Published online Jan 15, 2024. doi: 10.4239/wjd.v15.i1.92
Peer-review started: September 7, 2023
First decision: November 17, 2023
Revised: November 27, 2023
Accepted: December 25, 2023
Article in press: December 25, 2023
Published online: January 15, 2024
Processing time: 126 Days and 20.4 Hours
Diabetic kidney disease (DKD), characterized by increased urinary microalbumin levels and decreased renal function, is the primary cause of end-stage renal di
To determine if urinary exosomal miRNAs from diabetic patients can serve as noninvasive biomarkers for early DKD diagnosis.
Type 2 diabetic mellitus (T2DM) patients were recruited from the Second Hospital of Hebei Medical University and were divided into two groups: DM, diabetic pa
Urinary exosomal expression of miR-145-5p and miR-27a-3p was more upregulated in the DKD group than in the DM group (miR-145-5p: 4.54 ± 1.45 vs 1.95 ± 0.93, P < 0.001; miR-27a-3p: 2.33 ± 0.79 vs 1.71 ± 0.76, P < 0.05) and the NC group (miR-145-5p: 4.54 ± 1.45 vs 1.55 ± 0.83, P < 0.001; miR-27a-3p: 2.33 ± 0.79 vs 1.10 ± 0.51, P < 0.001). The exosomal miR-145-5p and miR-27a-3p positively correlated with albuminuria and serum creatinine and negatively correlated with the estimated glomerular filtration rate. miR-27a-3p was also closely related to blood glucose, gly
Urinary exosomal miR-145-5p and miR-27a-3p may serve as novel noninvasive diagnostic biomarkers or promising therapeutic targets for DKD.
Core Tip: Diabetic kidney disease (DKD) is a serious complication of diabetes mellitus (DM). Novel biomarkers and effective therapeutic targets for DKD are needed in clinical settings. In this study, urinary exosomal microRNA-145-5p (miR-145-5p) and miR-27a-3p from patients with DKD were associated with kidney injury progression in type 2 DM patients. MiR-145-5p was highly specific and sensitive to DKD. It may be involved in the signaling pathways related to the pathological processes of DKD. Urinary exosomal miR-145-5p and miR-27a-3p may serve as novel noninvasive diagnostic biomarkers and therapy targets for DKD.
- Citation: Han LL, Wang SH, Yao MY, Zhou H. Urinary exosomal microRNA-145-5p and microRNA-27a-3p act as noninvasive diagnostic biomarkers for diabetic kidney disease. World J Diabetes 2024; 15(1): 92-104
- URL: https://www.wjgnet.com/1948-9358/full/v15/i1/92.htm
- DOI: https://dx.doi.org/10.4239/wjd.v15.i1.92
Diabetic kidney disease (DKD) is a serious complication of diabetes mellitus (DM). It is characterized by increased urinary microalbumin levels and decreased estimated glomerular filtration rate (eGFR), resulting in rapid progression to end-stage renal disease[1]. According to a recent report, the number of adults with diabetes worldwide is approximately 0.537 billion, and the total number may rise to 0.643 billion by 2030. Notably, approximately 20%-40% of DM patients progress to DKD[2]. The pathological mechanisms of DKD, including abnormal glucose metabolism, advanced glycation end product generation, inflammation, oxidative stress, and renal hemodynamic changes that eventually lead to renal injury, are complicated and multifactorial[3,4]. Podocytes are the key cell type damaged early in DKD due to their highly differentiated postmitotic phenotype with restricted abilities for self-repair and renewal. Hyperglycemia-induced podocyte injury can lead to glomerular filtration dysfunction and proteinuria[5]. Early diagnosis and specific treatment can prevent DKD progression; however, renal biopsy, the golden standard for DKD diagnosis, cannot be widely used because it is invasive, and microalbuminuria is often unable to reflect early renal injury or kidney dysfunction pro
Exosome are lipid bilayer cup-shaped extracellular vesicles with 40-160 nm in diameter. Many cell types can excrete exosome; Hence, they can be detected in various body or tissue fluids, such as blood, urine, and saliva[8,9]. The bioactive cargo derived from parental cells, which includes proteins, metabolites, and genetic information, is delivered by the exosome to adjacent or distant cells that regulate the phenotypes or functions of recipient cells[10]. Circulating exosome cannot cross the glomerular filtration barrier. Urinary exosome are generally derived from diverse renal cells in nephron segments and are not easily confounded by circulating exosome. Urinary exosome have specific responses to the renal pathological changes[11]. Exosome can carry genetic information. MicroRNAs (miRNAs) are the most abundant exosomal RNAs, comprising roughly 22 nucleotides. miRNAs participate in post-transcriptional gene silencing or target mRNA degrading[12,13]. Several miRNAs play vital roles in the pathological processes of DKD[12]. MiR-145-5p, miRNA-27a, and miR-29c are associated with podocyte injury[14]. miRNAs contained in urinary exosome can avoid degradation by nuclease. They are remarkably stable, independent of urine, highly tissue-specific for the kidney, and can be collected in large quantities noninvasively[15-17]. Therefore, urinary exosomal miRNAs are better candidates for diagnostic markers than free miRNAs[18]. Ghai et al[17] showed that miR-31-5p and miR-200c-3p were upregulated in urinary exosome from DKD patients compared with patients with normoalbuminuria. Park et al[19] identified 22 differentially expressed miRNAs in urinary exosome from DKD patients; of these, 14 differentially expressed miRNAs were associated with renal inflammation and glomerular injury. Zhao et al[20] found that the urinary exosomal miR-4534 was signi
In the present study, we extracted urinary exosome from T2DM patients to determine the expression of miR-145-5p, miR-27a-3p, and miR-29c-3p and evaluate their sensitivity and specificity in diagnosing DKD. Moreover, the molecular biological function of exosomal miR-145-5p was predicted by conducting a bioinformatics analysis to seek novel noninvasive diagnostic biomarkers and potential therapy targets for DKD.
The Ethics Committee of the Second Hospital of Hebei Medical University approved the trial protocols (approval number: 2022-R059). Written informed consent was obtained from each participant before the study. The diagnosis of T2DM and DKD was based on the criteria of the American Diabetes Association[22,23]. All T2DM patients were recruited from the Endocrinology Department of the Second Hospital of Hebei Medical University from February 2022 to May 2022. The age of the enrolled patients was 18-75 year, and their eGFR was ≥ 60 mL/min/1.73 m2. T2DM patients with normoalbuminuria [urinary albumin to creatinine ratio (UACR) < 30 mg/g] were included in the DM group (n = 20), and T2DM patients with albuminuria (UACR ≥ 30 mg/g) were included in the DKD group (n = 20). In the DKD group, five patients were confirmed via kidney biopsy. Twenty healthy volunteers from the Medical Examination Department were included in the normal control (NC) group.
The exclusion criteria of the patients were as follows: (1) Those with T1DM and other specific types of diabetes; (2) Those with severe metabolic disorder or infectious disease within the last 1 mo; (3) Those with severe cardiovascular and cerebrovascular diseases within the last 3 mo; (4) Those with proliferative retinopathy or diabetic foot; (5) Those with nondiabetic renal diseases, kidney stones, and urinary tract infection; and (6) Those with a malignant tumor or auto
The general and clinical data, including age, gender, body mass index (BMI), fasting blood glucose (FBG), glycosylated hemoglobin A1c (HbA1c), fasting insulin (FINS), serum C-peptide (C-P), total cholesterol (TC), triglyceride (TG), low-density lipoprotein cholesterol (LDL-C), blood urea nitrogen (BUN), serum creatinine (Scr) and UACR of all participants were obtained from the medical record system. Fasting urine samples were collected for urinary exosome separation and exosomal miRNAs detection. The eGFR was calculated using the Chronic Kidney Disease Epidemiology Collaboration 2021 formula[24].
Each participant provided 200 mL of fasting urine, which was centrifuged at 4 °C and 3000 × g for 20 min. The super
The urinary exosome particle sizes were measured via nanoparticle tracking analysis (NTA) using ZetaView PMX 110 (Particle Metrix, Germany). An exosome suspension of 50 μL was appropriately diluted using 1× phosphate buffer saline. NTA was recorded and analyzed at pH 7.0. The entered conductivity was 15000 μS/cm sensed.
Total proteins were separated from exosome using radioimmunoprecipitation assay lysis buffer (Solarbio, China). Protein concentrations were estimated using a bicinchoninic acid protein analysis kit (Solarbio, China) according to our previous study[25]. Denatured proteins, 30 μg from each sample, were separated via 10% SDS-polyacrylamide gel electrophoresis (Epizyme, China) and transferred to polyvinylidene fluoride membranes (Millipore, United States). The membranes were blocked with 5% skim milk (BioFroxx, Germany) for 2 h at 24 °C and then incubated with the primary antibodies CD63 (1:1000, Abcam), CD9 (1:1000, Abcam), and TSG101 (1:1000, Abcam) at 4 °C overnight. The next day, the membranes were incubated with the secondary antibody (1:5000, Affinity) for 1 h at 24 °C. The protein bands were detected using chemiluminescence reagents (Sharebio, China) and quantified using ImageJ software (Bio-Rad, United States).
Real-time quantitative polymerase chain reaction (RT-qPCR) was performed according to a previous study[26]. Total RNA in the exosome was isolated using an RNA-easy isolation reagent (Vazyme, China). Complementary DNA (cDNA) was reverse transcribed from 1 μg total RNA using miRNA 1st Strand cDNA Synthesis Kit (Vazyme, China). RT-qPCR was performed on a CFX96 PCR system (Bio-Rad, United States) using GoTaq® qPCR Master Mix (Promega, United States). The relative expression of miRNAs were normalized to that of the internal reference U6 and then calculated using the 2-ΔΔCt method.
The following primers were used for RT-qPCR: hsa-miR-145-5p: F-5’-AAGCGACCGTCCAGTTTTCCC-3’, R-5’-ATCCAGTGCAGGGTCCGAGG-3’; hsa-miR-27a-3p: F-5’-AATCGGCGTTCACAGTGGCTAA-3’, R-5’-ATCCAGTGCAGGGTCCGAGG-3’; hsa-miR-29c-3p: F-5’-CGCGGCATAGCACCATTTGAAA-3’, R-5’-ATCCAGTGCAGGGTCCGAGG-3’; U6: F-5’-CTCGCTTCGGCAGCACA-3’, R-5’-AACGCTTCACGAATTTGCGT-3’.
The potential target genes of miR-145-5p were predicted using three different gene databases: TargetScan7.2 (http://wwtargetscan.org/vert_72/), miRDB (https://mirdb.org/mirdb/index.html), and miRTarBase (https://mirtarbase.cuhk.edu.cn). Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were implemented on the Database for Annotation, Visualization, and Integrated Discovery (DAVID) 6.8 (https://david.ncifcrf.gov/). GO analysis elucidated detailed biological functions of potential target genes of miR-145-5p from the molecular function (MF), biological process (BP), and cellular component (CC) aspects. A P-value and a false discovery rate (FDR) of < 0.05 were considered statistically different. -Log10 (P-value) and FDR also represented the enrichment degree of gene function and signaling pathway.
All data were processed by GraphPad Prism 8.0 software (La Jolla, United States). A normality test was first performed in each group. Normally distributed data are presented as mean ± SD and analyzed using one-way analysis of variance (ANOVA), followed by the Tukey test. The heterogeneity of the variance data was detected using Welch’s ANOVA test. Pearson correlation analysis was used to analyze the correlation between two normally distributed parameters. Receiver operating characteristic (ROC) curves were used to assess the diagnostic efficiency of urinary exosomal miRNAs in DKD. A P value of < 0.05 was considered statistically significant.
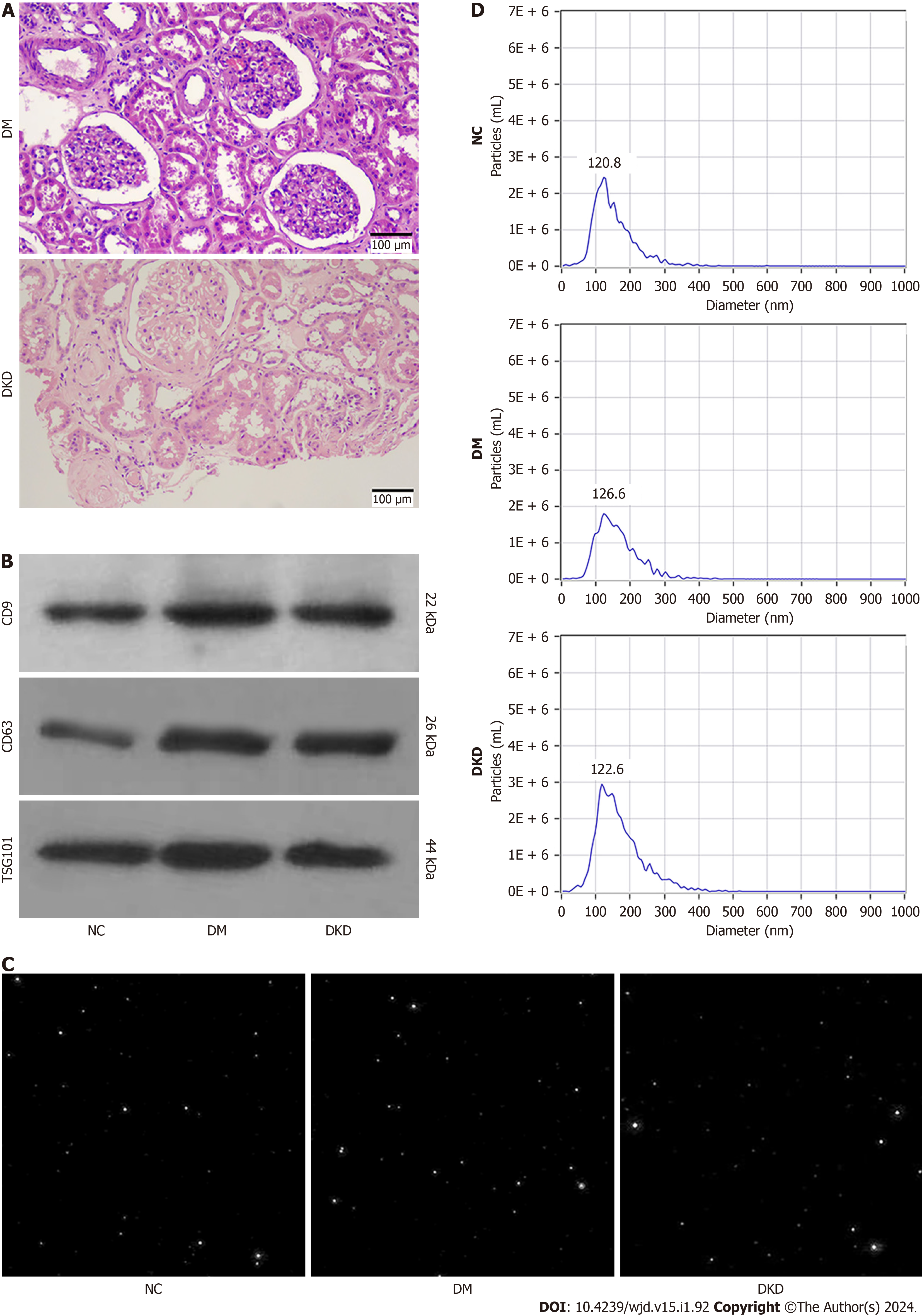
The general and clinical data of participants are shown in Table 1. There were no differences in age, gender, BMI, and C-P levels among the three groups (P > 0.05). FBG, HbA1c, FINS, TC, and LDL-C in the DM and DKD groups were higher than those in the NC group (P < 0.05), but there were no differences in these indexes between the DM and DKD groups (P > 0.05). Scr and UACR were higher and eGFR was lower in the DKD group than in the DM group (P < 0.001). Patients with DKD exhibited mesangial cell proliferation, extracellular matrix accumulation, and basement membrane thickening (Figure 1A).
| Biochemical and clinical data | NC (n = 20) | DM (n = 20) | DKD (n = 20) |
| Age (yr) | 45.80 ± 14.69 | 50.15 ± 13.50 | 48.79 ± 15.14 |
| Gender (female/male) | 12/8 | 9/11 | 7/13 |
| BMI (kg/m2) | 22.94 ± 2.32 | 24.95 ± 2.47 | 24.81 ± 3.9 |
| FBG (mmol/L) | 5.30 ± 0.38 | 8.90 ± 2.28c | 11.78 ± 5.12c |
| HbA1c (%) | 5.33 ± 0.24 | 8.74 ± 2.23c | 8.95 ± 1.58c |
| FINS (μIU/mL) | 9.58 ± 3.01 | 14.43 ± 8.12a | 19.63 ± 13.13b |
| C-P (ng/mL) | 1.98 ± 0.57 | 2.27 ± 1.07 | 2.80 ± 1.60 |
| TC (mmol/L) | 3.12 ± 0.50 | 4.53 ± 0.94c | 5.14 ± 1.49c |
| TG (mmol/L) | 1.43 ± 0.81 | 2.24 ± 1.45 | 3.49 ± 2.24b |
| LDL-C (mmol/L) | 2.71 ± 0.75 | 3.64 ± 1.15a | 4.32 ± 1.57c |
| BUN (mmol/L) | 4.61 ± 1.39 | 5.33 ± 1.47 | 6.05 ± 2.31a |
| Scr (μmol/L) | 61.50 ± 6.623 | 67.69 ± 7.76 | 97.02 ± 15.46c,d |
| eGFR (mL/min/1.73m²) | 114.10 ± 13.05 | 108.00 ± 10.59 | 76.94 ± 18.06c,d |
| UACR (mg/g) | 9.45 ± 4.76 | 13.37 ± 7.20 | 389.50 ± 311.70c,d |
Western blot analysis verified the expression of exosome marker proteins, including CD9, CD63, and TSG101, in the particles from the NC, DM, and DKD groups (Figure 1B). NTA screen capture showed that abundant nanoparticles existed in the urine specimens of the three groups (Figure 1C). The particle diameters at peak concentrations were 120.8 nm (91.9% of the total), 126.6 nm (95.5% of the total), and 122.6 nm (85.4% of the total) for the NC, DM, and DKD groups, respectively. The average diameter sizes of exosome were 135.3 ± 1.59 nm, 149.2 ± 1.81 nm, and 147.7 ± 10.55 nm, re
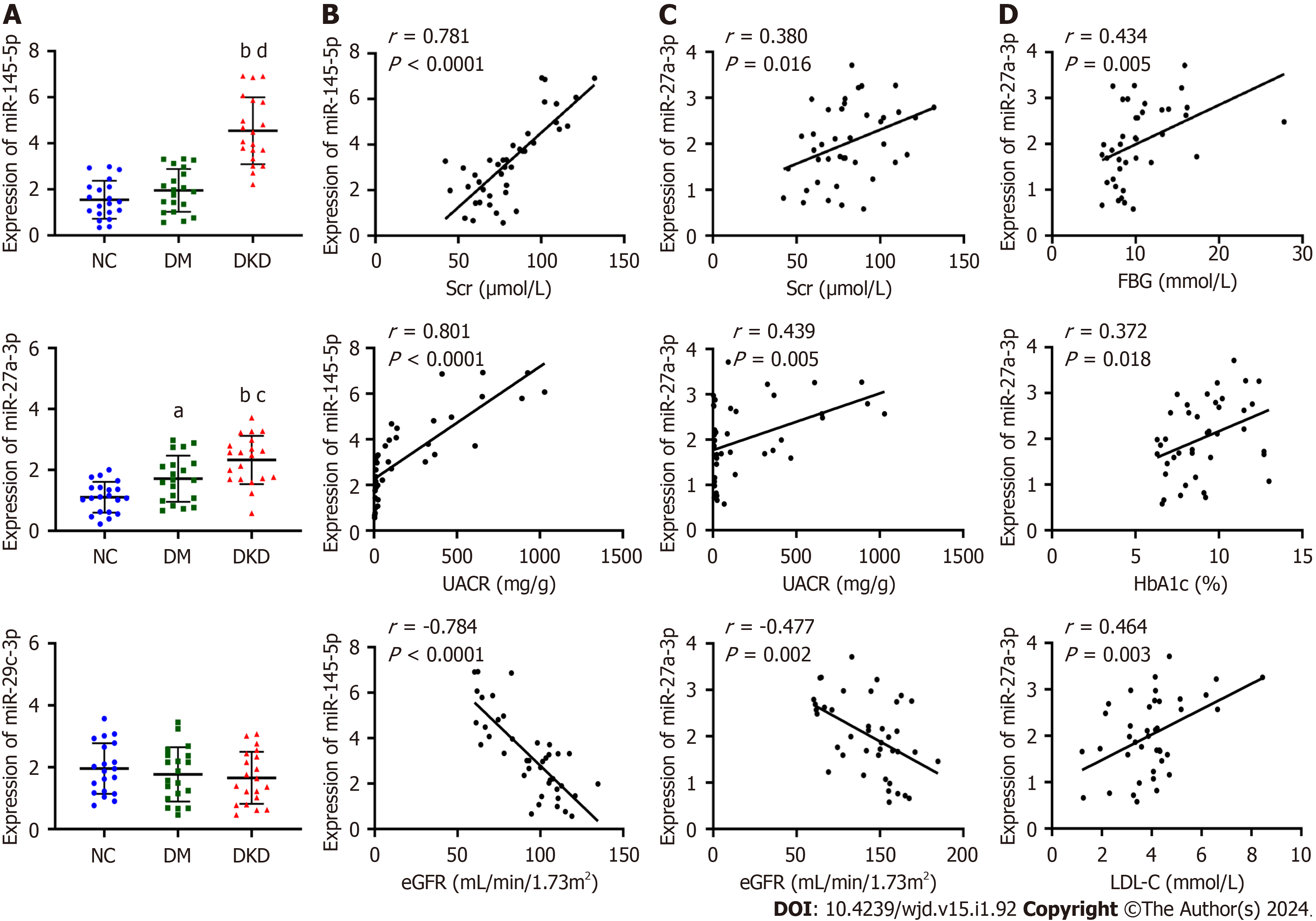
The relative expression of the urinary exosomal miR-145-5p, miR-27a-3p, and miR-29c-3p in the three groups was mea
Exosomal miR-145-5p was found to positively correlate with Scr (r = 0.781, P < 0.0001) and UACR (r = 0.801, P < 0.0001) and negatively correlate with eGFR (r = -0.784, P < 0.0001) (Figure 2B). Similarly, miR-27a-3p positively correlated with Scr (r = 0.380, P = 0.016) and UACR (r = 0.439, P = 0.005) and negatively correlated with eGFR (r = -0.477, P = 0.002) (Figure 2C). Moreover, miR-27a-3p positively correlated with glycolipid metabolism indexes, including FBG, HbA1c, and LDL-C (Figure 2D).
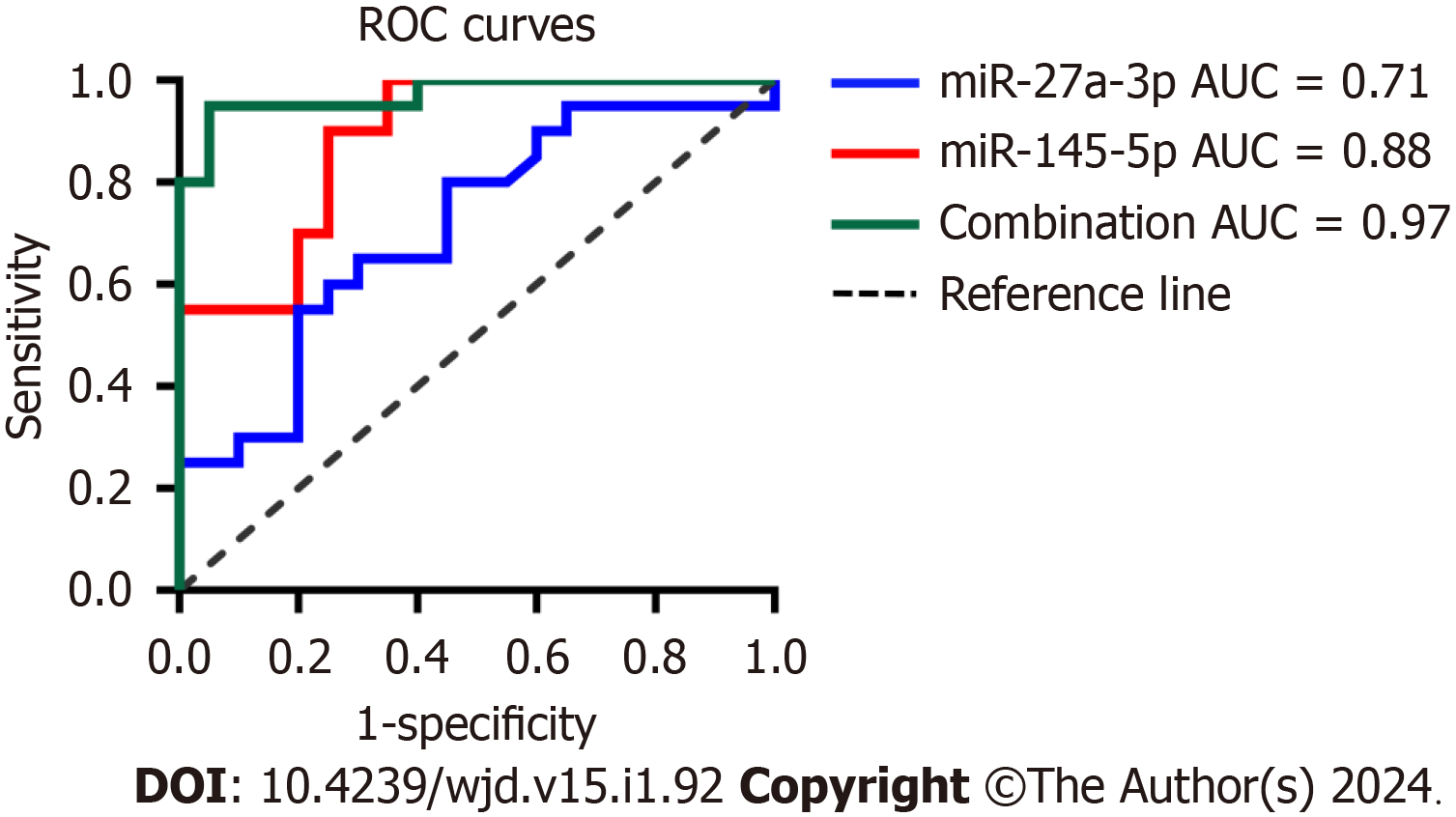
ROC analyses defined the diagnostic potential of urinary exosomal miR-145-5p and miR-27a-3p in DKD. MiR-145-5p had a better area under the curve (AUC) of 0.88 [95% confidence interval (CI): 0.784-0.985, P < 0.0001] than miR-27a-3p with an AUC of 0.71 (95%CI: 0.547-0.871, P = 0.0239) in DKD patients (Figure 3). For DKD diagnosis, exosomal miR-145-5p exhibited a higher sensitivity of 90% and a specificity of 75% at the best cutoff value of 2.67 than miR-27a-3p with a sensitivity of 65% and a specificity of 70% at the optimal cutoff value of 2.12. The combination of miR-145-5p and miR-27a-3p contributed to an increased AUC of 0.97 (95%CI: 0.927-1.000, P < 0.0001) with a sensitivity of 95% and a specificity of 90% for DKD diagnosis. Urinary exosomal miR-145-5p and miR-27a-3p may serve as potential biomarkers of early DKD diagnosis, especially miR-145-5p.
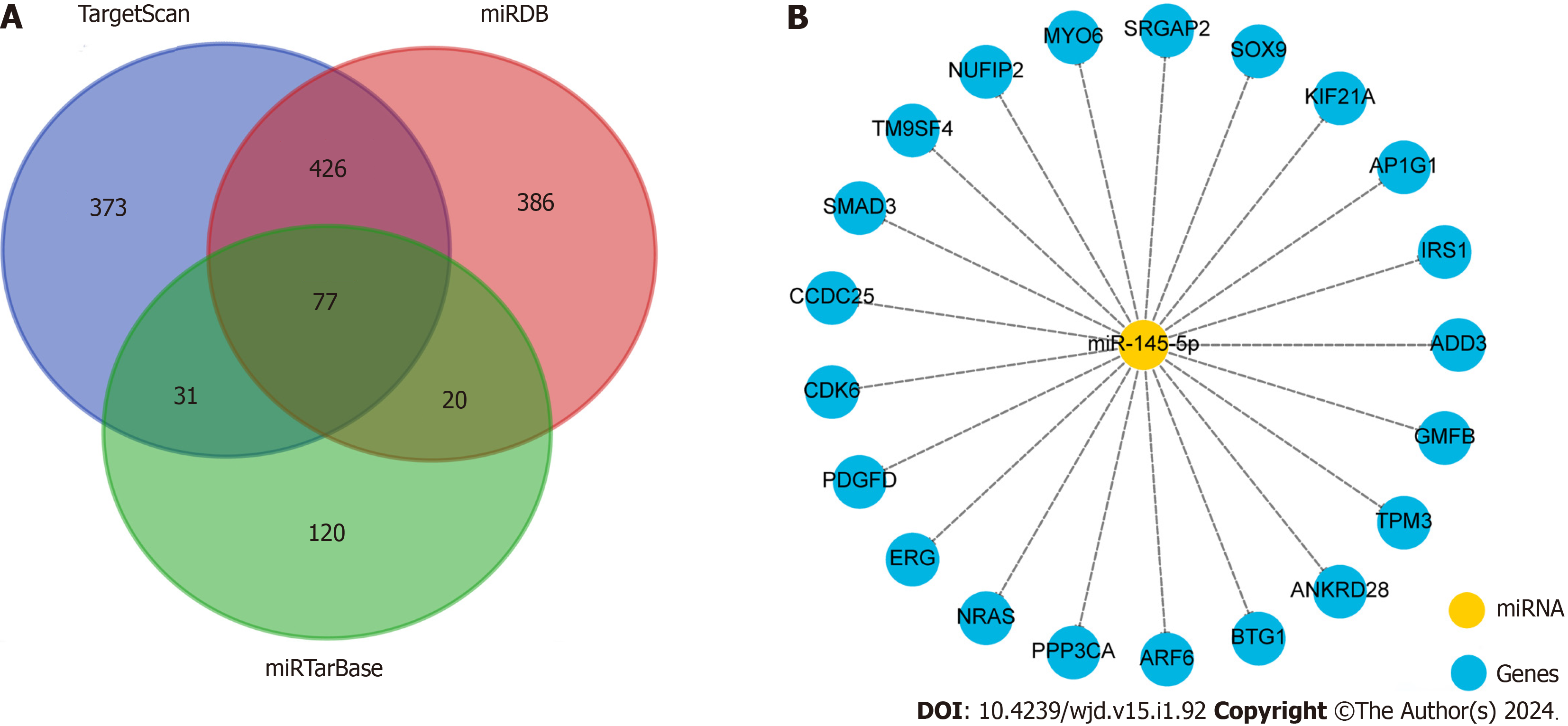
Since urinary exosomal miR-145-5p has the potential to serve as a promising noninvasive diagnostic biomarker of DKD, its biological function was further explored using bioinformatics analysis. In total, 907, 909, and 248 potential target mRNAs of miR-145-5p were predicted using TargetScan, miRDB, and miRTarBase, respectively. A total of 77 mRNAs were detected simultaneously using the three gene databases (Figure 4A). We listed some gene names according to the target score, such as SMAD3, SOX9, and SRGAP2, which may be involved in the pathophysiological processes of DM and DKD[27-29] (Figure 4B).
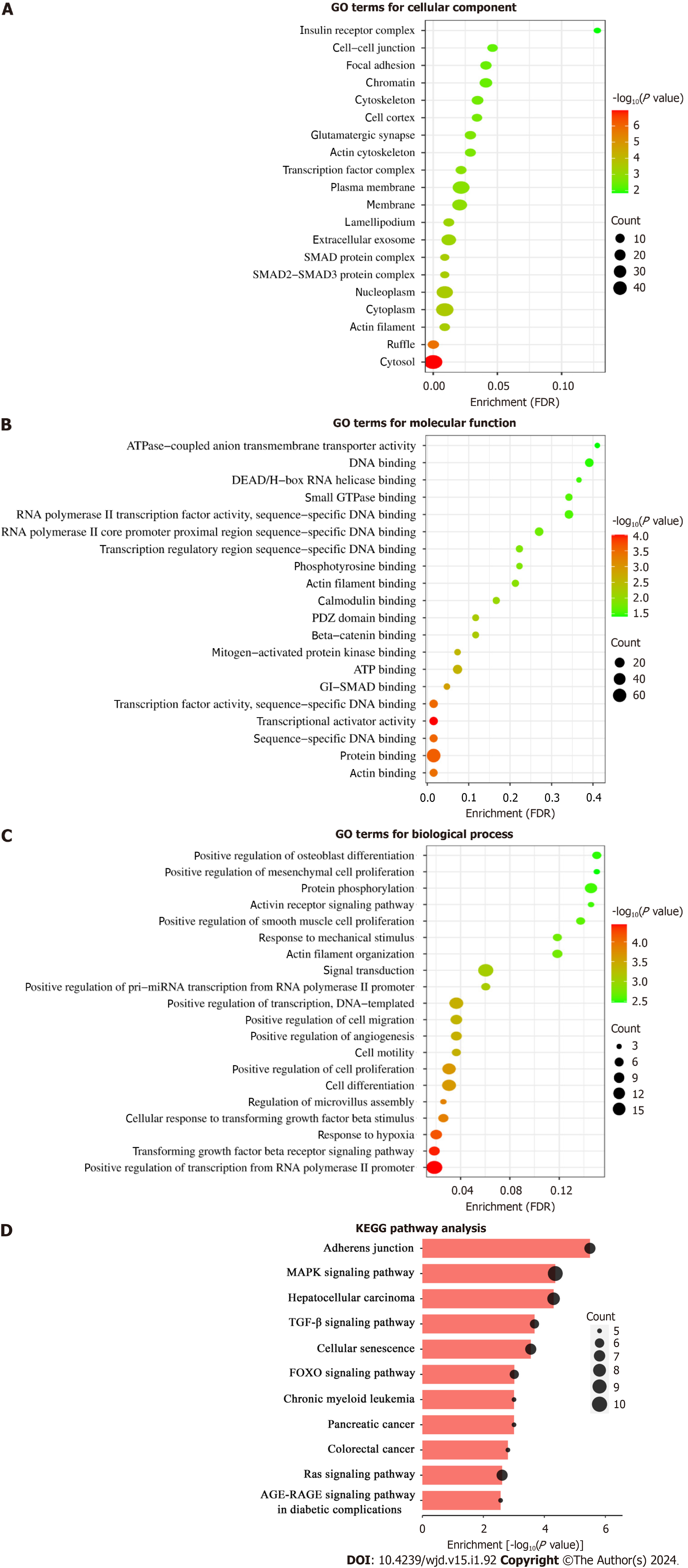
GO analysis classified and described these target genes on CC, MF, and BP aspects. The CC catalog contained various cell locations, including the cytosol (count: 42, FDR = 1.97E-05), nucleoplasm (count: 28, FDR = 0.009056116), SMAD protein complex (count: 3, FDR = 0.009056116), extracellular exosome (count: 19, FDR = 0.012172645), actin cytoskeleton (count: 6, FDR = 0.029013556), and actin filament (count: 5, FDR = 0.009056116) (Figure 5A). The terms including actin binding, protein binding, sequence-specific DNA binding, SMAD binding, mitogen-activated protein kinase (MAPK) binding, and small GTPase binding were enriched in the MF catalog (Figure 5B). Regarding the biological regulatory processes, terms such as transforming growth factor β (TGF-β) receptor signaling pathway, cellular response to TGF-β stimulus, response to hypoxia, positive regulation of cell proliferation, cell differentiation, cell motility, and actin filament organization were listed in the BP catalog (Figure 5C).
KEGG pathway enrichment analysis manifested the target genes of miR-145-5p. These genes were mainly enriched in 11 signaling pathways (FDR < 0.05), such as the MAPK signaling pathway (FDR = 0.002477523), TGF-β signaling pathway (FDR = 0.00784279), forkhead box O (FOXO) signaling pathway (FDR = 0.018431373), Ras signaling pathway (FDR= 0.036227759), and advanced glycosylation end products-the accumulation of their receptors (AGE-RAGE) signaling pathway in diabetic complications (FDR = 0.036960173), which may be involved in the pathological processes of DKD[3,30]. Besides, the pathways related to adherens junction, cellular senescence, and cancer were included (Figure 5D).
DKD is the primary cause of end-stage renal disease and seriously threatens the lives of patients with DM due to its irreversible and progressive evolution[1]. Early identification of high-risk patients may prevent DKD progression; However, sensitive and noninvasive diagnostic biomarkers for DKD are scarce.
miRNA dysregulation participates in various pathological processes of diabetic kidney injury and possesses great potential in the early diagnosis of DKD. A previous study showed that serum miR-145-5p was significantly lower in T1DM patients with nephropathy than T1DM controls[31]. MiR-145-5p overexpression suppressed podocyte apoptosis under high glucose (HG) conditions by inhibiting the Notch signaling pathway[32]. MiR-27a-3p was reported to be higher in the serum of T2DM patients than in the serum of non-diabetic individuals[33]. Upregulation of miR-27a acce
In the present study, urinary exosomal miR-145-5p was remarkably upregulated in T2DM patients with DKD. Barutta et al[38] demonstrated that urinary exosomal miR-145 was higher in T1DM patients with microalbuminuria than in normoalbuminuric and nondiabetic subjects. Moreover, miR-145 was enriched in both urinary exosome and their glo
ROC analysis revealed that exosomal miR-145-5p had a better AUC with higher sensitivity and specificity than miR-27a-3p in determining diabetic kidney damage in T2DM patients. Combining the two exosomal miRNAs led to the improvement of diagnostic efficiency and were expected to serve as novel markers for early identification and diagnosis of DKD. The prediction and biological functional analysis of the target genes of miR-145-5p can guide future research on their pathological effects on DKD. GO analysis revealed the cell locations where the genes acted. The terms “actin fila
This study had some limitations. First, it was a cross-sectional, observational study with a small sample size without clinical follow-up and evaluation of these biomarkers in the middle or long term. Second, data on global miRNA se
Our results show that urinary exosomal miR-145-5p and miR-27a-3p were markedly increased in DKD patients and were associated with the progression of kidney injury in T2DM patients. These findings imply that urinary exosomal miR-145-5p and miR-27a-3p are promising noninvasive biomarkers for DKD diagnosis. In particular, miR-145-5p was highly specific and sensitive to DKD. MiR-145-5p may be involved in signaling pathways, including MAPK, TGF-β, FOXO, Ras, and AGE-RAGE pathways, which are related to the pathological processes, including inflammation, apoptosis, and fibrosis of DKD. These hint that miR-145-5p is a potential therapeutic target for DKD.
Diabetic kidney disease (DKD) is the primary cause of end-stage renal disease due to its irreversible and rapidly progressive evolution. DKD remains a serious threat to the lives of diabetic patients.
Early diagnosis and specific treatment can prevent DKD progression. Urinary exosomal microRNAs (miRNAs) are generally derived from renal cells and directly mirror the pathological changes in the kidney. Urinary exosomal miRNAs are remarkably stable and highly tissue-specific for the kidney and may act as promising biomarkers for DKD.
To explore whether urinary exosomal miRNAs from diabetic patients can serve as noninvasive biomarkers for the early diagnosis of DKD.
Patients with type 2 diabetes mellitus (T2DM) were enrolled and divided into a DM group, diabetic patients without albuminuria, and a DKD group, diabetic patients with a urinary albumin to creatinine ratio of ≥ 30 mg/g. Healthy subjects were included in the normal control group. The relative expressions of urinary exosomal miR-145-5p, miR-27a-3p, and miR-29c-3p were detected using real-time quantitative polymerase chain reaction. Correlation analysis, receiver operating characteristic analysis, and bioinformatics analysis were used to explore the potential of urinary exosomal miR-145-5p and miR-27a-3p as DKD biomarkers.
The expression of urinary exosomal miR-145-5p and miR-27a-3p was significantly upregulated in the DKD group. They were closely related to kidney damage and abnormal glycolipid metabolism in T2DM patients. Exosomal miR-145-5p had higher sensitivity and specificity for diagnosing DKD; combining miR-145-5p and miR-27a-3p increased their diagnostic efficiency. Bioinformatics analysis suggested that miR-145-5p regulated various molecular biological functions and sig
Urinary exosomal miR-145-5p and miR-27a-3p may serve as novel noninvasive diagnostic biomarkers for DKD.
Urinary exosomal miR-145-5p and miR-27a-3p may complement traditional DKD diagnostic methods. They may also be effective therapeutic targets for DKD cell-free therapy in the future.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Endocrinology and metabolism
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C, C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Arumugam VA, India; Cai L, United States; Ding H, China; Martinez-Castelaoa A, Spain S-Editor: Wang JJ L-Editor: A P-Editor: Xu ZH
| 1. | Alicic RZ, Rooney MT, Tuttle KR. Diabetic Kidney Disease: Challenges, Progress, and Possibilities. Clin J Am Soc Nephrol. 2017;12:2032-2045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1257] [Cited by in RCA: 1796] [Article Influence: 224.5] [Reference Citation Analysis (0)] |
| 2. | Ma X, Ma J, Leng T, Yuan Z, Hu T, Liu Q, Shen T. Advances in oxidative stress in pathogenesis of diabetic kidney disease and efficacy of TCM intervention. Ren Fail. 2023;45:2146512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 66] [Reference Citation Analysis (0)] |
| 3. | Kanwar YS, Wada J, Sun L, Xie P, Wallner EI, Chen S, Chugh S, Danesh FR. Diabetic nephropathy: mechanisms of renal disease progression. Exp Biol Med (Maywood). 2008;233:4-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 447] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 4. | Xiao L, Wang M, Yang S, Liu F, Sun L. A glimpse of the pathogenetic mechanisms of Wnt/β-catenin signaling in diabetic nephropathy. Biomed Res Int. 2013;2013:987064. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 5. | Audzeyenka I, Bierżyńska A, Lay AC. Podocyte Bioenergetics in the Development of Diabetic Nephropathy: The Role of Mitochondria. Endocrinology. 2022;163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 49] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 6. | Retnakaran R, Cull CA, Thorne KI, Adler AI, Holman RR; UKPDS Study Group. Risk factors for renal dysfunction in type 2 diabetes: U.K. Prospective Diabetes Study 74. Diabetes. 2006;55:1832-1839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 646] [Cited by in RCA: 696] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 7. | Yamazaki T, Mimura I, Tanaka T, Nangaku M. Treatment of Diabetic Kidney Disease: Current and Future. Diabetes Metab J. 2021;45:11-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 147] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 8. | Wang C, Li Z, Liu Y, Yuan L. Exosomes in atherosclerosis: performers, bystanders, biomarkers, and therapeutic targets. Theranostics. 2021;11:3996-4010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 125] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 9. | Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3378] [Cited by in RCA: 3537] [Article Influence: 321.5] [Reference Citation Analysis (0)] |
| 10. | Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6920] [Cited by in RCA: 6572] [Article Influence: 1314.4] [Reference Citation Analysis (0)] |
| 11. | Lu Y, Liu D, Feng Q, Liu Z. Diabetic Nephropathy: Perspective on Extracellular Vesicles. Front Immunol. 2020;11:943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 85] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 12. | Lv J, Wu Y, Mai Y, Bu S. Noncoding RNAs in Diabetic Nephropathy: Pathogenesis, Biomarkers, and Therapy. J Diabetes Res. 2020;2020:3960857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 13. | Yu X, Odenthal M, Fries JW. Exosomes as miRNA Carriers: Formation-Function-Future. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 299] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 14. | Ishii H, Kaneko S, Yanai K, Aomatsu A, Hirai K, Ookawara S, Ishibashi K, Morishita Y. MicroRNAs in Podocyte Injury in Diabetic Nephropathy. Front Genet. 2020;11:993. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 15. | Vitorino R, Ferreira R, Guedes S, Amado F, Thongboonkerd V. What can urinary exosomes tell us? Cell Mol Life Sci. 2021;78:3265-3283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 16. | Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1443] [Cited by in RCA: 1559] [Article Influence: 155.9] [Reference Citation Analysis (0)] |
| 17. | Ghai V, Wu X, Bheda-Malge A, Argyropoulos CP, Bernardo JF, Orchard T, Galas D, Wang K. Genome-wide Profiling of Urinary Extracellular Vesicle microRNAs Associated With Diabetic Nephropathy in Type 1 Diabetes. Kidney Int Rep. 2018;3:555-572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 18. | Cho NJ, Kim DY, Kwon SH, Ha TW, Kim HK, Lee MR, Chun SW, Park S, Lee EY, Gil HW. Urinary exosomal microRNA profiling in type 2 diabetes patients taking dipeptidyl peptidase-4 inhibitor compared with sulfonylurea. Kidney Res Clin Pract. 2021;40:383-391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 19. | Park S, Lee K, Park IB, Kim NH, Cho S, Rhee WJ, Oh Y, Choi J, Nam S, Lee DH. The profiles of microRNAs from urinary extracellular vesicles (EVs) prepared by various isolation methods and their correlation with serum EV microRNAs. Diabetes Res Clin Pract. 2020;160:108010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 20. | Zhao Y, Shen A, Guo F, Song Y, Jing N, Ding X, Pan M, Zhang H, Wang J, Wu L, Ma X, Feng L, Qin G. Urinary Exosomal MiRNA-4534 as a Novel Diagnostic Biomarker for Diabetic Kidney Disease. Front Endocrinol (Lausanne). 2020;11:590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 21. | Zang J, Maxwell AP, Simpson DA, McKay GJ. Differential Expression of Urinary Exosomal MicroRNAs miR-21-5p and miR-30b-5p in Individuals with Diabetic Kidney Disease. Sci Rep. 2019;9:10900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 78] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 22. | American Diabetes Association. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2021. Diabetes Care. 2021;44:S15-S33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1098] [Cited by in RCA: 1884] [Article Influence: 471.0] [Reference Citation Analysis (0)] |
| 23. | American Diabetes Association. 11. Microvascular Complications and Foot Care: Standards of Medical Care in Diabetes-2021. Diabetes Care. 2021;44:S151-S167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 246] [Article Influence: 61.5] [Reference Citation Analysis (1)] |
| 24. | Burballa C, Crespo M, Redondo-Pachón D, Pérez-Sáez MJ, Mir M, Arias-Cabrales C, Francés A, Fumadó L, Cecchini L, Pascual J. MDRD or CKD-EPI for glomerular filtration rate estimation in living kidney donors. Nefrologia (Engl Ed). 2018;38:207-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 25. | Han L, Wang S, Li J, Zhao L, Zhou H. Urinary exosomes from patients with diabetic kidney disease induced podocyte apoptosis via microRNA-145-5p/Srgap2 and the RhoA/ROCK pathway. Exp Mol Pathol. 2023;134:104877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 26. | Wang T, Li N, Yuan L, Zhao M, Li G, Chen Y, Zhou H. MALAT1/miR-185-5p mediated high glucose-induced oxidative stress, mitochondrial injury and cardiomyocyte apoptosis via the RhoA/ROCK pathway. J Cell Mol Med. 2023;27:2495-2506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 27. | Tuleta I, Frangogiannis NG. Diabetic fibrosis. Biochim Biophys Acta Mol Basis Dis. 2021;1867:166044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 124] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 28. | Kaur S, Mirza AH, Overgaard AJ, Pociot F, Størling J. A Dual Systems Genetics Approach Identifies Common Genes, Networks, and Pathways for Type 1 and 2 Diabetes in Human Islets. Front Genet. 2021;12:630109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 29. | . In This Issue of Diabetes. Diabetes. 2018;67:535-536. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 30. | Kato M, Yuan H, Xu ZG, Lanting L, Li SL, Wang M, Hu MC, Reddy MA, Natarajan R. Role of the Akt/FoxO3a pathway in TGF-beta1-mediated mesangial cell dysfunction: a novel mechanism related to diabetic kidney disease. J Am Soc Nephrol. 2006;17:3325-3335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 157] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 31. | Barutta F, Bellini S, Guarrera S, Matullo G, Schalkwijk C, Stehouwer CD, Chaturvedi N, Soedamah-Muthu SS, Durazzo M, Gruden G. Association of serum MicroRNA-145-5p levels with microvascular complications of type 1 Diabetes: The EURODIAB prospective complications study. Diabetes Res Clin Pract. 2022;190:109987. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 32. | Wei B, Liu YS, Guan HX. MicroRNA-145-5p attenuates high glucose-induced apoptosis by targeting the Notch signaling pathway in podocytes. Exp Ther Med. 2020;19:1915-1924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 33. | Dahlmans D, Houzelle A, Jörgensen JA, Phielix E, Lindeboom L, Hesselink MKC, Schrauwen P, Hoeks J. Evaluation of Muscle microRNA Expression in Relation to Human Peripheral Insulin Sensitivity: A Cross-Sectional Study in Metabolically Distinct Subject Groups. Front Physiol. 2017;8:711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 34. | Zhou Z, Wan J, Hou X, Geng J, Li X, Bai X. MicroRNA-27a promotes podocyte injury via PPARγ-mediated β-catenin activation in diabetic nephropathy. Cell Death Dis. 2017;8:e2658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 85] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 35. | Guo J, Li J, Zhao J, Yang S, Wang L, Cheng G, Liu D, Xiao J, Liu Z, Zhao Z. MiRNA-29c regulates the expression of inflammatory cytokines in diabetic nephropathy by targeting tristetraprolin. Sci Rep. 2017;7:2314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 36. | Medeiros T, Myette RL, Almeida JR, Silva AA, Burger D. Extracellular Vesicles: Cell-Derived Biomarkers of Glomerular and Tubular Injury. Cell Physiol Biochem. 2020;54:88-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 37. | Delić D, Wiech F, Urquhart R, Gabrielyan O, Rieber K, Rolser M, Tsuprykov O, Hasan AA, Krämer BK, Baum P, Köhler A, Gantner F, Mark M, Hocher B, Klein T. Linagliptin and telmisartan induced effects on renal and urinary exosomal miRNA expression in rats with 5/6 nephrectomy. Sci Rep. 2020;10:3373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 38. | Barutta F, Tricarico M, Corbelli A, Annaratone L, Pinach S, Grimaldi S, Bruno G, Cimino D, Taverna D, Deregibus MC, Rastaldi MP, Perin PC, Gruden G. Urinary exosomal microRNAs in incipient diabetic nephropathy. PLoS One. 2013;8:e73798. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 211] [Cited by in RCA: 254] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 39. | Zhang S, Wu J, Zhu X, Song H, Ren L, Tang Q, Xu X, Liu C, Zhang J, Hu W, Liu Z, Shi S. A novel approach to identify the mechanism of miR-145-5p toxicity to podocytes based on the essential genes targeting analysis. Mol Ther Nucleic Acids. 2021;26:749-759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 40. | Castaño C, Kalko S, Novials A, Párrizas M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc Natl Acad Sci U S A. 2018;115:12158-12163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 293] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 41. | Yu Y, Bai F, Qin N, Liu W, Sun Q, Zhou Y, Yang J. Non-Proximal Renal Tubule-Derived Urinary Exosomal miR-200b as a Biomarker of Renal Fibrosis. Nephron. 2018;139:269-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 42. | Blaine J, Dylewski J. Regulation of the Actin Cytoskeleton in Podocytes. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 87] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 43. | Kanwar YS, Sun L, Xie P, Liu FY, Chen S. A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annu Rev Pathol. 2011;6:395-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 439] [Cited by in RCA: 586] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 44. | Zhou H, Li YJ. Rho kinase inhibitors: potential treatments for diabetes and diabetic complications. Curr Pharm Des. 2012;18:2964-2973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 45. | Zhang Y, Jin D, Kang X, Zhou R, Sun Y, Lian F, Tong X. Signaling Pathways Involved in Diabetic Renal Fibrosis. Front Cell Dev Biol. 2021;9:696542. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 103] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 46. | Li H, Dong A, Li N, Ma Y, Zhang S, Deng Y, Chen S, Zhang M. Mechanistic Study of Schisandra chinensis Fruit Mixture Based on Network Pharmacology, Molecular Docking and Experimental Validation to Improve the Inflammatory Response of DKD Through AGEs/RAGE Signaling Pathway. Drug Des Devel Ther. 2023;17:613-632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |