Published online Feb 15, 2024. doi: 10.4239/wjd.v15.i2.275
Peer-review started: October 2, 2023
First decision: November 9, 2023
Revised: November 22, 2023
Accepted: January 9, 2024
Article in press: January 9, 2024
Published online: February 15, 2024
Processing time: 124 Days and 19.2 Hours
Adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 (APPL1) plays a crucial role in regulating insulin signaling and glucose metabolism. Mutations in the APPL1 gene have been associated with the development of maturity-onset diabetes of the young type 14 (MODY14). Cur
To assess the pathogenicity of APPL1 gene mutations in diabetic patients and to characterize the functional role of the APPL1 domain.
Patients exhibiting clinical signs and a medical history suggestive of MODY were screened for the study. Whole exome sequencing was performed on the patients as well as their family members. The pathogenicity of the identified APPL1 variants was predicted on the basis of bioinformatics analysis. In addition, the pathogenicity of the novel APPL1 variant was preliminarily evaluated through in vitro functional experiments. Finally, the impact of these variants on APPL1 protein expression and the insulin pathway were assessed, and the potential mechanism underlying the interaction between the APPL1 protein and the insulin receptor was further explored.
A total of five novel mutations were identified, including four missense mutations (Asp632Tyr, Arg633His, Arg532Gln, and Ile642Met) and one intronic mutation (1153-16A>T). Pathogenicity prediction analysis revealed that the Arg532Gln was pathogenic across all predictions. The Asp632Tyr and Arg633His variants also had pathogenicity based on MutationTaster. In addition, multiple alignment of amino acid sequences showed that the Arg532Gln, Asp632Tyr, and Arg633His variants were conserved across different species. Moreover, in in vitro functional experiments, both the c.1894G>T (at Asp632Tyr) and c.1595G>A (at Arg532Gln) mutations were found to downregulate the expression of APPL1 on both protein and mRNA levels, indicating their pathogenic nature. Therefore, based on the patient’s clinical and family history, combined with the results from bioinformatics analysis and functional experiment, the c.1894G>T (at Asp632Tyr) and c.1595G>A (at Arg532Gln) mutations were classified as pathogenic mutations. Importantly, all these mutations were located within the phosphotyrosine-binding domain of APPL1, which plays a critical role in the insulin sensitization effect.
This study provided new insights into the pathogenicity of APPL1 gene mutations in diabetes and revealed a potential target for the diagnosis and treatment of the disease.
Core Tip: We identified five new mutations in the adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 (APPL1) gene, a critical regulator of insulin signaling and glucose metabolism, in maturity-onset diabetes of the young type 14 patients. We conducted bioinformatics and functional experiments and showed that two mutations were pathogenic, resulting in reduced expression of the APPL1 protein and mRNA. All mutations were in the phosphotyrosine-binding domain of APPL1, which is important for its insulin-sensitizing effect. This study gave new insights into the pathogenicity of APPL1 mutations in diabetes and revealed potential targets for improved diagnosis and treatment strategies.
- Citation: Shi P, Tian Y, Xu F, Liu LN, Wu WH, Shi YZ, Dai AQ, Fang HY, Li KX, Xu C. Assessment of pathogenicity and functional characterization of APPL1 gene mutations in diabetic patients. World J Diabetes 2024; 15(2): 275-286
- URL: https://www.wjgnet.com/1948-9358/full/v15/i2/275.htm
- DOI: https://dx.doi.org/10.4239/wjd.v15.i2.275
Maturity-onset diabetes of the young (MODY) is a rare form of hereditary monogenic diabetes caused by single gene mutations, constituting approximately 1%-2% of all diabetes cases[1,2]. A total of 14 MODY phenotypes have been identified, exhibiting significant heterogeneity in their clinical presentations. Notably, approximately 80% of MODY cases are initially misdiagnosed as either type 1 diabetes mellitus (T1DM) or T2DM[3,4].
MODY14, characterized by mutations in the adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 (APPL1) gene, represents one of the least known MODY subtypes. Currently, only two related mutations have been reported, namely [c.1655T>A (p.Leu552*) and c.281G>A p.(Asp94Asn)][5-7]. To date, our understanding of MODY14 remains limited. To enhance our comprehension of MODY14 and APPL1 mutations, it is crucial to identify additional cases, conduct comprehensive research, and consolidate knowledge in this field. By doing so, we can gain a deeper understanding of the underlying mechanisms and clinical implications of MODY14, ultimately paving the way for improved diagnostic and therapeutic strategies.
The APPL1 gene, situated on chromosome 3p14.3, has 23 exons[8]. It exhibits widespread expression in numerous human tissues, such as pancreas, liver, adipose tissue, brain, and muscle[9,10]. APPL1 serves as a multifunctional adaptor protein, playing an important role in distinct signal transduction and membrane trafficking pathways. Structurally, it contains three primary domains: A Bin-Amphiphysin-Rvs (BAR) domain; a pleckstrin homology (PH) domain; and a phosphotyrosine-binding (PTB) domain[11]. These domains facilitate interactions with various signaling molecules and receptors, thereby regulating intracellular signaling pathways. The BAR domain can recognize and deform membranes with curvature and regulate intracellular trafficking and vesicle formation[12]. The PH domain can bind to phosphoinositides, such as phosphatidylinositol-3,4,5-trisphosphate, and target APPL1 to the plasma membrane, where it participates in various signaling pathways[13]. Meanwhile, the PTB domain can interact with adiponectin receptors 1/2, tropomyosin receptor kinase A, and other molecules, mediating intracellular signal transduction[14-16].
Current studies indicate that APPL1 is an important mediator of insulin sensitization. APPL1 can facilitate the binding of insulin receptor (IR) substrates (IRS) to IR, thereby activating PI3K/Akt signaling pathway and augmenting insulin sensitivity[17]. Notably, in this process, the PTB domain can interact with IR and promote insulin signal transduction. In addition, APPL1 participates in adiponectin signaling by binding to adiponectin receptors, thereby enhancing lipid oxidation and glucose uptake[18,19]. In summary, further exploration of the interaction and regulatory network of APPL1 with other signaling molecules is warranted. Further, more clinical evidence is required to elucidate the precise role and underlying of APPL1 in diabetes and other metabolic diseases.
In this study, we identified five novel APPL1 mutations, including four missense mutations and one intron mutation. To enhance our understanding of MODY14, we performed bioinformatics analysis and in vitro experiments to characterize the functional impact of these mutations. Based on the experimental results and literature review, we discussed their implications for diagnosis, treatment, and molecular pathogenesis. Notably, this article was the first to report cases of MODY14 in Asia on an international scale. Moreover, our study has identified the largest number of APPL1 mutations, providing important data for APPL1 mutation research. By enriching the gene database of MODY14, our discoveries provide new insights into the molecular mechanism and clinical management of this rare diabetes, ultimately guiding optimal treatment strategies, prognosis predictions, and genetic counseling for affected families.
The purpose of this study was to determine the pathogenic status of suspected MODY diabetes in patients and evaluate the effects of novel APPL1 mutations on disease development. We performed whole-exome sequencing (WES) to identify patients carrying APPL1 gene mutations and conducted bioinformatics analysis of these mutations. Then, we conducted in vitro experiments to verify the pathogenicity of these mutations. Finally, the molecular mechanisms and signaling pathways involved in MODY pathogenesis were elucidated in this study.
This study adhered to the ethical principles outlined in the Declaration of Helsinki of 1964, along with its subsequent revisions and equivalent ethical standards. Prior to participation, informed consent was obtained from each participating patient or their legal guardian. The Ethics Committee of Shandong Provincial Hospital approved this study.
The study cohort consisted of 5 patients from five pedigrees. Patients meeting any of the following criteria were enrolled for WES: Younger than 30-years-old; had a family history of diabetes; or had negative insulin antibodies. Then, the clinical history and blood samples of patients were collected for further pathogenicity analysis.
To assess the potential pathogenicity of the identified mutations, MutationTaster (https://www.mutationtaster.org/), Poly Phen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml), Revel (https://sites.google.com/site/revelgeno
Genomic DNA was extracted from blood leucocytes from all study participants using Tiangen Biotech DNA kit. We performed WES on blood DNA and applied the SeqCap EZ MedExome Target Enrichment Kit (Roche NimbleGen) to capture human exons and adjacent introns after fragmenting, ligating, amplifying, and purifying genomic DNA. DNA sequencing was carried out using Illumina HiSeq platform, and the resulting data were aligned to the Hg19 reference genome. Mutation calls were made using NextGENe. The identified mutations were further verified by Sanger sequencing.
WT and mutant human APPL1 plasmids (transcript ID: NM_012096.3) were generated by the transient overexpression vector GV141 (GeneChem, China)[20]. HEK293 cells were transfected with the plasmids and cultured in complete medium supplemented with 10% fetal bovine serum (Excell, FSD500, South America), penicillin, and streptomycin. Cells were seeded in six-well plates once they reached 80%-90% confluence. Transfection was performed when the degree of cell fusion reached 70%-90%. We added 2 μg of corresponding plasmids to each well of the six-well plate and transfected them into HEK293 cells. The transfection operation was performed followed the instructions of the Lipofectamine 3000 (Invitrogen, American) transfection kit. To ensure optimal transfection efficiency, the process was carried out on a sterile bench (SW-CJ-IC dual person purification workbench).
After transfection, cells were collected after 24 h and lysed with Trizol (TaKaRa, Japan). Chloroform was added to separate RNA from DNA and proteins. The RNA was precipitated with isopropanol and washed several times with 75% ethanol. The RNA concentration was measured by nanodrop software after extraction. To convert mRNA into complementary DNA, reverse transcription was performed following the instructions of the reverse transcription kit manual (TaKaRa, Japan) using Mastercycler5333 PCR instrument (Eppendorf, Germany). Next, Bestar SybrGreen qPCR mastermix, PCR Forward Primer, PCR Reverse Primer, DNA template, and ddH2O were mixed well in a 96-well plate. Finally, qPCR was performed on a real-time fluorescence quantitative PCR instrument (Roche, United States).
RIPA and PMSF (Shanghai Shenneng Gaming Company, China) were mixed at a ratio of 99:1 in the six-well plate after 48 h of transfection. Lysis buffer was added to each well, and the cells were scraped with a cell scraper. The lysate was transferred to EP tubes and incubated on ice for 20 min. The lysate was centrifuged for 15 min to extract protein. The protein concentration was determined using an enzyme-linked immunosorbent assay. Then, loading buffer was added to the protein samples and boiled for 10 min. Proteins were separated with different molecular weights by electrophoresis on a 10% SDS-polyacrylamide gel. The membrane was transferred and blocked in 5% milk (skimmed milk powder purchased from Yili Group, China) for 1 h. Next, primary antibodies (Flag mouse anti 1:1000, β-actin mouse anti 1:7500) were added overnight at 4 °C. After recovering the primary antibodies, the membrane was washed with TBST for 10 min × 3 times. The secondary antibodies (mouse anti 1:5000) were added and incubated for 1 h. After washing the film, it was developed under the Alpha Fluorochem Q imaging analysis system (United States).
The experimental data was analyzed using SPSS software (Version 25.0). Measurement data were presented as mean ± SD and analyzed using an independent samples t-test. Statistical significance was defined as a P value < 0.05.
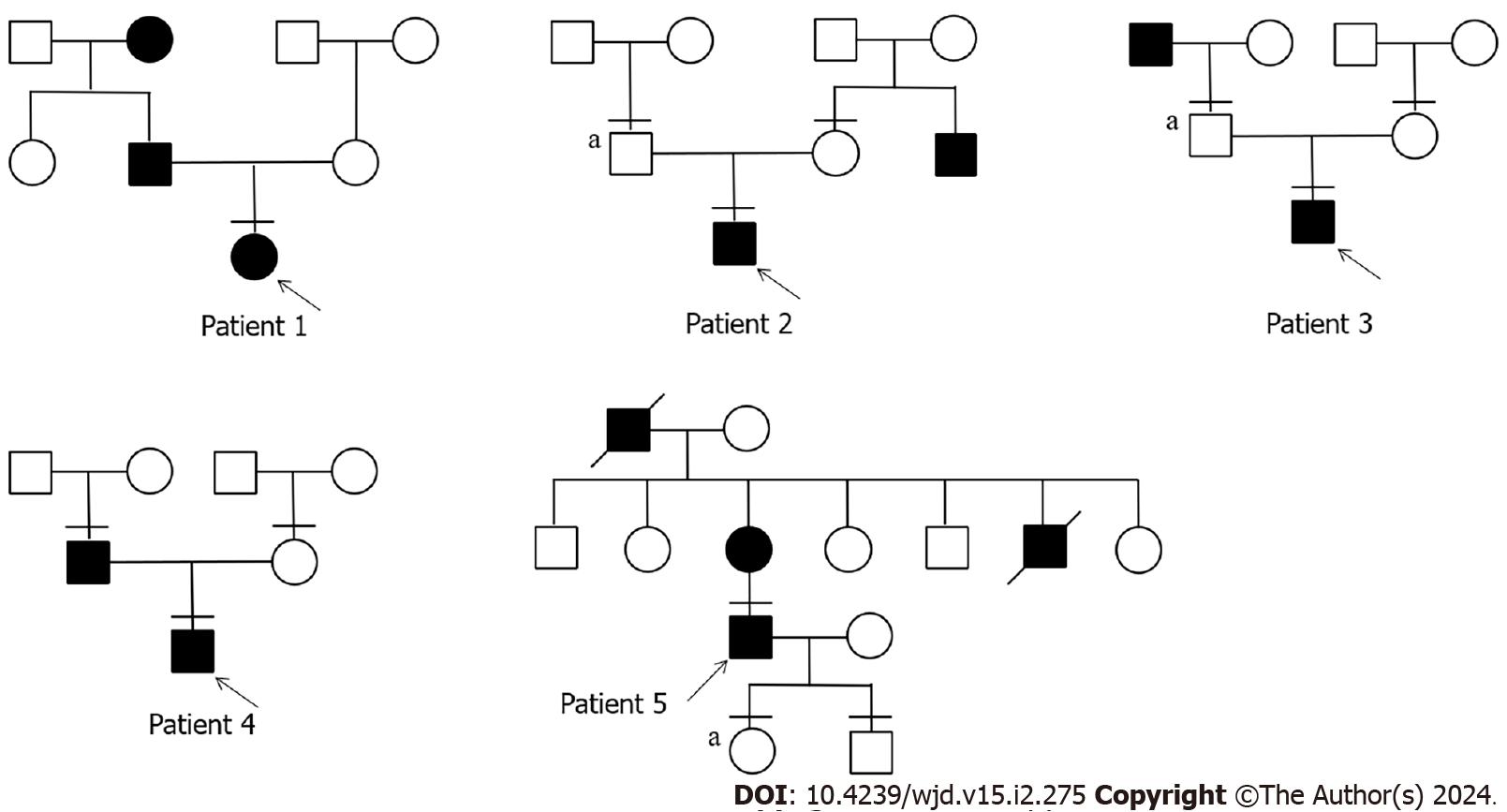
We studied 5 patients with APPL1 variants, four of whom had experienced elevated fasting blood glucose before the age of 25, while the fifth patient developed diabetes at the age of 39 (Table 1). Patient 1 was diagnosed the diabetes at the age of 13, with a family history of diabetes in both his grandmother and father (Figure 1). His grandmother continued taking oral hypoglycemic drugs, while his father managed diabetes through diet control, resulting in normalized blood glucose level. Patient 1 had obvious polyuria, polydipsia, polyphagia, and diabetic ketoacidosis at the onset of the disease. Insulin antibody testing yielded negative results. The patient used insulin therapy after diagnosis, and then his insulin autoantibodies and islet cell antibodies turned positive after 8 years of insulin therapy.
| Patient | APPL1 variant | Sex | Age of onset in yr | BMI in kg/m2 | HbA1C 4%-6% | FBG 4.4-6.1 mmol/L |
| 1 | c.1894G>T (p. Asp632Tyr) | Female | 13 | 14.98 | 11 | 8.25 |
| 2 | c.1898G>A (p. Arg633His) | Male | 21 | 36.63 | 6.9 | 8.3 |
| 3 | c.1595G>A (p. Arg532Gln) | Male | 13 | 20.62 | 14.2 | 11.69 |
| 4 | c.1926A>G (p. Ile642Met) | Male | 12 | 20.52 | 8.3 | 8.46 |
| 5 | c.1153-16A>T (p.?) | Male | 39 | 25.91 | 6.1 | 7.66 |
Patient 2 developed diabetes at the age of 21. WES of his family revealed that his father had no diabetes but also carried the same mutation. Only his uncle had diabetes in his family. Additionally, patient 2 presented with obesity and ketoacidosis at the onset of the disease. After the ketoacidosis subsided, the patient shifted to diet control.
Patient 3 developed diabetes at the age of 13 and had a diabetic grandfather. Although the patient’s father carried the same mutation, he remained unaffected by the disease. The patient also had ketoacidosis when he developed diabetes. He stopped taking medication after 2 mo of treatment with insulin combined with oral drugs and now only controls glucose by diet.
Patient 4 developed diabetes at the age of 12, and only his father had diabetes in his family. It is worth noting that the patient’s blood glucose reached 19.81 mmol/L 2 h after a meal, accompanied by hyperinsulinemia (insulin > 300.00 mU/L 2 h after a meal). The patient relied on oral medication at the onset of the disease and transitioned to glucose control through diet and increased exercise.
Patient 5 developed diabetes at the age of 39. Both his mother and grandfather had diabetes. Patient 5 had a son and a daughter. His daughter carried the variant but as of the writing of this article had not shown any symptoms of diabetes. Patient 5 had been taking oral hypoglycemic drugs since he was diagnosed with diabetes.
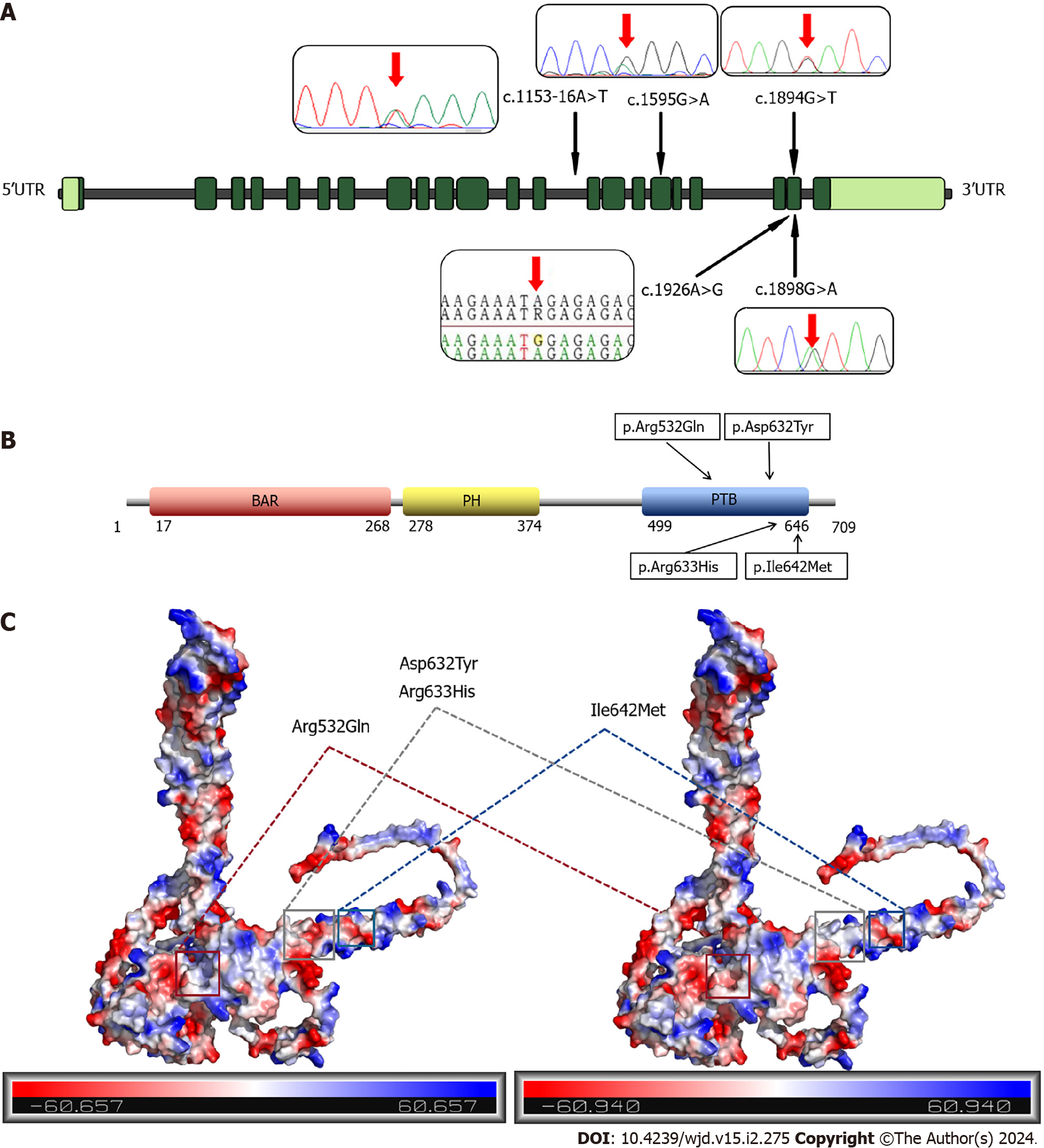
We identified five variants, of which Asp632Tyr, Arg633His, Arg532Gln and Ile642Met mutations are missense mutations, and 1153-16A>T is an intron mutation (Figure 2A). The Asp632Tyr, Arg633His, and Ile642Met variants are located in exon 21, while the Arg532Gln variant is located in exon 17. The 1153-16A>T intronic mutation is upstream of exon 14. These four missense mutations are located in the PTB domain of APPL1, which can bind to the IR and regulate the insulin signaling pathway (Figure 2B). The Arg532Gln, Asp632Tyr, and Arg633His variants all caused changes in the surface potential of APPL protein, while the Ile642Met variant had no obvious abnormality. Among them, the Arg532Gln and Arg633His variants resulted in a decrease in positive surface potential, while the Asp632Tyr variant led to the elimination of negative surface potential (Figure 2C). These changes in potential indicate that the mutation may disrupt the interaction of APPL1 with other macromolecules. Moreover, amino acid mutations can influence protein function and folding by altering hydrophilicity (Supplementary Figure 1).
To assess the pathogenicity of the four missense mutations, we employed MutationTaster, PolyPhen-2, and Revel for prediction analysis. Remarkably, all three software tools consistently predicted the Arg532Gln variant as pathogenic, with MutationTaster indicating a high likelihood of pathogenicity (Table 2). The three prediction outcomes for the Ile642Met variant were all benign. Moreover, the Asp632Tyr and Asp632Tyr variants were both pathogenic in MutationTaster and benign in PolyPhen-2 and Revel. Multiple alignments of amino acid sequences demonstrated that the residues Asp632Tyr, Arg633His, and Arg532Gln were conserved across various species. This implies that they may exert a detrimental impact on the structure and function of the protein, reinforcing their potential pathogenicity. However, we noticed that in multiple species, the amino acid at position 642 of APPL1 was not isoleucine, but methionine, as in our patients’ mutation, indicating that this site may not have a significant influence on the function or structure of the protein (Figure 3). Additionally, the pathogenicity analysis of the intronic mutations showed that the prediction results of MutationTaster and IntSplice were not pathogenic.
| Patient | APPL1 variant | MutationTaster pathogenicity and probability value | PolyPhen-2 pathogenicity and score | Revel pathogenicity and score |
| 1 | c.1894G>T (p. Asp632Tyr) | Disease causing (0.995) | Benign (0.285) | Benign (0.120) |
| 2 | c.1898G>A (p. Arg633His) | Disease causing (0.999) | Benign (0.052) | Benign (0.140) |
| 3 | c.1595G>A (p. Arg532Gln) | Disease causing (1.000) | Probably damaging (1.000) | Probably damaging (0.477) |
| 4 | c.1926A>G (p. Ile642Met) | Polymorphism (0.998) | Benign (0.007) | Benign (0.038) |
| 5 | c.1153-16A>T (p.?) | Polymorphism (1) | - | - |
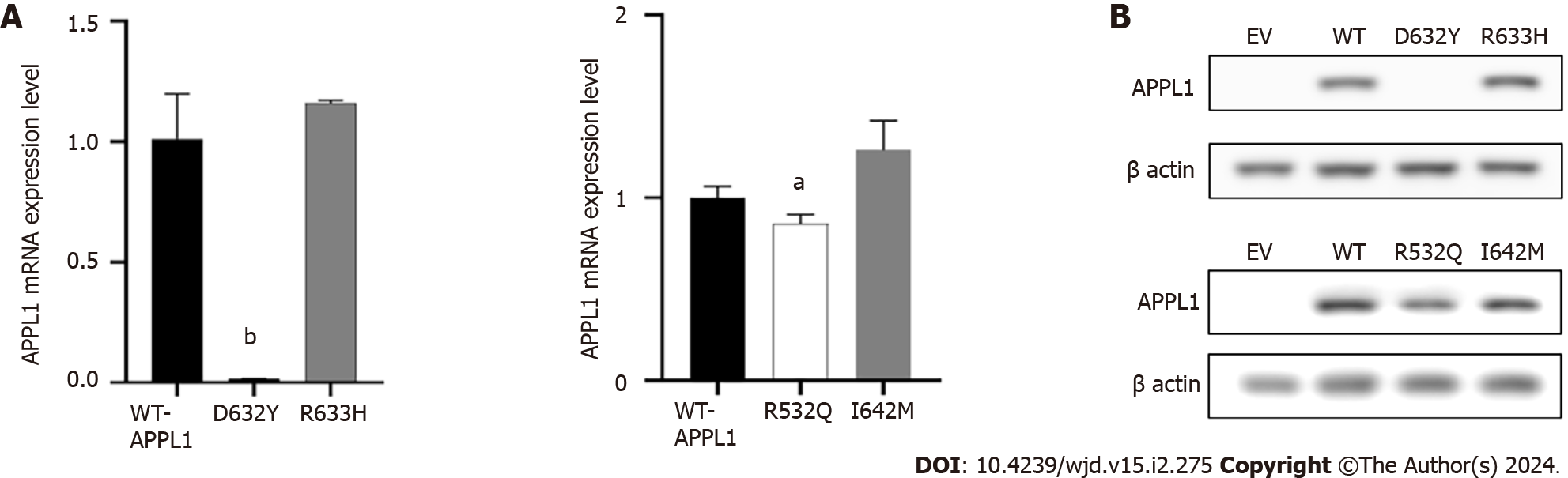
The experiment was used to confirm the pathogenicity of four missense mutations. As shown in Figure 4A, the mRNA expression level of APPL1 Asp632Tyr variant decreased by 98% (P = 0.001) compared with WT-APPL1, while the Arg532Gln variant decreased by 14% (P = 0.035), indicating that both mutations at these sites resulted in reduced expression of APPL1 mRNA. The Arg633His and Ile642Met variants did not cause significant changes in APPL1 mRNA expression. In the experiment, we also observed that the expression of mutant proteins was consistent with mRNA expression. Compared with WT APPL1, the protein band of the Asp632Tyr-APPL1 variant disappeared, indicating that this variant would prevent the expression of APPL1 (Figure 4B). In addition, the protein expression of the Arg532Gln variant was significantly reduced compared with WT APPL1, indicating that this mutation also inhibited the expression of APPL1 protein. There was no significant change in APPL1 protein expression when mutated at Arg633His or Ile642Met, suggesting that these two mutations may not affect the expression of APPL1 protein.
To further elucidate the role of APPL1, we searched for APPL1-related protein pathways in the STRING database. Our analysis revealed that the insulin-related pathway protein AKT had the highest binding affinity with APPL1 (Figure 5A). In the AKT pathway, APPL1 could also bind to IR, which plays a role in insulin sensitization by interacting with the PTB domain where our four missense mutations were located. As shown in the Figure 5B, the NPEY motif of IR (with TYR-999 as the phosphorylation site) might interact with the amino acids between β5 and C-terminal helix of the PTB domain. Notably, the Asp632Tyr mutation is in the closest proximity to this binding site. Based on this observation, we speculate that this site might be associated with the interaction between APPL1 and IR.
MODY14 is an extremely rare form of inherited diabetes caused by mutations in the APPL1 gene. So far, only two variants [c.1655T>A (p.Leu552*) and c.281G>A p.(Asp94Asn)] of APPL1 were found to be associated with MODY14. In this study, we identified four novel missense mutations and one intronic mutation in APPL1, representing the largest number of de novo APPL1 mutations reported so far. For the first time, we demonstrated that two missense mutations [c.1894G>T (p.Asp632Tyr) and c.1595G>A (p.Arg532Gln)] in APPL1 are pathogenic. Bioinformatics analysis provided compelling evidence for their deleterious effects. In addition, we further investigated the role of APPL1 in insulin signaling and elucidated its potential molecular mechanisms.
To determine whether the APPL1 variants of the patients were related to their diabetes symptoms, we performed a comprehensive analysis combining their clinical manifestations and in vitro functional experiments. Preliminary experimental validation showed that the mutations carried by patients 1 (carrying mutation Asp632Tyr) and 3 (carrying mutation Asp632Tyr) were pathogenic. Considering their age of onset, family history, and the results of bioinformatics analysis, these 2 patients were diagnosed with MODY14. Interestingly, patient 1 had some insulin antibodies turned positive after many years of insulin therapy, suggesting a subsequent development of T1DM. On the other hand, the father of patient 3 carried the mutation but did not develop the disease, indicating potential incomplete penetrance of the mutation. Patients 2 and 4 were young at the onset of the disease and only needed medication or diet control. However, their family history did not align well with the autosomal dominant inheritance pattern. Pathogenicity prediction and functional testing both indicated that their APPL1 variants were non-pathogenic. Therefore, based on the comprehensive analysis, we speculated that patients 2 and 4 were more supportive of the diagnosis of T2DM, especially patient 4, who was overweight at the onset of the disease. We hypothesized that overeating and obesity may contribute to an earlier onset of T2DM in this patient. In addition, patient 5 had a more obvious family history of diabetes but an older age of onset. Taking the pathogenicity analysis into consideration, we propose that patient 5 aligns more closely with the diagnosis of T2DM. Therefore, among the five mutations, c.1894G>T (p.Asp632Tyr) and c.1595G>A (p.Arg532Gln) were pathogenic mutations, and patients carrying these mutations had MODY14.
Among the three major domains of the APPL1 protein, we found that the four missense mutations were all located in the PTB domain, which can bind to both AKT and IR (mainly). We considered that the pathogenicity of the mutation sites was related to the reduced sensitizing effect of APPL1 in the insulin pathway. After insulin binds to its receptor, APPL1 carries IRS1 and IRS2 to the IR and promotes the binding of the IR and IRS by directly interacting with the IR through its PTB domain[17,21] (Figure 6). The peptide binding site in most PTB domains is located between strand β5 of the central β sandwich and the C-terminal helix[22,23]. When we docked APPL1 and the IR, we found that the β subunit of the IR also contained an NPXY motif (NPEY), then the PTB domain docked with it to facilitate the subsequent transmission of insulin signals. Therefore, the mutation of pathogenic sites in the PTB domain of APPL1 may affect the binding of the IR and IRS, leading to an impaired insulin signaling pathway as well as increased blood glucose and insulin resistance. Furthermore, despite the adjacent location of the Asp632Tyr and Arg633His variants, their pathogenicity differs. This observation suggests that the Asp632 site may play a crucial role in binding to proteins associated with the insulin pathway.
In addition, the BAR domain of APPL1 can also enhance insulin-stimulated AKT phosphorylation by directly binding to AKT and competitively inhibiting Tribbles homolog 3 (mainly), thereby achieving the effects of lowering blood glucose (activating AKT to inhibit glucagon-induced hepatic glucose production, promoting glucose transporter type 4 translocation and cellular glucose uptake) and insulin resistance[24-27]. It is noteworthy that all the MODY14 patients we identified had mutations located in the PTB domain. Among the previously reported MODY14 patients, the c.1655T>A (p.L552*) mutation was also located in the PTB domain, indicating a high aggregation of mutations in the PTB domain[7]. This suggests that compared to the BAR domain, the PTB domain may play a more significant role in insulin pathway signal transduction.
Although mutations in APPL1 are relatively rare, recent advancements in exploring its molecular mechanisms and physiological functions have highlighted its key role in regulating glucose metabolism. Through its PTB domain, APPL1 interacts with AdipoR1 and AdipoR2, facilitating the transmission of adiponectin-stimulated signals to downstream targets[28]. In addition, APPL1 may provide a way of communication between the adiponectin and insulin signaling pathways, mediating the sensitization effect of insulin on muscle glucose disposal[18,19]. A study showed that APPL1 can counteract the high-fat diet-induced insulin resistance and hepatic glucose metabolism disorder, and improve blood glucose levels and insulin sensitivity in mice. Therefore, APPL1 may serve as a potential target for treating diabetes[11]. However, it is worth noting that a study reported that the expression of APPL1 in the muscle of T2DM rats was reduced, leading to weakened insulin-induced AKT signal activation[29]. To some extent, this consolidates the key role of APPL1 in regulating muscle insulin signaling and metabolism, but in this study, patients 2 and 4 who likely had T2DM did not show a reduction in APPL1 expression in the in vitro functional experiments.
This study also had some limitations. First, the functional experiments did not fully replicate the real physiological environment and conditions. Hence, we cannot completely rule out the possibility that these mutations might impact the interaction of APPL1 with other proteins or small molecules. Additionally, we did not verify the pathogenicity of the intronic mutation through functional experiments. We also failed to obtain blood samples from some family members for genetic testing. This may result in imprecise estimates of the mode of inheritance and penetrance of the mutation, and the existence of potential epistatic or modifier factors cannot be definitively determined. In the future, a broader range of cell lines or animal models are needed for in vitro and in vivo experiments to further investigate the impact of APPL1 gene mutations on the insulin signaling pathway and other metabolic pathways. Our study only serves as an initial investigation of the pathogenic mechanism of MODY14. At the protein level, aberrantly folded proteins can be degraded by the ubiquitin system, while RNA silencing or nonsense-mediated decay can diminish the level or stability of mRNA encoding aberrant proteins, thereby attenuating the production of aberrant proteins. These aspects warrant further investigation in subsequent research. Future studies should encompass more extensive epidemiological and statistical analysis, as well as detailed investigations in molecular biology and systems biology, to reveal the role and regulation of APPL1 gene mutations in the complex metabolic network.
In summary, this study identified five novel APPL1 variants, of which c.1894G>T (p.Asp632Tyr) and c.1595G>A (p.Arg532Gln) were pathogenic variants for MODY14. We believe that the PTB domain of APPL1 plays a significant role in the insulin signaling pathway in MODY14 patients. This study provided novel insights and evidence for further elucidating the pathogenicity of the APPL1 gene in MODY14 and provided new targets and strategies for developing new diagnostic and therapeutic methods.
Mutations in the adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 (APPL1) gene have been associated with the development of maturity-onset diabetes of the young type 14 (MODY14), a rare form of monogenic diabetes. So far, only two mutations [c.1655T>A (p.Leu552*) and c.281G>A p.(Asp94Asn)] have been found to be related to this disease. Due to the limited knowledge of MODY14, it is necessary to identify more cases and conduct a comprehensive study of MODY14 and APPL1 mutations. In this study, we discovered five new APPL1 gene mutations by whole exome sequencing (WES) and bioinformatics analysis, of which two were confirmed to be pathogenic mutations by in vitro functional assays. These mutations were all located in the phosphotyrosine binding (PTB) domain of APPL1, which has a significant impact on insulin sensitivity.
This study aimed to identify the pathogenicity and functional role of APPL1 gene mutations in diabetes. It mainly identified and evaluated the pathogenicity of APPL1 gene mutations and explored the effects of these mutations on APPL1 protein expression and insulin signaling pathway. This will provide potential targets for the diagnosis and treatment of MODY14 and will provide new clues for the interaction mechanism of the APPL1 protein and insulin receptor.
The main objective of this study was to evaluate the pathogenicity of APPL1 gene mutations in diabetic patient and to characterize the functional role of APPL1 domains. By WES and bioinformatics analysis, five novel APPL1 gene mutations were identified, among which c.1894G>T (at Asp632Tyr) and c.1595G>A (at Arg532Gln) were confirmed as pathogenic mutations by in vitro functional experiments.
This study used WES to sequence all the exons in the genome that encode proteins, thus discovering variants associated with diseases. Then, bioinformatics analysis was used to align and predict the sequencing results, thus evaluating the pathogenicity and conservation of the variants. The pathogenicity was further verified by in vitro functional experiments.
Our study identified five novel APPL1 gene mutations, among which c.1894G>T (at Asp632Tyr) and c.1595G>A (at Arg532Gln) were confirmed as pathogenic mutations by in vitro functional experiments. Both mutations are located in the PTB domain of APPL1, which has an important impact on insulin sensitivity. The results showed that the mutations can reduce the expression level of APPL1 protein, thus affecting the activation of the insulin signaling pathway and the regulation of glucose metabolism.
APPL1 gene mutations c.1894G>T (at Asp632Tyr) and c.1595G>A (at Arg532Gln) are pathogenic in diabetes, and these mutations are located in the PTB domain of APPL1, which has an important impact on insulin sensitivity.
In the future, the structure and function of APPL1 protein can be further studied, especially the mechanism of action of the PTB domain and the binding mode and regulatory effect of APPL1 protein with the insulin receptor. In addition, the effect of APPL1 gene mutations on the clinical manifestations and treatment response of diabetic patients can be verified. More effective methods and criteria for the diagnosis and treatment of MODY14 can be provided.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Endocrinology and metabolism
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Horowitz M, Australia; Mohammadi S, Iran; Tamemoto H, Japan S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Yu HG
| 1. | Samadli S, Zhou Q, Zheng B, Gu W, Zhang A. From glucose sensing to exocytosis: takes from maturity onset diabetes of the young. Front Endocrinol (Lausanne). 2023;14:1188301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 2. | Delvecchio M, Pastore C, Giordano P. Treatment Options for MODY Patients: A Systematic Review of Literature. Diabetes Ther. 2020;11:1667-1685. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 3. | Ivanoshchuk DE, Shakhtshneider EV, Rymar OD, Ovsyannikova AK, Mikhailova SV, Fishman VS, Valeev ES, Orlov PS, Voevoda MI. The Mutation Spectrum of Maturity Onset Diabetes of the Young (MODY)-Associated Genes among Western Siberia Patients. J Pers Med. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 4. | Nkonge KM, Nkonge DK, Nkonge TN. The epidemiology, molecular pathogenesis, diagnosis, and treatment of maturity-onset diabetes of the young (MODY). Clin Diabetes Endocrinol. 2020;6:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 99] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 5. | Broome DT, Pantalone KM, Kashyap SR, Philipson LH. Approach to the Patient with MODY-Monogenic Diabetes. J Clin Endocrinol Metab. 2021;106:237-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 6. | Kant R, Davis A, Verma V. Maturity-Onset Diabetes of the Young: Rapid Evidence Review. Am Fam Physician. 2022;105:162-167. [PubMed] |
| 7. | Prudente S, Jungtrakoon P, Marucci A, Ludovico O, Buranasupkajorn P, Mazza T, Hastings T, Milano T, Morini E, Mercuri L, Bailetti D, Mendonca C, Alberico F, Basile G, Romani M, Miccinilli E, Pizzuti A, Carella M, Barbetti F, Pascarella S, Marchetti P, Trischitta V, Di Paola R, Doria A. Loss-of-Function Mutations in APPL1 in Familial Diabetes Mellitus. Am J Hum Genet. 2015;97:177-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 8. | Ivanoshchuk DE, Shakhtshneider EV, Rymar OD, Ovsyannikova AK, Mikhailova SV, Orlov PS, Ragino YI, Voevoda MI. Analysis of APPL1 Gene Polymorphisms in Patients with a Phenotype of Maturity Onset Diabetes of the Young. J Pers Med. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 9. | Cheng KK, Lam KS, Wu D, Wang Y, Sweeney G, Hoo RL, Zhang J, Xu A. APPL1 potentiates insulin secretion in pancreatic β cells by enhancing protein kinase Akt-dependent expression of SNARE proteins in mice. Proc Natl Acad Sci U S A. 2012;109:8919-8924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 10. | Wu KKL, Long K, Lin H, Siu PMF, Hoo RLC, Ye D, Xu A, Cheng KKY. The APPL1-Rab5 axis restricts NLRP3 inflammasome activation through early endosomal-dependent mitophagy in macrophages. Nat Commun. 2021;12:6637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 11. | Cheng KK, Iglesias MA, Lam KS, Wang Y, Sweeney G, Zhu W, Vanhoutte PM, Kraegen EW, Xu A. APPL1 potentiates insulin-mediated inhibition of hepatic glucose production and alleviates diabetes via Akt activation in mice. Cell Metab. 2009;9:417-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 111] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 12. | Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1353] [Cited by in RCA: 1343] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 13. | Mitsuuchi Y, Johnson SW, Sonoda G, Tanno S, Golemis EA, Testa JR. Identification of a chromosome 3p14.3-21.1 gene, APPL, encoding an adaptor molecule that interacts with the oncoprotein-serine/threonine kinase AKT2. Oncogene. 1999;18:4891-4898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 167] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 14. | Ye S, Luo Y, Lu W, Jones RB, Linhardt RJ, Capila I, Toida T, Kan M, Pelletier H, McKeehan WL. Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs. Biochemistry. 2001;40:14429-14439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 119] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 15. | Zeng L, Kuti M, Mujtaba S, Zhou MM. Structural insights into FRS2α PTB domain recognition by neurotrophin receptor TrkB. Proteins. 2014;82:1534-1541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 16. | Liu Z, Xiao T, Peng X, Li G, Hu F. APPLs: More than just adiponectin receptor binding proteins. Cell Signal. 2017;32:76-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 17. | Ryu J, Galan AK, Xin X, Dong F, Abdul-Ghani MA, Zhou L, Wang C, Li C, Holmes BM, Sloane LB, Austad SN, Guo S, Musi N, DeFronzo RA, Deng C, White MF, Liu F, Dong LQ. APPL1 potentiates insulin sensitivity by facilitating the binding of IRS1/2 to the insulin receptor. Cell Rep. 2014;7:1227-1238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 100] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 18. | Deepa SS, Dong LQ. APPL1: role in adiponectin signaling and beyond. Am J Physiol Endocrinol Metab. 2009;296:E22-E36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 219] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 19. | Artimani T, Najafi R. APPL1 as an important regulator of insulin and adiponectin-signaling pathways in the PCOS: A narrative review. Cell Biol Int. 2020;44:1577-1587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Wu H, Shu M, Liu C, Zhao W, Li Q, Song Y, Zhang T, Chen X, Shi Y, Shi P, Fang L, Wang R, Xu C. Identification and characterization of novel carboxyl ester lipase gene variants in patients with different subtypes of diabetes. BMJ Open Diabetes Res Care. 2023;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 21. | Saito T, Jones CC, Huang S, Czech MP, Pilch PF. The interaction of Akt with APPL1 is required for insulin-stimulated Glut4 translocation. J Biol Chem. 2007;282:32280-32287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 102] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 22. | Li J, Mao X, Dong LQ, Liu F, Tong L. Crystal structures of the BAR-PH and PTB domains of human APPL1. Structure. 2007;15:525-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Uhlik MT, Temple B, Bencharit S, Kimple AJ, Siderovski DP, Johnson GL. Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J Mol Biol. 2005;345:1-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 198] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 24. | Prudente S, Sesti G, Pandolfi A, Andreozzi F, Consoli A, Trischitta V. The mammalian tribbles homolog TRIB3, glucose homeostasis, and cardiovascular diseases. Endocr Rev. 2012;33:526-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 94] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 25. | Schenck A, Goto-Silva L, Collinet C, Rhinn M, Giner A, Habermann B, Brand M, Zerial M. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell. 2008;133:486-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 318] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 26. | Mîinea CP, Sano H, Kane S, Sano E, Fukuda M, Peränen J, Lane WS, Lienhard GE. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem J. 2005;391:87-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 384] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 27. | Saito T, Okada S, Shimoda Y, Tagaya Y, Osaki A, Yamada E, Shibusawa R, Nakajima Y, Ozawa A, Satoh T, Mori M, Yamada M. APPL1 promotes glucose uptake in response to mechanical stretch via the PKCζ-non-muscle myosin IIa pathway in C2C12 myotubes. Cell Signal. 2016;28:1694-1702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 28. | Mao X, Kikani CK, Riojas RA, Langlais P, Wang L, Ramos FJ, Fang Q, Christ-Roberts CY, Hong JY, Kim RY, Liu F, Dong LQ. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat Cell Biol. 2006;8:516-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 525] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 29. | Kido K, Ato S, Yokokawa T, Sato K, Fujita S. Resistance training recovers attenuated APPL1 expression and improves insulin-induced Akt signal activation in skeletal muscle of type 2 diabetic rats. Am J Physiol Endocrinol Metab. 2018;314:E564-E571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |