT, Yao JY. Illuminating diabetes via multi-omics: Unraveling disease mechanisms and advancing personalized therapy. World J Diabetes 2025; 16(7): 106218
- URL: https://www.wjgnet.com/1948-9358/full/v16/i7/106218.htm
- DOI: https://dx.doi.org/10.4239/wjd.v16.i7.106218
Diabetes mellitus (DM) is a common metabolic disorder typically categorized as type 1 DM (T1DM), type 2 DM (T2DM), or gestational DM (GDM) apart from various uncommon types such as monogenic diabetes, syndromic diabetes and secondary diabetes. T1DM frequently stems from autoimmune destruction of pancreatic β-cells[1-3], whereas T2DM is characterized by insulin resistance alongside varying degrees of β-cell dysfunction[4-9]. GDM, in turn, develops during pregnancy and adversely impacts both maternal and fetal health[10-16]. Despite their distinct etiologies, all DM subtypes share the feature - sustained hyperglycemia, culminating in micro-/macrovascular complications such as diabetic kidney disease (DKD) and diabetic retinopathy (DR)[17-21].
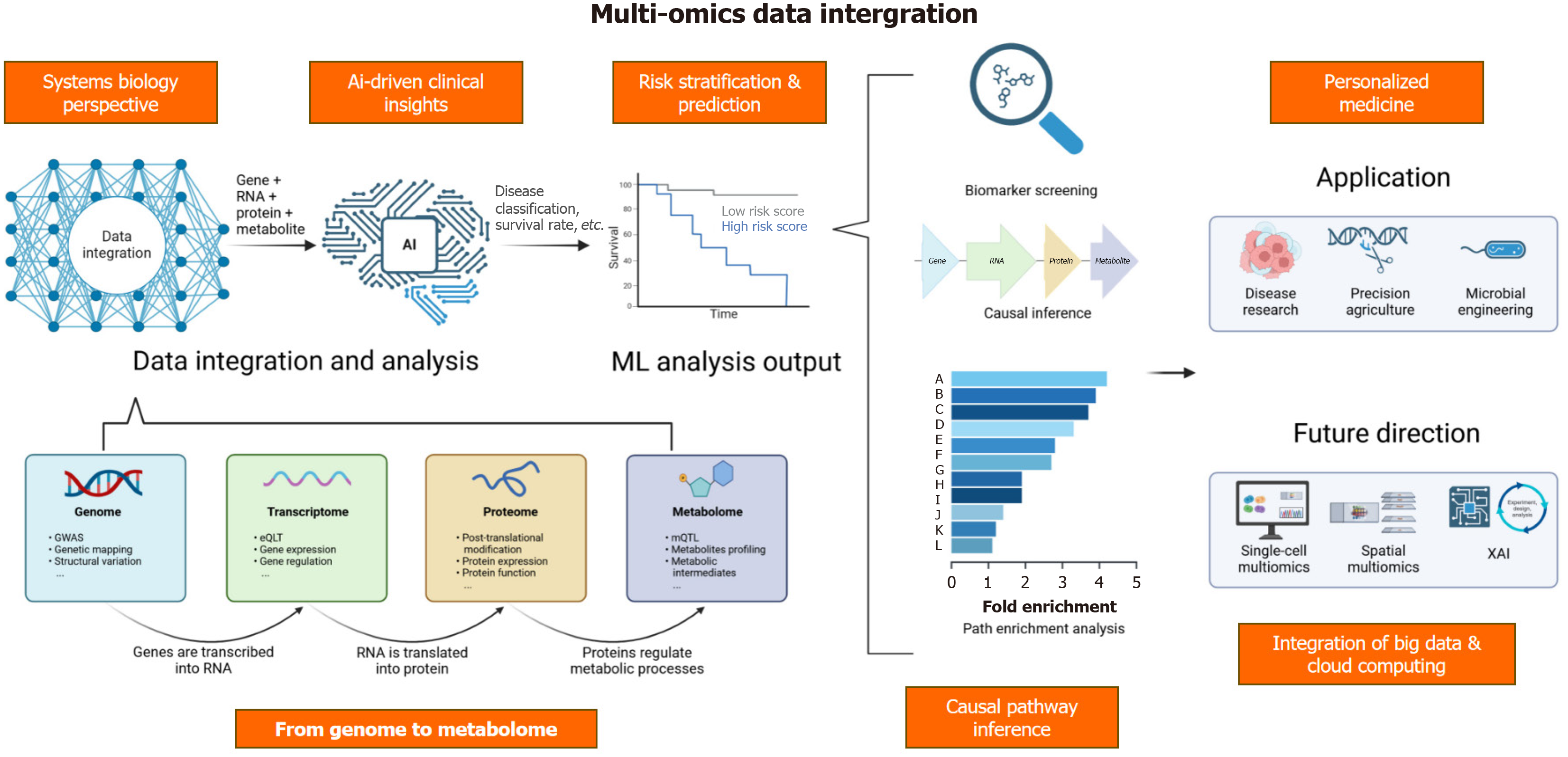
‘Which multi-omics findings are most clinically translatable for T1DM, T2DM, and GDM?’ Addressing this question demands clearer delineation among subtypes-T1DM is largely driven by autoimmunity, T2DM by metabolic overload and insulin resistance, and GDM by pregnancy-related hormonal imbalances. Metabolomics has been instrumental in uncovering each subtype’s metabolic disruption[5,10-15,22,23]. However, no single omics layer can fully capture the disease’s complexities. Consequently, multi-omics integration-encompassing genomics, transcriptomics, proteomics, and epigenomics - provides a more comprehensive lens through which gene-protein-metabolite-environment interactions are elucidated[1,4-9,24-29].
By combining multiple omics layers, researchers have greatly expanded our understanding of β-cell dysfunction, insulin resistance, and diabetic complications. For instance, coupling transcriptomics with metabolomics reveals how inflammatory pathways correlate with lipid metabolic shifts[30-38], while proteogenomic analyses highlight potential therapeutic targets in gluconeogenesis or glucose uptake[19,20,39-43]. Through these approaches, a refined under
We systematically searched PubMed and Web of Science for the period spanning January 2020 to December 2025. Our search strings included “(multi-omics OR genomics OR proteomics OR metabolomics OR microbiomics OR epigenomics) AND (diabetes OR T1DM OR T2DM OR GDM)”. We included original studies, reviews, and meta-analyses offering mechanistic or translational insights into the main DM subtypes. Studies focusing solely on non-diabetic metabolic disorders or lacking clear multi-omics integration were excluded. We prioritized large-cohort or consortium-based evidence as well as innovative single-cell and machine learning (ML)-based studies.
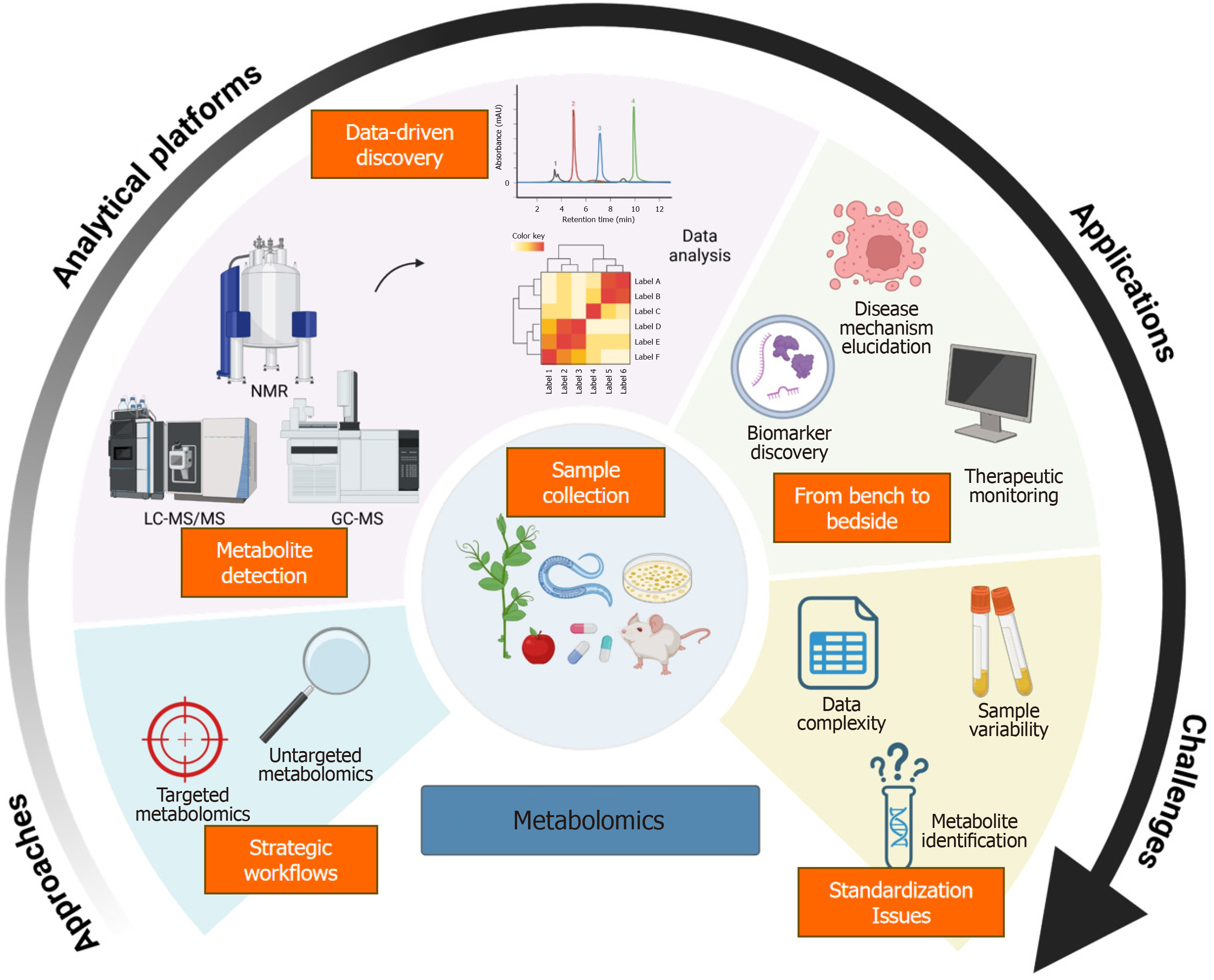
Metabolomics provides a critical view of small-molecule metabolites (e.g., amino acids, lipids, carbohydrates), clarifying the metabolic differences in T1DM, T2DM, GDM, and advanced complications like DKD or DR[5,7,10-12,14,16,18,26,43,51,52].
Autoimmune-mediated β-cell destruction and insulin deficiency yield specific pro-inflammatory metabolites[1-3].
T2DM is characterized by insulin resistance and elevated BCAAs or lipid imbalances[5,8,22,23,27-32,35-37,41,47,53-55], distinct from T1DM’s primary autoimmunity.
GDM results from pregnancy-related hormonal fluctuations that disrupt glucose/lipid homeostasis[10-16]. Metabolomic profiles often reveal dysregulated bile acids and lipids, potentially allowing earlier diagnosis.
Although chronic hyperglycemia predisposes all DM subtypes to renal or retinal damage, multi-omics suggests that T1DM, T2DM, and GDM may follow subtype-specific pathways. Recent metabolomic studies document amino acids, lipids, and inflammatory markers tied to complication severity[17-20].
With liquid chromatography-mass spectrometry, gas chromatography-mass spectrometry, nuclear magnetic resonance, and rigorous computational pipelines, metabolomics offers fresh insights into hyperglycemia’s pathophysiology and pinpoints promising biomarkers or therapeutic targets[4,6,9,21,24-26,33,34,42-46,48,50,52,56,57]. By capturing each pa
While metabolomics uncovers vital biochemical shifts, multi-omics integration extends to genomics, transcriptomics, proteomics, and microbiomics[58-61]. Genomics unearths hereditary predispositions; transcriptomics highlights gene-expression changes induced by hyperglycemia or insulin resistance[1-3,7-9,26-28]. Proteomics analyzes post-translational modifications, enzyme expression, and critical signaling events[5,6,19,20,24,25,29,43,52].
Microbiomics, focusing on 16S rRNA sequencing or shotgun metagenomics, further reveals how gut dysbiosis may exacerbate insulin resistance, particularly in T2DM and GDM[62-64]. Integrating these layers offers a holistic perspective on how DM progresses from normoglycemia to overt hyperglycemia.
For example, genome-wide association studies identify DM susceptibility loci, while transcriptomics verify gene-expression alterations in islet, muscle, or liver tissues[4,8,27]. Proteomics further confirm protein-level changes relevant to insulin pathways[19,20,24]. Together with metabolomics, these integrated data illuminate regulators of gluconeogenesis, adipogenesis, and inflammation (Figure 2)[65-67].
Table 1 outlines representative multi-omics investigations in DM. Beyond familiar markers such as BCAAs and certain lipid profiles, integrated omics efforts uncover new signatures distinguishing DM subtypes or correlating with disease severity. For instance, elevated glycerophospholipids correlate with insulin resistance in T2DM, whereas atypical amino acid pathways may foreshadow DKD or cognitive decline[21,30,32,38,45,47,54,55].
| Dm subtype/focus | Omics approach(s) | Primary objective/key findings | Ref. |
| T1DM (human cohorts) | Genomics (HLA region); metabolomics (LC-MS); epigenomics (RRBS) | Investigate autoimmune-driven β-cell destruction and identify early T1DM biomarkers | [1-3] |
| T2DM (human & animal) | Transcriptomics (RNA-Seq); proteomics (LC-MS/MS); metabolomics (NMR) | Elucidate insulin resistance, islet dysfunction; discover novel therapeutic targets | [8,9] |
| GDM (human studies) | Metabolomics (GC-MS); microbiomics (16S rRNA) | Uncover pregnancy-specific metabolite and microbiome signatures predictive of GDM | [10,12-15] |
| DKD/DR complications | Proteomics (targeted); metabolomics (untargeted); ML-based integration | Correlate inflammatory and fibrotic biomarkers with organ damage; early detection in both T1DM and T2DM | [17-20] |
| GI complications | Multi-omics synergy (transcript + metabolite) | Reveal changes in gut motility, microbiome composition, disease progression in T2DM | [25,36,42] |
| Maternal hyperglycemia | Metabolomics (LC-MS); lipidomics | Assess how hyperglycemia in pregnant sows influences neonatal hepatic metabolism | [43] |
| Microbiome in T1DM/ T2DM/GDM | Microbiomics (16S rRNA, shotgun metagenomics); Metabolomics (SCFAs) | Examine gut dysbiosis related to insulin resistance, inflammation, and distinct metabolic phenotypes | [62-64] |
Mechanistically, these analyses link genotypes or gene-expression changes to imbalances in lipid and glucose metabolism, clarifying how genetic risk factors translate into clinical phenotypes. Proteomics corroborates the enzyme-level or post-translational modifications that shape metabolic flux. In T1DM, multi-omics shows how autoimmune-associated gene variants converge with metabolic dysfunction, identifying potential early interventions[1-3]. Meanwhile, GDM-focused metabolomics surveys serum and placental samples to detect abnormal energy substrates predictive of future diabetic complications[10,12-15]. Multi-omics approaches also spotlight pathophysiological networks underlying DR, diabetic cardiomyopathy (DCM), and microvascular damage, enabling more precise patient stratification and potential disease-modifying strategies[24,25,36,52,54].
By bridging molecular perturbations and clinical endpoints, multi-omics surpasses single-marker paradigms. These findings (Table 1) further support biomarker discovery and highlight new therapeutic levers for DM detection, risk stratification, and treatment.
Single-cell multi-omics deciphers the cellular heterogeneity masked by bulk analyses[68-70]. In DR or DCM, single-cell profiling reveals specific endothelial or immune cell subsets that drive tissue injury[71,72]. Such detailed views may identify novel therapeutic avenues, including immunomodulation in T1DM or targeted interventions for T2DM-related cardiovascular complications.
ML excels at integrating and analyzing large-scale multi-omics datasets[73,74]. Techniques ranging from graph neural networks to autoencoders improve biomarker identification and DM classification accuracy[75,76]. However, model optimization varies across T1DM, T2DM, and GDM due to their distinct disease drivers. Collaborations among data-sharing platforms (e.g., MetaboLights) facilitate reproducibility and drive multi-omics biomarkers toward clinical implementation[77-79].
Multi-omics research is shifting from conceptual breakthroughs to real-world clinical utility[80,81]. By jointly examining genomic, transcriptomic, and metabolic data in patient samples, clinicians can more accurately identify subphenotypes beyond basic metrics (e.g., body mass index, fasting glucose)[77-79,82-87]. In T2DM, multi-omics biomarkers can flag individuals at elevated risk for nephropathy or cardiomyopathy, enabling earlier, more focused interventions[21,24,27,29,43-46,48,53,88,89]. Rare metabolic disorders such as maple syrup urine disease also benefit from multi-omics, which locates hidden epigenetic or transcriptomic anomalies[90,91]. The same logic applies to autoimmune diseases and cancers, where integrated cell-free DNA methylation and fragmentation analyses provide noninvasive diagnostic options with higher specificity[77-79,83,85,92-94].
ML-based clinical decision tools can now predict drug responses or therapy resistance by merging transcriptomic, metabolomic, and proteomic data[32,34,57,95]. In non-alcoholic fatty liver disease/metabolic dysfunction-associated fatty liver disease and hepatocellular carcinoma, multi-omics uncovers asymptomatic molecular changes and druggable pathways[86,88,89]. Collectively, these advances highlight how multi-omics can tailor interventions across DM subtypes, immunomodulation in T1DM, metabolic-targeting strategies in T2DM, or fetal-protective regimens in GDM (Table 2).
| Focus/theme | Key multi-omics finding | Potential clinical application |
| T1DM: Early autoimmune risk assessment | Genomics/epigenomics highlight HLA variants & methylation changes; Metabolomics reveals pro-inflammatory signatures | Identifying high-risk individuals for early intervention[1-3] |
| T2DM: Insulin resistance & metabolic overload | Elevated BCAAs/lipids from metabolomics [5,8,23,30-38]; Transcriptomics pinpoints insulin signaling defects [24,25] | Improved patient stratification; Tailored dietary or pharmacological interventions targeting dysregulated pathways |
| GDM: Pregnancy-specific biomarkers & interventions | Metabolomics + microbiomics [10,12-15] reveal distinct lipid/bile acid profiles, gut flora changes | Early screening of at-risk mothers; Nutritional or probiotic therapies to minimize fetal impact |
| Diabetic complications (DKD, DR) | Proteomics + metabolomics identify inflammation/fibrosis [17-20]; Single-cell multi-omics links endothelial dysfunction to hyperglycemia | Risk stratification for DKD, DR; Earlier monitoring and targeted therapy |
Despite significant progress, implementing multi-omics in routine practice faces several hurdles:
Analyzing heterogeneous datasets (DNA, epigenetics, RNA, proteins, metabolites) demands expert bioinformatics, secure data management, and specialized hardware[83,95,96]. Large or rare cohorts produce massive, high-dimensional data requiring advanced ML algorithms[82,83,95].
Discrepancies in sample collection, instrumentation, and preprocessing hamper reproducibility. Although attempts to harmonize data exist, cross-study consistency remains a concern[89,91,97,98].
Generating multi-omics data for sizable cohorts is expensive and time-intensive, especially for acute or rare diseases[84,87,97]. Specialized instrumentation and expertise further restrict broader adoption.
Findings from omics-based analyses demand laboratory confirmation (e.g., CRISPR, siRNA)[1,20,43,90]. In DR studies, single-cell transcriptomics plus metabolomics may identify key endothelial subsets behind vascular dysfunction, but robust in vitro/in vivo validation is crucial[24,25,36,52,54].
Applicability of this review is limited by the intrinsic heterogeneity of multi-omics studies, which vary in methodology, cohort size, and data standardization. Moreover, routine multi-omics adoption encounters barriers involving cost, infrastructure, and ethical concerns around large-scale data sharing. Future research should focus on establishing unified protocols, recruiting diverse populations, and leveraging emerging single-cell/spatial omics and artificial intelligence (AI) frameworks for maximal clinical impact.
Accessing large-scale patient omics data typically requires stringent institutional review board approvals and robust informed consent. Varying privacy regulations across institutions or countries can complicate data harmonization, underscoring the need for standardized ethical frameworks and global data-sharing strategies.
AI offers powerful analytics but also poses risks such as unauthorized access or commercialization of sensitive patient data. Transparent governance and robust cybersecurity measures are paramount to protect patient confidentiality.
Many multi-omics databases underrepresent certain ethnic or socioeconomic groups, leading to potential biases in disease diagnosis, risk prediction, and treatment. Incorporating diversity in cohort recruitment and cross-institutional collaborations can help minimize disparities and enhance the real-world relevance of multi-omics findings.
Although multi-omics results are largely consistent across diverse data layers, variations in patient ethnicity, sample size constraints, and diverging analytical pipelines introduce inconsistencies. Validating these findings demands large-scale longitudinal studies that explore causality and reduce confounders. Major open questions include how epigenetic modifications differ among T1DM, T2DM, and GDM at single-cell resolution; which multi-omics signatures best predict complications like DCM; and whether cost-effective ML pipelines can be deployed for widespread clinical screening.
By harmonizing genomics, transcriptomics, proteomics, and metabolomics, multi-omics has substantially reshaped our view of DM’s complexity. Compared to single-omics, multi-omics supplies a more granular depiction of disease heterogeneity, enabling earlier biomarker discovery and improved therapeutic targeting-ranging from T2DM-specific in
Even with these advancements, fully embedding multi-omics into diabetes care mandates sustained interdisciplinary collaboration, standardized methodologies, and powerful computational tools. Data-sharing consortia, open-science initiatives, and single-cell/spatial omics innovations promise to illuminate cellular heterogeneity, especially in diabetic complications. Overcoming these challenges can usher in a new era of diabetes management-one anchored by me
As multi-omics evolves, it yields unparalleled opportunities for transforming diabetes management into a predictive, preventive, and patient-centered framework. With concerted efforts in biomarker development, therapeutic innovation, and translational science, the vision of personalized diabetes treatment moves ever closer to reality.
By uniting genomics, transcriptomics, proteomics, and microbiomics, multi-omics research has revolutionized our understanding of DM’s pathophysiology and reshaped its clinical management. In the near term, AI-enhanced pipelines that integrate multi-omics data are poised to improve early detection and enable precision therapies. Establishing standardized protocols and more diverse patient cohorts will be essential to ensure a seamless transition of these breakthroughs from the laboratory to everyday diabetes care. With sustained progress in biomarker discovery, the
| 1. | Song C, Zheng W, Song C, Zhou H, Yao J. Protective Effects of Food-Derived Kaempferol on Pancreatic β-Cells in Type 1 Diabetes Mellitus. Foods. 2024;13:3797. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 2. | Alcazar O, Hernandez LF, Nakayasu ES, Nicora CD, Ansong C, Muehlbauer MJ, Bain JR, Myer CJ, Bhattacharya SK, Buchwald P, Abdulreda MH. Parallel Multi-Omics in High-Risk Subjects for the Identification of Integrated Biomarker Signatures of Type 1 Diabetes. Biomolecules. 2021;11:383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 3. | Bonnell J, Alcazar O, Watts B, Buchwald P, Abdulreda MH, Ogihara M. Supervised Parametric Learning in the Identification of Composite Biomarker Signatures of Type 1 Diabetes in Integrated Parallel Multi-Omics Datasets. Biomedicines. 2024;12:492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 4. | Zhang F, Chen X, Yang M, Shen X, Wang Y, Zhong D, Zeng F, Jin R. Metabolic impairments associated with type 2 diabetes mellitus and the potential effects of exercise therapy: An exploratory randomized trial based on untargeted metabolomics. PLoS One. 2024;19:e0300593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 5. | Wang YX, Pi JC, Yao YF, Peng XP, Li WJ, Xie MY. Hypoglycemic effects of white hyacinth bean polysaccharide on type 2 diabetes mellitus rats involvement with entero-insular axis and GLP-1 via metabolomics study. Int J Biol Macromol. 2024;281:136489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 6. | Tobias DK, Hamaya R, Clish CB, Liang L, Deik A, Dennis C, Bullock K, Zhang C, Hu FB, Manson JE. Type 2 diabetes metabolomics score and risk of progression to type 2 diabetes among women with a history of gestational diabetes mellitus. Diabetes Metab Res Rev. 2024;40:e3763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 7. | Wu G, Zhao J, Zhao J, Song N, Zheng N, Zeng Y, Yao T, Zhang J, Weng J, Yuan M, Zhou H, Shen X, Li H, Zhang W. Exploring biological basis of Syndrome differentiation in coronary heart disease patients with two distinct Syndromes by integrated multi-omics and network pharmacology strategy. Chin Med. 2021;16:109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 8. | Wigger L, Barovic M, Brunner AD, Marzetta F, Schöniger E, Mehl F, Kipke N, Friedland D, Burdet F, Kessler C, Lesche M, Thorens B, Bonifacio E, Legido-Quigley C, Barbier Saint Hilaire P, Delerive P, Dahl A, Klose C, Gerl MJ, Simons K, Aust D, Weitz J, Distler M, Schulte AM, Mann M, Ibberson M, Solimena M. Multi-omics profiling of living human pancreatic islet donors reveals heterogeneous beta cell trajectories towards type 2 diabetes. Nat Metab. 2021;3:1017-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 95] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 9. | Chen D, Zhao X, Sui Z, Niu H, Chen L, Hu C, Xuan Q, Hou X, Zhang R, Zhou L, Li Y, Yuan H, Zhang Y, Wu J, Zhang L, Wu R, Piao HL, Xu G, Jia W. A multi-omics investigation of the molecular characteristics and classification of six metabolic syndrome relevant diseases. Theranostics. 2020;10:2029-2046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 10. | Zhou J, Yu J, Ren J, Ren Y, Zeng Y, Wu Y, Zhang Q, Xiao X. Association of maternal blood metabolomics and gestational diabetes mellitus risk: a systematic review and meta-analysis. Rev Endocr Metab Disord. 2025;26:205-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 11. | Yao D, Shen C, Zhang X, Tang J, Yu J, Tu M, Panpipat W, Chaijan M, Zhang H, Xu X, Liu Y, Cheong LZ. Untargeted metabolomics study of mature human milk from women with and without gestational diabetes mellitus. Food Chem. 2024;460:140663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 12. | Wen J, Liu Q, Geng S, Shi X, Wang J, Yao X, Hu L. Impact of imidacloprid exposure on gestational hyperglycemia: A multi-omics analysis. Ecotoxicol Environ Saf. 2024;280:116561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 13. | Jiang Z, Ye X, Cao D, Xiang Y, Li Z. Association of Placental Tissue Metabolite Levels with Gestational Diabetes Mellitus: a Metabolomics Study. Reprod Sci. 2024;31:569-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Cai X, Pan S, Li M, Lu P, Guo X, Zheng S. Non-Targeted Metabolomics Analysis of Mother and Infant in Gestational Diabetes Mellitus and Neonatal Clinical Characterization. Clin Lab. 2024;70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 15. | Aleidi SM, Al Fahmawi H, AlMalki RH, Al Mogren M, Alwahsh M, Mujammami M, Costanzo M, Abdel Rahman A. Untargeted metabolomics profiling of gestational diabetes mellitus: insights into early diagnosis and metabolic pathway alterations. Front Mol Biosci. 2024;11:1485587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 16. | Luo SS, Zou KX, Zhu H, Cheng Y, Yan YS, Sheng JZ, Huang HF, Ding GL. Integrated Multi-Omics Analysis Reveals the Effect of Maternal Gestational Diabetes on Fetal Mouse Hippocampi. Front Cell Dev Biol. 2022;10:748862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 17. | Xu K, Zhang L, Wang T, Yu T, Zhao X, Yu N, Zhang Y. Investigating the mechanism of supraspinatus tendinopathy induced by type 2 diabetes mellitus in rats using untargeted metabolomics analysis. BMC Musculoskelet Disord. 2024;25:920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 18. | Mo C, Zhao J, Liang J, Chen Y, Wang H, Dai Y, Huang G. Effects of Zhuang medicine compound Xiancao Granule on diabetic kidney disease: A multi-omics analysis. J Ethnopharmacol. 2024;321:117517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 19. | Li Y, Wang L, Zhang J, Xu B, Zhan H. Integrated multi-omics and bioinformatic methods to reveal the mechanisms of sinomenine against diabetic nephropathy. BMC Complement Med Ther. 2023;23:287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 20. | Xu Z, Xiang X, Su S, Zhu Y, Yan H, Guo S, Guo J, Shang EX, Qian D, Duan JA. Multi-omics analysis reveals the pathogenesis of db/db mice diabetic kidney disease and the treatment mechanisms of multi-bioactive compounds combination from Salvia miltiorrhiza. Front Pharmacol. 2022;13:987668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 21. | Lucio-Gutiérrez JR, Cordero-Pérez P, Ávila-Velázquez JL, Torres-González L, Farías-Navarro IC, Govea-Torres G, Sánchez-Martínez C, García-Hernández PA, Coello-Bonilla J, Pérez-Trujillo M, Parella T, Waksman-Minsky NH, Saucedo AL. Targeted and untargeted serum NMR metabolomics to reveal initial kidney disease in diabetes mellitus. J Pharm Biomed Anal. 2024;247:116240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 22. | Park S, Kim EK. Machine Learning-Based Plasma Metabolomics in Liraglutide-Treated Type 2 Diabetes Mellitus Patients and Diet-Induced Obese Mice. Metabolites. 2024;14:483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 23. | Benabdelkamel H, Sebaa R, AlMalki RH, Masood A, Alfadda AA, Abdel Rahman AM. Untargeted metabolomics reveals the impact of Liraglutide treatment on metabolome profiling and metabolic pathways in type-2 diabetes mellitus. Saudi Pharm J. 2024;32:102172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 24. | Zou J, Song Q, Shaw PC, Zuo Z. Dendrobium officinale regulate lipid metabolism in diabetic mouse liver via PPAR-RXR signaling pathway: Evidence from an integrated multi-omics analysis. Biomed Pharmacother. 2024;173:116395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 25. | Zhang Y, Zhang Y, Yin R, Fang X, Miao R, Guan H, Yao Y, Tian J. Multi-omics characterization of type 2 diabetes mellitus-induced gastroenteropathy in the db/db mouse model. Front Cell Dev Biol. 2024;12:1417255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 26. | Ballanti M, Antonetti L, Mavilio M, Casagrande V, Moscatelli A, Pietrucci D, Teofani A, Internò C, Cardellini M, Paoluzi O, Monteleone G, Lefebvre P, Staels B, Mingrone G, Menghini R, Federici M. Decreased circulating IPA levels identify subjects with metabolic comorbidities: A multi-omics study. Pharmacol Res. 2024;204:107207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 27. | Yousri NA, Albagha OME, Hunt SC. Integrated epigenome, whole genome sequence and metabolome analyses identify novel multi-omics pathways in type 2 diabetes: a Middle Eastern study. BMC Med. 2023;21:347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 28. | Lee H, Gao Y, Ko E, Lee J, Lee HK, Lee S, Choi M, Shin S, Park YH, Moon HB, Uppal K, Kim KT. Nonmonotonic response of type 2 diabetes by low concentration organochlorine pesticide mixture: Findings from multi-omics in zebrafish. J Hazard Mater. 2021;416:125956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 29. | Backman M, Flenkenthaler F, Blutke A, Dahlhoff M, Ländström E, Renner S, Philippou-Massier J, Krebs S, Rathkolb B, Prehn C, Grzybek M, Coskun Ü, Rothe M, Adamski J, de Angelis MH, Wanke R, Fröhlich T, Arnold GJ, Blum H, Wolf E. Multi-omics insights into functional alterations of the liver in insulin-deficient diabetes mellitus. Mol Metab. 2019;26:30-44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 30. | Luo T, Jiang X, Xu N, Zhao X, Xie X, Xia X, Bian X, Liu H. Risk factors and metabolomics of mild cognitive impairment in type 2 diabetes mellitus. Front Mol Biosci. 2024;11:1341290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 31. | Li S, Song Z, Fan C, Zhang W, Ma T, Li X, Zhang Q, Zhao M, Yu T, Li S. Potential of FGF21 in type 2 diabetes mellitus treatment based on untargeted metabolomics. Biochem Pharmacol. 2024;225:116306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 32. | Arslan AK, Yagin FH, Algarni A, Karaaslan E, Al-Hashem F, Ardigò LP. Enhancing type 2 diabetes mellitus prediction by integrating metabolomics and tree-based boosting approaches. Front Endocrinol (Lausanne). 2024;15:1444282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 33. | Okamura T, Hamaguchi M, Kobayashi G, Ichikawa T, Hasegawa Y, Miyoshi T, Senmaru T, Nakanishi N, Sasano R, Fukui M. A multi-omics approach to overeating and inactivity-induced muscle atrophy in db/db mice. J Cachexia Sarcopenia Muscle. 2024;15:2030-2045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 34. | Du NH, Sinturel F, Nowak N, Gosselin P, Saini C, Guessous I, Jornayvaz FR, Philippe J, Rey G, Dermitzakis ET, Zenobi R, Dibner C, Brown SA. Multi-omics correlates of insulin resistance and circadian parameters mapped directly from human serum. Eur J Neurosci. 2024;60:5487-5504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 35. | Qi J, Lv Y, Zhong NE, Han WQ, Gou QL, Sun CF. Multi-omics analysis identifies potential mechanisms by which high glucose accelerates macrophage foaming. Mol Cell Biochem. 2023;478:665-678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 36. | Guo H, Ding Q, Huang Y, Guo Z, Ding F, Zhang H, Zheng Z, Zhang X, Weng S. Multi-omics Analysis Reveals the Crucial Mediators of DJB in the Treatment of Type 2 Diabetes. Obes Surg. 2023;33:1676-1686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 37. | Al Bataineh MT, Künstner A, Dash NR, Alsafar HS, Ragab M, Schmelter F, Sina C, Busch H, Ibrahim SM. Uncovering the relationship between gut microbial dysbiosis, metabolomics, and dietary intake in type 2 diabetes mellitus and in healthy volunteers: a multi-omics analysis. Sci Rep. 2023;13:17943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 38. | Liu L, Liang YB, Liu XL, Wang HQ, Qi YF, Wang M, Chen BX, Zhou QB, Tong WX, Zhang Y. Untargeted metabolomics combined with pseudotargeted lipidomics revealed the metabolite profiles of blood-stasis syndrome in type 2 diabetes mellitus. Heliyon. 2024;10:e39554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 39. | Lv K, Ying H, Hu G, Hu J, Jian Q, Zhang F. Integrated multi-omics reveals the activated retinal microglia with intracellular metabolic reprogramming contributes to inflammation in STZ-induced early diabetic retinopathy. Front Immunol. 2022;13:942768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 40. | Kaur S, Kumari P, Singh G, Joshi N, Kaur T, Dhiman V, Singh G, Sachdeva N, Kumar D, Barnwal RP, Bhadada SK. Unveiling novel metabolic alterations in postmenopausal osteoporosis and type 2 diabetes mellitus through NMR-based metabolomics: A pioneering approach for identifying early diagnostic markers. J Proteomics. 2024;302:105200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 41. | Wang Y, Chen J, Ni Y, Liu Y, Gao X, Tse MA, Panagiotou G, Xu A. Exercise-changed gut mycobiome as a potential contributor to metabolic benefits in diabetes prevention: an integrative multi-omics study. Gut Microbes. 2024;16:2416928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 42. | Tong A, Li Z, Liu X, Ge X, Zhao R, Liu B, Zhao L, Zhao C. Laminaria japonica polysaccharide alleviates type 2 diabetes by regulating the microbiota-gut-liver axis: A multi-omics mechanistic analysis. Int J Biol Macromol. 2024;258:128853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 43. | Shashikadze B, Valla L, Lombardo SD, Prehn C, Haid M, Riols F, Stöckl JB, Elkhateib R, Renner S, Rathkolb B, Menche J, Hrabĕ de Angelis M, Wolf E, Kemter E, Fröhlich T. Maternal hyperglycemia induces alterations in hepatic amino acid, glucose and lipid metabolism of neonatal offspring: Multi-omics insights from a diabetic pig model. Mol Metab. 2023;75:101768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 44. | Liu Z, Ma Z, Jin L, Nizhamuding X, Zeng J, Zhang T, Zhang J, Wang J, Zhao H, Zhou W, Zhang C. Altered neopterin and IDO in kynurenine metabolism based on LC-MS/MS metabolomics study: Novel therapeutic checkpoints for type 2 diabetes mellitus. Clin Chim Acta. 2024;557:117859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 45. | Xiong RQ, Li YP, Lin LP, Yao JY. Identification of potential biomarkers for diabetic cardiomyopathy using LC-MS-based metabolomics. Endocr Connect. 2024;13:e230384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 46. | Li DK, Smith LE, Rookyard AW, Lingam SJ, Koay YC, McEwen HP, Twigg SM, Don AS, O'Sullivan JF, Cordwell SJ, White MY. Multi-omics of a pre-clinical model of diabetic cardiomyopathy reveals increased fatty acid supply impacts mitochondrial metabolic selectivity. J Mol Cell Cardiol. 2022;164:92-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 47. | Jiang L, Zhang J, Fang M, Qin Y, Huang Y, Tao R. Analysis of subgingival micro-organisms based on multi-omics and Treg/Th17 balance in type 2 diabetes with/without periodontitis. Front Microbiol. 2022;13:939608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 48. | Yang Z, Yang D, Tan F, Wong CW, Yang JY, Zhou D, Cai Z, Lin SH. Multi-Omics Comparison of the Spontaneous Diabetes Mellitus and Diet-Induced Prediabetic Macaque Models. Front Pharmacol. 2021;12:784231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 49. | Gallego-Paüls M, Hernández-Ferrer C, Bustamante M, Basagaña X, Barrera-Gómez J, Lau CE, Siskos AP, Vives-Usano M, Ruiz-Arenas C, Wright J, Slama R, Heude B, Casas M, Grazuleviciene R, Chatzi L, Borràs E, Sabidó E, Carracedo Á, Estivill X, Urquiza J, Coen M, Keun HC, González JR, Vrijheid M, Maitre L. Variability of multi-omics profiles in a population-based child cohort. BMC Med. 2021;19:166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 50. | Takahashi S, Saito K, Jia H, Kato H. An integrated multi-omics study revealed metabolic alterations underlying the effects of coffee consumption. PLoS One. 2014;9:e91134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 51. | Zhang F, Shan S, Fu C, Guo S, Liu C, Wang S. Advanced Mass Spectrometry-Based Biomarker Identification for Metabolomics of Diabetes Mellitus and Its Complications. Molecules. 2024;29:2530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 52. | Scisciola L, Chianese U, Caponigro V, Basilicata MG, Salviati E, Altucci L, Campiglia P, Paolisso G, Barbieri M, Benedetti R, Sommella E. Multi-omics analysis reveals attenuation of cellular stress by empagliflozin in high glucose-treated human cardiomyocytes. J Transl Med. 2023;21:662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 53. | Yuan F, Zhang T, Jia S, Zhao J, Wan B, Liu G. Fine mapping-based multi-omics analysis interprets the gut-lung axis function of SGLT2 inhibitors. Front Cell Infect Microbiol. 2024;14:1447327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 54. | Shi M, Zhao B, Cai W, Yuan H, Liang X, Li Z, Liu X, Jin Y, Liu X, Wei C. Multi-omics mechanical analysis of gut microbiota, carboxylic acids, and cardiac gene expression interaction triggering diabetic cardiomyopathy. mSystems. 2025;10:e0145024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 55. | Chen L, Chen B, Dai Y, Sun Q, Wu J, Zheng D, Vgontzas AN, Tang X, Li Y. The association of objective daytime sleepiness with impaired glucose metabolism in patients with obstructive sleep apnea: a multi-omics study. Sleep. 2025;48:zsae240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 56. | Huang Y, Liang M, Liao Y, Ji Z, Lin W, Pu X, Wang L, Wang W. Investigating the Mechanisms of 15-PGDH Inhibitor SW033291 in Improving Type 2 Diabetes Mellitus: Insights from Metabolomics and Transcriptomics. Metabolites. 2024;14:509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 57. | Ďásková N, Modos I, Krbcová M, Kuzma M, Pelantová H, Hradecký J, Heczková M, Bratová M, Videňská P, Šplíchalová P, Králová M, Heniková M, Potočková J, Ouřadová A, Landberg R, Kühn T, Cahová M, Gojda J. Multi-omics signatures in new-onset diabetes predict metabolic response to dietary inulin: findings from an observational study followed by an interventional trial. Nutr Diabetes. 2023;13:7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 58. | Wang S, Yong H, He XD. Multi-omics: Opportunities for research on mechanism of type 2 diabetes mellitus. World J Diabetes. 2021;12:1070-1080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 59. | Hu C, Jia W. Multi-omics profiling: the way towards precision medicine in metabolic diseases. J Mol Cell Biol. 2021;13:576-593. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 60. | Soares NP, Magalhaes GC, Mayrink PH, Verano-Braga T. Omics to Unveil Diabetes Mellitus Pathogenesis and Biomarkers: Focus on Proteomics, Lipidomics, and Metabolomics. Adv Exp Med Biol. 2024;1443:211-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 61. | Gutierrez Reyes CD, Alejo-Jacuinde G, Perez Sanchez B, Chavez Reyes J, Onigbinde S, Mogut D, Hernández-Jasso I, Calderón-Vallejo D, Quintanar JL, Mechref Y. Multi Omics Applications in Biological Systems. Curr Issues Mol Biol. 2024;46:5777-5793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 62. | Go D, Yeon GH, Park SJ, Lee Y, Koh HG, Koo H, Kim KH, Jin YS, Sung BH, Kim J. Integration of metabolomics and other omics: from microbes to microbiome. Appl Microbiol Biotechnol. 2024;108:538. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 63. | Zhang S, Cai Y, Meng C, Ding X, Huang J, Luo X, Cao Y, Gao F, Zou M. The role of the microbiome in diabetes mellitus. Diabetes Res Clin Pract. 2021;172:108645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 64. | Merkevičius K, Kundelis R, Maleckas A, Veličkienė D. Microbiome Changes after Type 2 Diabetes Treatment: A Systematic Review. Medicina (Kaunas). 2021;57:1084. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 65. | Sun YV, Liu C, Staimez L, Ali MK, Chang H, Kondal D, Patel S, Jones D, Mohan V, Tandon N, Prabhakaran D, Quyyumi AA, Narayan KMV, Agrawal A. Cardiovascular disease risk and pathophysiology in South Asians: can longitudinal multi-omics shed light? Wellcome Open Res. 2020;5:255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 66. | Tayanloo-Beik A, Roudsari PP, Rezaei-Tavirani M, Biglar M, Tabatabaei-Malazy O, Arjmand B, Larijani B. Diabetes and Heart Failure: Multi-Omics Approaches. Front Physiol. 2021;12:705424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 67. | Tiwari A, Rathor P, Trivedi PK, Ch R. Multi-Omics Reveal Interplay between Circadian Dysfunction and Type2 Diabetes. Biology (Basel). 2023;12:301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 68. | Wu X, Yang X, Dai Y, Zhao Z, Zhu J, Guo H, Yang R. Single-cell sequencing to multi-omics: technologies and applications. Biomark Res. 2024;12:110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 27] [Reference Citation Analysis (0)] |
| 69. | Zhao Z, D'Oliveira Albanus R, Taylor H, Tang X, Han Y, Orchard P, Varshney A, Zhang T, Manickam N, Erdos M, Narisu N, Taylor L, Saavedra X, Zhong A, Li B, Zhou T, Naji A, Liu C, Collins F, Parker SC, Chen S. An integrative single-cell multi-omics profiling of human pancreatic islets identifies T1D associated genes and regulatory signals. Res Sq. 2023;rs.3.rs-3343318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 70. | Fasolino M, Schwartz GW, Patil AR, Mongia A, Golson ML, Wang YJ, Morgan A, Liu C, Schug J, Liu J, Wu M, Traum D, Kondo A, May CL, Goldman N, Wang W, Feldman M, Moore JH, Japp AS, Betts MR; HPAP Consortium, Faryabi RB, Naji A, Kaestner KH, Vahedi G. Single-cell multi-omics analysis of human pancreatic islets reveals novel cellular states in type 1 diabetes. Nat Metab. 2022;4:284-299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 95] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 71. | Li X, Dong X, Zhang W, Shi Z, Liu Z, Sa Y, Li L, Ni N, Mei Y. Multi-omics in exploring the pathophysiology of diabetic retinopathy. Front Cell Dev Biol. 2024;12:1500474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 72. | Su Q, Huang W, Huang Y, Dai R, Chang C, Li QY, Liu H, Li Z, Zhao Y, Wu Q, Pan DG. Single-cell insights: pioneering an integrated atlas of chromatin accessibility and transcriptomic landscapes in diabetic cardiomyopathy. Cardiovasc Diabetol. 2024;23:139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Reference Citation Analysis (0)] |
| 73. | Burghelea D, Moisoiu T, Ivan C, Elec A, Munteanu A, Tabrea R, Antal O, Kacso TP, Socaciu C, Elec FI, Kacso IM. The use of metabolomics and machine learning algorithms to predict post-transplant diabetes mellitus in renal transplant patients on Tacrolimus therapy. Med Pharm Rep. 2024;97:467-476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 74. | Ballard JL, Wang Z, Li W, Shen L, Long Q. Deep learning-based approaches for multi-omics data integration and analysis. BioData Min. 2024;17:38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 75. | Shao X, Gao S, Bai P, Yang Q, Lin Y, Pang M, Wu W, Wang L, Li Y, Zhou S, Liu H, Yu P. Machine learning-based multi-omics models for diagnostic classification and risk stratification in diabetic kidney disease. Clin Transl Med. 2025;15:e70133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 76. | Ghebrehiwet I, Zaki N, Damseh R, Mohamad MS. Revolutionizing personalized medicine with generative AI: a systematic review. Artif Intell Rev. 2024;57:128. [DOI] [Full Text] |
| 77. | Zhang W, Ye B, Song Y, Yang P, Si W, Jing H, Yang F, Yuan D, Wu Z, Lyu J, Peng K, Zhang X, Wang L, Li Y, Liu Y, Wu C, Hao X, Zhang Y, Qi W, Wang J, Dong F, Zhao Z, Jing H, Li Y. Integrating multi-omics features enables non-invasive early diagnosis and treatment response prediction of diffuse large B-cell lymphoma. Clin Transl Med. 2025;15:e70174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 78. | Yang Y, Long H, Feng Y, Tian S, Chen H, Zhou P. A multi-omics method for breast cancer diagnosis based on metabolites in exhaled breath, ultrasound imaging, and basic clinical information. Heliyon. 2024;10:e32115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 79. | Rouzbahani AK, Khalili-Tanha G, Rajabloo Y, Khojasteh-Leylakoohi F, Garjan HS, Nazari E, Avan A. Machine learning algorithms and biomarkers identification for pancreatic cancer diagnosis using multi-omics data integration. Pathol Res Pract. 2024;263:155602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 80. | Ahmed Z. Practicing precision medicine with intelligently integrative clinical and multi-omics data analysis. Hum Genomics. 2020;14:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 81. | Anwardeen NR, Naja K, Elrayess MA. Advancements in precision medicine: multi-omics approach for tailored metformin treatment in type 2 diabetes. Front Pharmacol. 2024;15:1506767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 82. | Zhang Z, Huang J, Zhang Z, Shen H, Tang X, Wu D, Bao X, Xu G, Chen S. Application of omics in the diagnosis, prognosis, and treatment of acute myeloid leukemia. Biomark Res. 2024;12:60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 83. | Rashid MM, Hamano M, Iida M, Iwata M, Ko T, Nomura S, Komuro I, Yamanishi Y. Network-based identification of diagnosis-specific trans-omic biomarkers via integration of multiple omics data. Biosystems. 2024;236:105122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 84. | Lunke S, Bouffler SE, Patel CV, Sandaradura SA, Wilson M, Pinner J, Hunter MF, Barnett CP, Wallis M, Kamien B, Tan TY, Freckmann ML, Chong B, Phelan D, Francis D, Kassahn KS, Ha T, Gao S, Arts P, Jackson MR, Scott HS, Eggers S, Rowley S, Boggs K, Rakonjac A, Brett GR, de Silva MG, Springer A, Ward M, Stallard K, Simons C, Conway T, Halman A, Van Bergen NJ, Sikora T, Semcesen LN, Stroud DA, Compton AG, Thorburn DR, Bell KM, Sadedin S, North KN, Christodoulou J, Stark Z. Integrated multi-omics for rapid rare disease diagnosis on a national scale. Nat Med. 2023;29:1681-1691. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 74] [Reference Citation Analysis (0)] |
| 85. | Zhou Q, Xie Z, He L, Sun G, Meng H, Luo Z, Feng Y, Chu X, Li L, Zhang J, Hao Y, Geng M, Zhang X, Chen S. Multi-omics profiling reveals peripheral blood biomarkers of multiple sclerosis: implications for diagnosis and stratification. Front Pharmacol. 2024;15:1458046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 86. | Perakakis N, Stefanakis K, Mantzoros CS. The role of omics in the pathophysiology, diagnosis and treatment of non-alcoholic fatty liver disease. Metabolism. 2020;111S:154320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (1)] |
| 87. | Sinan ÜY, Unlu S. [The role of -omics technology in the pathophysiology, diagnosis, and treatment of cardiovascular diseases]. Turk Kardiyol Dern Ars. 2017;45:470-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 88. | Wu XY, Geng N, Chen QQ, Li J. [Application of omics in the diagnosis of metabolic dysfunction-associated fatty liver disease]. Zhonghua Gan Zang Bing Za Zhi. 2023;31:1245-1249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 89. | Chen F, Wang J, Wu Y, Gao Q, Zhang S. Potential Biomarkers for Liver Cancer Diagnosis Based on Multi-Omics Strategy. Front Oncol. 2022;12:822449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 90. | Tejedor JR, Soriano-Sexto A, Beccari L, Castejón-Fernández N, Correcher P, Sainz-Ledo L, Alba-Linares JJ, Urdinguio RG, Ugarte M, Fernández AF, Rodríguez-Pombo P, Fraga MF, Pérez B. Integration of multi-omics layers empowers precision diagnosis through unveiling pathogenic mechanisms on maple syrup urine disease. J Inherit Metab Dis. 2025;48:e12829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 91. | Wanders RJA, Vaz FM, Ferdinandusse S, van Kuilenburg ABP, Kemp S, van Karnebeek CD, Waterham HR, Houtkooper RH. Translational Metabolism: A multidisciplinary approach towards precision diagnosis of inborn errors of metabolism in the omics era. J Inherit Metab Dis. 2019;42:197-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 92. | Zhao G, Jiang R, Shi Y, Gao S, Wang D, Li Z, Zhou Y, Sun J, Wu W, Peng J, Kuang T, Rong Y, Yuan J, Zhu S, Jin G, Wang Y, Lou W. Circulating cell-free DNA methylation-based multi-omics analysis allows early diagnosis of pancreatic ductal adenocarcinoma. Mol Oncol. 2024;18:2801-2813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 93. | Zhang Z, Lan H, Zhao S. Analysis of the Value of Quantitative Features in Multimodal MRI Images to Construct a Radio-Omics Model for Breast Cancer Diagnosis. Breast Cancer (Dove Med Press). 2024;16:305-318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 94. | Sousa P, Silva L, Câmara JS, Guedes de Pinho P, Perestrelo R. Integrating OMICS-based platforms and analytical tools for diagnosis and management of pancreatic cancer: a review. Mol Omics. 2025;21:108-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 95. | Wu W, Wang S, Zhang Y, Yin W, Zhao Y, Pang S. MOSGAT: Uniting Specificity-Aware GATs and Cross Modal-Attention to Integrate Multi-Omics Data for Disease Diagnosis. IEEE J Biomed Health Inform. 2024;28:5624-5637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 96. | Xiao H, Wang J, Wan S. WIMOAD: Weighted Integration of Multi-Omics data for Alzheimer's Disease (AD) Diagnosis. bioRxiv. 2024;2024.09.25.614862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 97. | Aydin B, Caliskan A, Arga KY. Overview of omics biomarkers in pituitary neuroendocrine tumors to design future diagnosis and treatment strategies. EPMA J. 2021;12:383-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 98. | Usha Rani G, Kadali S, Kurma Reddy B, Shaheena D, Naushad SM. Application of machine learning tools and integrated OMICS for screening and diagnosis of inborn errors of metabolism. Metabolomics. 2023;19:49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 99. | Zhang J, Zhang N, Mai Q, Zhou C. The frontier of precision medicine: application of single-cell multi-omics in preimplantation genetic diagnosis. Brief Funct Genomics. 2024;23:726-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 100. | Samare-Najaf M, Razavinasab SA, Samareh A, Jamali N. Omics-based novel strategies in the diagnosis of endometriosis. Crit Rev Clin Lab Sci. 2024;61:205-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 10.0] [Reference Citation Analysis (0)] |