Published online Jun 15, 2022. doi: 10.4239/wjd.v13.i6.434
Peer-review started: January 27, 2022
First decision: April 18, 2022
Revised: April 24, 2022
Accepted: May 28, 2022
Article in press: May 28, 2022
Published online: June 15, 2022
Processing time: 131 Days and 5.6 Hours
Endothelin (ET)-traps are Fc-fusion proteins with a design based on the physiological receptors of ET-1. Previous work has shown that use of the selected ET-traps potently and significantly reduces different markers of diabetes pathology back to normal, non-disease levels.
To demonstrate the selected ET-traps potently and significantly bind to ET-1.
We performed phage display experiments to test different constructs of ET-traps, and conducted bio-layer interferometry binding assays to verify that the selected ET-traps bind specifically to ET-1 and display binding affinity in the double-digit picomolar range (an average of 73.8 rM, n = 6).
These experiments have confirmed our choice of the final ET-traps and provided proof-of-concept for the potential use of constructs as effective biologics for diseases associated with pathologically elevated ET-1.
There is increased need for such therapeutics as they could help save millions of lives around the world.
Core Tip: This study verified the specificity of endothelin (ET)-traps, which are an Fc-based fusion protein that acts as a potential therapeutic for various cET-1 related disorders, including diabetes and chronic kidney disease. ET-traps, unlike ET receptor antagonists, do not completely block the ET system and hence have minimal side effects. ET-traps would help save millions of lives around the world.
- Citation: Jain A, Bozovicar K, Mehrotra V, Bratkovic T, Johnson MH, Jha I. Investigating the specificity of endothelin-traps as a potential therapeutic tool for endothelin-1 related disorders. World J Diabetes 2022; 13(6): 434-441
- URL: https://www.wjgnet.com/1948-9358/full/v13/i6/434.htm
- DOI: https://dx.doi.org/10.4239/wjd.v13.i6.434
Endothelin-1 (ET-1) is a vasoactive peptide synthesized and secreted by a diverse range of cells, and thus implicated in signaling events in a wide variety of target tissues[1]. ET-1 plays a key role in physiological functions. However, supraphysiological levels of ET-1 induce pathology and are implicated in a host of different diseases, including cardiovascular disease[2-4], neurodegenerative disorders[5-8], chronic kidney disease, different cancers, such as prostate cancer[1,9,10], pregnancy disorders like preeclampsia[7,11], as well as diabetes[8,12,13]. Given that a key feature of these diseases is elevated ET-1 Levels, one proposed strategy of therapeutic intervention is to target the increased levels of ET-1. To this end, we have created ET-traps, molecular constructs that bind and sequester increased levels of endogenous ET-1.
Diabetes is a serious metabolic complication that affects about more than 7% of the world population[14]. An increase in different extracellular matrix (ECM) proteins has been found to be a key pathological factor of diabetes[15,16]. The study by Jain et al[12] found an increase in collagen 4α1 and fibronectin both at the mRNA and protein levels. This increase was found in heart and kidney tissues and was found to be ET-1 dependent[12]. In addition, the increase in ECM proteins due to high glucose levels was found to be mediated via ET-1[17]. ET-1 Levels are in fact increased in patients with diabetes compared with control subjects[18,19]. Accordingly, our previous in vitro work confirmed the ET-traps to have an efficacious effect on cells treated with a pathological dose of ET-1, as well as those treated with pathologically high glucose (25 mmol/L)[12]. We also established the proof-of-concept (PoC) for ET-traps as a therapeutic in the diabetes disease space at the in vivo level. The use of ET-traps gave a significant reduction in different markers of diabetes disease pathology, which suggested the ET-traps could be considered a therapeutic for diabetes with a novel mechanism of action. Importantly, the ET-traps were found to be non-toxic at the proposed therapeutic concentration both in vitro and in vivo[12,13].
ET-1 exerts its effects by binding to the endothelin A and B receptors, two highly homologous cell-surface proteins that belong to the G-protein-coupled receptor superfamily[20]. The two receptors share about 60% similarity at the level of primary structure[1], i.e. both receptors exhibit a high polypeptide sequence identity with each other. Nevertheless, the two receptors show a clear distinction in ligand binding selectivity based on their ligand-binding domains.
Orry et al[21] constructed a model of interaction of the ET-1 peptide with the endothelin A receptor, where ET-1 makes contacts with both the N-terminal receptor domain and two different extracellular loops (ECL).
Further, amino acids of the C-terminal and residues in the third intracellular loop are important for ET-1 binding[22]. In this study, we performed binding affinity experiments to ascertain that our selected ET-traps bind specifically just to ET-1.
The ETs are a family of potent vasoactive peptides. ET-1 has two paralogs in the ET family; ET-2 and ET-3[23]. ET-1 was identified by Yanagisawa et al[24] in 1988. A year later, 2 homologs of ET-1 were discovered; ET-2 and ET-3[25].
ET-2 is a peptide encoded by the EDN2 gene located on chromosome 1 in humans[26]. ET-2 has a key role in ovarian physiology[25]. Previous research findings have also revealed that ET-2 is critical for the growth and survival of postnatal mice and plays important roles in energy homeostasis, thermoregulation, and the maintenance of lung function[26].
ET-3 is a peptide that in humans is encoded by the EDN3 gene[27]. The active peptide is a ligand for ET receptor type B (EDNRB). The interaction of this ET with EDNRB is essential for development of neural crest-derived cell lineages, such as melanocytes and enteric neurons[28].
Therefore, both the ET-1 paralogs (ET-2 and ET-3) are essential for different physiological processes and so it is important that any ET-1 sequestering agent selectively targets ET-1 and hence the problems associated with increased expression of ET-1 to avoid disrupting the remaining processes of the ET system.
In this study, we first performed phage display experiments to ascertain the binding of the ET-traps to ET-1. Phage display is one of the most powerful and widely used laboratory techniques for the study of protein–protein, protein–peptide and protein–DNA interactions[29]. This technology is based on expressing the protein or peptide of interest on bacterial virus protein coat, allowing the study of molecular interaction between the virion-displayed ligand (in this case ET-traps) and an immobilized target (i.e. ET-1).
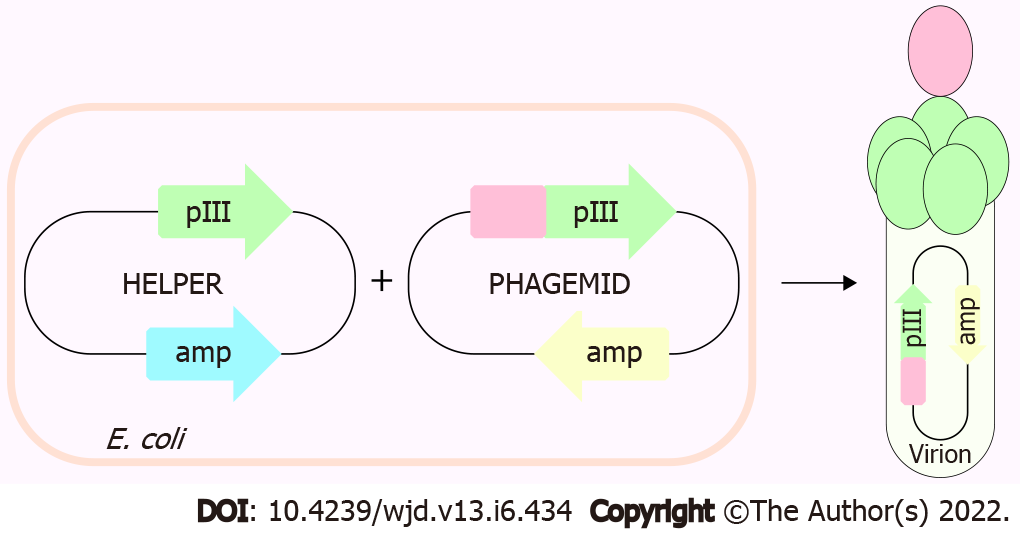
ET-traps constructs were cloned and displayed in a monovalent phage display system using the 3 + 3 display approach (Figure 1). Each codon-optimized construct (Genscript) was amplified by polymerase chain reaction from its parent plasmid pUC18 using forward and reverse primers with overhangs harboring NcoI and NotI restriction sites. Reaction mixtures were subjected to agarose gel electrophoresis, amplicons were purified with QIAEX II gel extraction kit (Qiagen), and subsequently digested with NcoI/NotI restriction enzymes alongside pIT2 phagemid vector.
Digested inserts were ligated into pIT2 phagemid vector and chemically competent Escherichia coli TG1 were transformed with the resulting recombinant phagemids with the heat-shock method. Phages were amplified and rescued by superinfection with KM13 helper phage. PEG/NaCl was used to precipitate and isolate phage clones which were spectrophotometrically quantified with NanoDrop 1000. Phage titers were calculated using equation 1 (derived by Day and Wiseman)[30] and subjected to phage enzyme-linked immunosorbent assay (ELISA).
MaxiSorp microtiter plate (Nunc) wells were coated with anti-cMyc antibodies (1 µg/mL in PBS) overnight at 4 °C. Wells were blocked with 5% skimmed milk and 100 µL of 5 × 1010 phage clone virions in 0.5% milk/0.1% PBST were added and incubated for 1 h with gentle agitation. After extensive washing, bound phages were detected with anti-M13 monoclonal antibodies conjugated with horseradish peroxidase (GE Healthcare) and chromogenic substrate (3,3’,5,5’- tetramethylbenzidine). Reaction was terminated with 2 M H2SO4 and absorbance was measured at 450 nm. The signals generated were later used for normalization of ET-1 binding activity.
To increase the adsorption surface area, and thus the detection signal, N-biotin-ET-1 (Phoenix pharma) was coupled to paramagnetic streptavidin beads (MyOne Streptavidin T1, Thermo Fisher Scientific; 10 mg/mL beads) as an alternative to the conventional phage ELISA on 96-well plates. 5 µL of beads were incubated with 2.5 × 1010 ET-traps-displaying phages for 1 h in 500 µL of 0.5% milk/0.1% PBST. After washing and detection reaction, paramagnetic beads were captured on a magnet, and the supernatants were transferred to a 96-well plate for absorbance A450 measurements. In parallel, binding of ET-trap constructs to streptavidin beads in absence of ET-1 was analyzed. Absorbance signals were blank-subtracted and normalized according to the relative display levels as determined in anti-cMyc phage ELISA assay.
The gene for the ET-traps was designed and optimized for expression in mammalian cells (HEK293) prior to being synthesised. The sequence was then sub-cloned into a cloning and expression vector for human Fc fusion proteins.
In brief, HEK293 cells were passaged to the optimum stage for transient transfection. Cells were transiently transfected with the appropriate expression vector and cultured for a further 6-14 d. An appropriate volume of cells was transfected with the aim of obtaining 1-5 mg of purified Fc fusion protein. Cultures were harvested and one-step purification performed using affinity chromatography. For this, culture supernatant containing Fc fusion protein was loaded onto a MabSelect SuRe Protein A column at 4 mL/min and washed with PBS pH 7.2. A step elution was performed with sodium citrate buffer (pH 3.0). Eluted protein was neutralised with 10% (v/v) Tris buffer (pH 9.0). Upon purification, the Fc fusion protein was buffer exchanged into PBS pH 7.4. The protein was analysed for purity by SDS-PAGE and concentration determined by UV spectroscopy (at 280 nm).
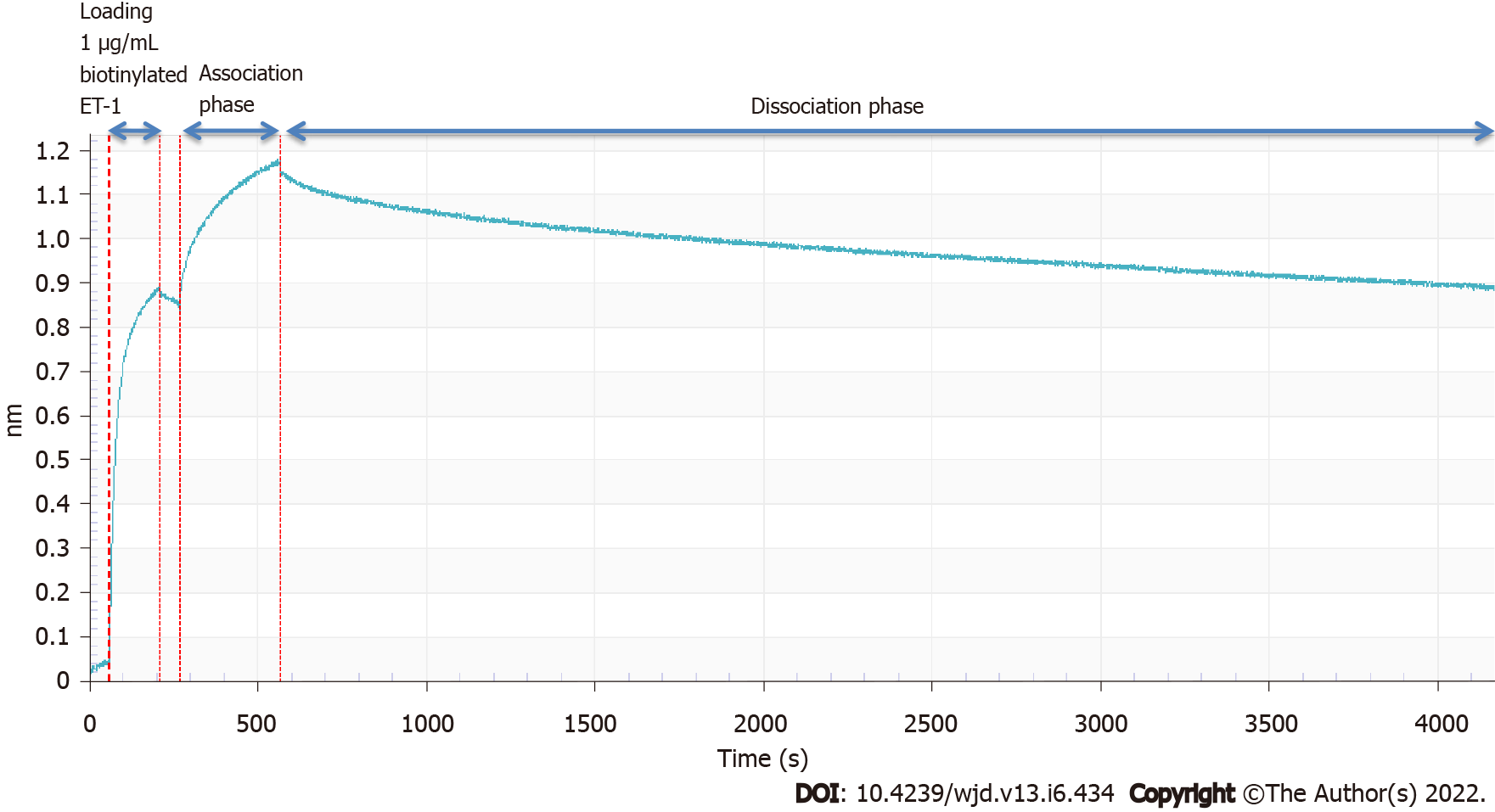
For the binding affinity measurement, we employed the use of the Octet Red96 system (Patel et al 2013). In brief, the kinetics of the selected ET-traps binding to biotinylated ET-1 (Phoenix Pharmaceuticals) was determined using the Octet Red96 system (ForteBio, Menlo Park, CA). The buffer for the assays was PBS with 0.01% (w/v) bovine serum albumin and 0.002% Tween20. The measurements were carried out at 30 °C. 1 µg/mL bio-ET-1 was captured on dip-and-read streptavidin sensors, followed by binding of the selected ET-traps at 500 nM concentration. The ForteBio Octet analysis software (ForteBio, Menlo Park, CA) was used to generate the sensorgram.
Previous work identified a strong binder to ET-1[31]. In this study, we tested different sequence combinations of ET-traps that could also bind ET-1. We performed phage display experiments to ascertain this.
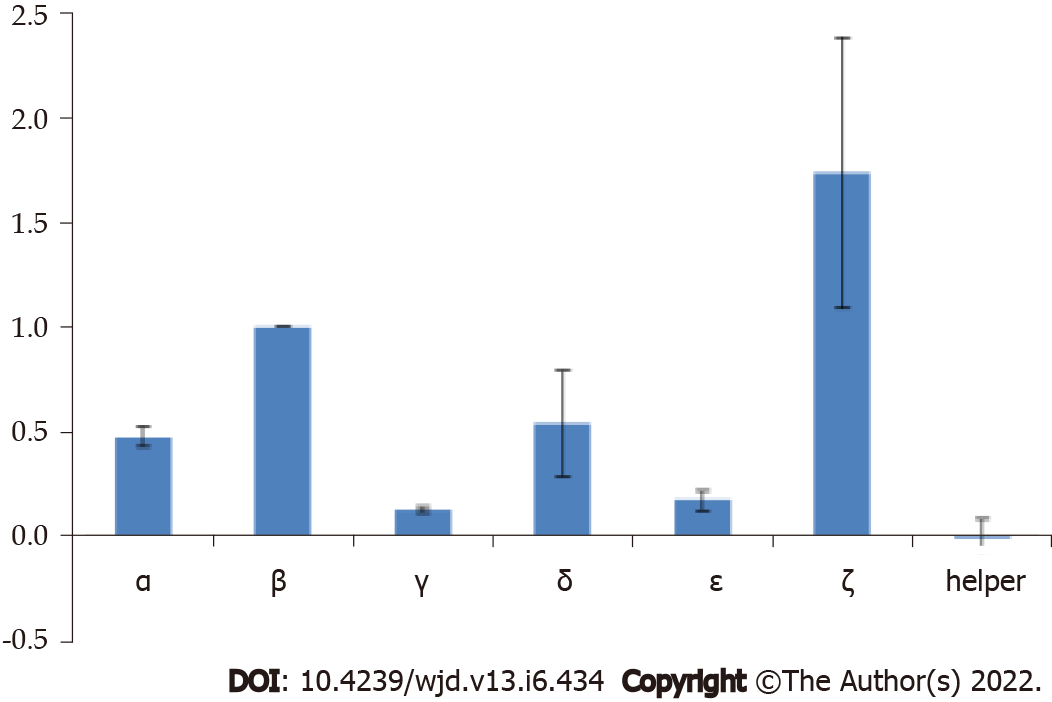
Phage display is a powerful technique commonly used today to identify different protein–protein interactions. We displayed individual ET-traps in a monovalent setting (i.e. 3 + 3 display type[32]) to prevent avidity effects on binding to ET-1. The cMyc-tag peptide present in the linker region that tethers ET-traps to the anchoring phage coat protein p3 allows for assessment of constructs’ display levels by phage ELISA against anti-cMyc antibody. These were, in turn, used to normalize signals from phage ELISA where binding of ET-traps to biotinylated ET-1 was analyzed (Figure 2).
The construct ζ gave strong binding to ET-1 in phage display experiments, but the Fc-fusion molecule was not stable and the results could not be replicated with the soluble fusion protein. The phage experiments confirmed that construct β indeed gave consistent, high binding. It was further observed that in the form of an Fc-fusion construct β showed high binding affinity to ET-1 consistently (Figure 3).
This paper discusses the characterization of ET-traps that might be useful in the treatment of ET-1 related diseases or disorders, such as preeclampsia, cardiovascular diseases, chronic kidney disease, diabetes or neurodegenerative disorders[11,18,19,33-36]. Previous work has shown that the use of the selected ET-traps gave a therapeutic effect on reducing different markers of diabetes-induced disease pathology[12,13]. The ET-traps helped reduce different markers of diabetes disease pathology, such as over-expression of ECM proteins, proteinuria and tissue damage to kidneys and heart. This effect was found to be statistically significant both in vitro and in vivo. The ET-traps were designed based on a previous study[22]. The purpose of this study was to ascertain our selection of the ET-traps. Both ECL2 and ECL3, including the flanking transmembrane regions, were found to play an important role in ligand selection[22]. Further, residues in the intracellular loop and of the C-terminus are important for ET-1 binding[22]. These domains were used to create the final ET-traps that gave an efficacious, therapeutic effect in our proof-of-concept studies done both at the in vitro and in vivo levels in the diabetes disease space[12,13]. The final design of our ET-traps ensured that the selected ET-traps do not bind the ET-1 paralogs, which is important for selective activity and thus fewer potential adverse effects.
We began with phage display experiments to test different combinations of potential ET-traps, including the previously selected ET-traps that were used to perform the proof of concept (PoC) work[12,13]. These experiments confirmed the selection of the most efficient ET-traps. Phage experiments were performed on different combinations of sequences (Figure 3). These experiments allowed us to confirm our final ET-traps selection and we then proceeded to test the binding of its cognate Fc fusion to ET-1.
As ET-1 is abundant in the body while ET-2 is almost undetectable, ET-1 was more convenient to research; this assumption has meant ET-2 is relatively under-researched[26]. However, recent research evidence suggests distinct roles and features of ET-2. In mice with the ET-2 gene knocked-out, the animals displayed growth retardation, and were hypothermic and hypoglycemic, which resulted in early mortality[37].
ET receptor antagonists (ERAs) can have a deleterious effect on physiological ET-2 functions by completely blocking the receptors and thereby inhibiting the physiological actions of ET-2. With our ET-traps, we would overcome this; the ET-traps specifically bind to ET-1 and do not block the receptors to effect ET-2 actions like an inhibitor to the ET system might do. The ET-traps have been designed to specifically bind ET-1.
Aberrations in the EDN3 gene that is responsible for producing ET-3 have been associated with congenital disorders involving neural crest-derived cells, like Hirschsprung disease and Waardenburg syndrome[38,39]. This shows that ET-3 is one of the important peptides of the ET family, which is involved in various developmental processes. Further, use of ERAs would essentially block the function of this molecule thereby potentially causing serious birth defects. This again precludes completely blocking the physiological functions of ET-3, as it is one of the important factors for essential developmental processes. Again, with the ET-traps, we would not completely be blocking the ET system, rather just targeting elevated ET-1 Levels upstream. As found in this study, the selected ET-traps bind ET-1 with a high binding affinity in the double-digit picomolar range (an average of 73.8 rM, n = 6). This was also previously found and reported by Jain et al[12] in their diabetes PoC study. This work showed that the selected ET-traps have an efficacious, therapeutic effect in ameliorating diabetes disease pathology[12,13]. This was not associated with any toxic effects as evinced by the toxicology data. This corroborates that the selected ET-traps are efficacious at the working concentration and specific to just ET-1.
The ET-traps were designed to specifically bind ET-1. The results of this study confirm that our selected ET-traps specifically bind to ET-1. This is in agreement with previous PoC studies that detected no toxic effects of the selected ET-traps at the working concentration. This is an important factor for the potential use of ET-traps as a therapeutic.
Endothelin (ET)-1 is a very potent vasoactive peptide that is significantly elevated in different diseases.
We wanted to develop a cure that would target this peptide and would help save millions of lives around the world.
To develop a tool that specifically targets ET-1.
We employed phage display and binding assays.
A very high binding affinity was observed for our selected tool.
Developed a potent tool targeting ET-1.
A new target in drug discovery and development.
This paper is dedicated to Dr. M.L. Mehrotra. We would also like to thank Mr. Ashok Jain for all his dedication and support during this difficult time.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Pharmacology and pharmacy
Country/Territory of origin: United Kingdom
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ahmed AA, Egypt; He Z, China A-Editor: Zhu JQ, China S-Editor: Zhang H L-Editor: A P-Editor: Zhang H
| 1. | Nelson J, Bagnato A, Battistini B, Nisen P. The endothelin axis: emerging role in cancer. Nat Rev Cancer. 2003;3:110-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 414] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 2. | Böhm F, Pernow J. The importance of endothelin-1 for vascular dysfunction in cardiovascular disease. Cardiovasc Res. 2007;76:8-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 353] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 3. | Wernly B, Jung C. Trapping endothelin-1 to hunt down cardiovascular disease? Drug Discov Today. 2019;24:2108-2110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 4. | Ergul A. Endothelin-1 and endothelin receptor antagonists as potential cardiovascular therapeutic agents. Pharmacotherapy. 2002;22:54-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 5. | Ranno E, D'Antoni S, Spatuzza M, Berretta A, Laureanti F, Bonaccorso CM, Pellitteri R, Longone P, Spalloni A, Iyer AM, Aronica E, Catania MV. Endothelin-1 is over-expressed in amyotrophic lateral sclerosis and induces motor neuron cell death. Neurobiol Dis. 2014;65:160-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Ferrari CC, Tarelli R. Parkinson's disease and systemic inflammation. Parkinsons Dis. 2011;2011:436813. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 7. | Jain A. Endothelin-1-induced endoplasmic reticulum stress in disease. J Pharmacol Exp Ther. 2013;346:163-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 8. | Jain A, Coffey C, Mehrotra V, Flammer J. Endothelin-1 traps as a potential therapeutic tool: from diabetes to beyond? Drug Discov Today. 2019;24:1937-1942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 9. | Nelson JB, Hedican SP, George DJ, Reddi AH, Piantadosi S, Eisenberger MA, Simons JW. Identification of endothelin-1 in the pathophysiology of metastatic adenocarcinoma of the prostate. Nat Med. 1995;1:944-949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 425] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 10. | Chiao JW, Moonga BS, Yang YM, Kancherla R, Mittelman A, Wu-Wong JR, Ahmed T. Endothelin-1 from prostate cancer cells is enhanced by bone contact which blocks osteoclastic bone resorption. Br J Cancer. 2000;83:360-365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 75] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 11. | Jain A. Endothelin-1: a key pathological factor in pre-eclampsia? Reprod Biomed Online. 2012;25:443-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Jain A, Chen S, Yong H, Chakrabarti S. Endothelin-1 traps potently reduce pathologic markers back to basal levels in an in vitro model of diabetes. J Diabetes Metab Disord. 2018;17:189-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Jain A, Mehrotra V, Jha I, Jain A. In vivo studies demonstrate that endothelin-1 traps are a potential therapy for type I diabetes. J Diabetes Metab Disord. 2019;18:133-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | Emerging Risk Factors Collaboration, Sarwar N, Gao P, Seshasai SR, Gobin R, Kaptoge S, Di Angelantonio E, Ingelsson E, Lawlor DA, Selvin E, Stampfer M, Stehouwer CD, Lewington S, Pennells L, Thompson A, Sattar N, White IR, Ray KK, Danesh J. Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. Lancet. 2010;375: 2215-2222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3857] [Cited by in RCA: 3452] [Article Influence: 230.1] [Reference Citation Analysis (0)] |
| 15. | Chen S, Mukherjee S, Chakraborty C, Chakrabarti S. High glucose-induced, endothelin-dependent fibronectin synthesis is mediated via NF-kappa B and AP-1. Am J Physiol Cell Physiol. 2003;284:C263-C272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 101] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 16. | Chen S, Feng B, Thomas AA, Chakrabarti S. miR-146a regulates glucose induced upregulation of inflammatory cytokines extracellular matrix proteins in the retina and kidney in diabetes. PLoS One. 2017;12:e0173918. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 17. | Law B, Fowlkes V, Goldsmith JG, Carver W, Goldsmith EC. Diabetes-induced alterations in the extracellular matrix and their impact on myocardial function. Microsc Microanal. 2012;18:22-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Seligman BG, Biolo A, Polanczyk CA, Gross JL, Clausell N. Increased plasma levels of endothelin 1 and von Willebrand factor in patients with type 2 diabetes and dyslipidemia. Diabetes Care. 2000;23:1395-1400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 77] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 19. | Schneider JG, Tilly N, Hierl T, Sommer U, Hamann A, Dugi K, Leidig-Bruckner G, Kasperk C. Elevated plasma endothelin-1 levels in diabetes mellitus. Am J Hypertens. 2002;15:967-972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 118] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 20. | Karet FE, Davenport AP. Endothelin and the human kidney: a potential target for new drugs. Nephrol Dial Transplant. 1994;9:465-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 21. | Orry AJ, Wallace BA. Modeling and docking the endothelin G-protein-coupled receptor. Biophys J. 2000;79:3083-3094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 22. | Adachi M, Hashido K, Trzeciak A, Watanabe T, Furuichi Y, Miyamoto C. Functional domains of human endothelin receptor. J Cardiovasc Pharmacol. 1993;22 Suppl 8:S121-S124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 23. | Davenport AP, Hyndman KA, Dhaun N, Southan C, Kohan DE, Pollock JS, Pollock DM, Webb DJ, Maguire JJ. Endothelin. Pharmacol Rev. 2016;68:357-418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 616] [Cited by in RCA: 565] [Article Influence: 62.8] [Reference Citation Analysis (0)] |
| 24. | Yanagisawa M, Kurihara H, Kimura S, Tomobe Y, Kobayashi M, Mitsui Y, Yazaki Y, Goto K, Masaki T. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature. 1988;332:411-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8220] [Cited by in RCA: 7980] [Article Influence: 215.7] [Reference Citation Analysis (0)] |
| 25. | Inoue A, Yanagisawa M, Kimura S, Kasuya Y, Miyauchi T, Goto K, Masaki T. The human endothelin family: three structurally and pharmacologically distinct isopeptides predicted by three separate genes. Proc Natl Acad Sci U S A. 1989;86:2863-2867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1654] [Cited by in RCA: 1632] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 26. | Ling L, Maguire JJ, Davenport AP. Endothelin-2, the forgotten isoform: emerging role in the cardiovascular system, ovarian development, immunology and cancer. Br J Pharmacol. 2013;168:283-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 27. | Hofstra RM, Osinga J, Tan-Sindhunata G, Wu Y, Kamsteeg EJ, Stulp RP, van Ravenswaaij-Arts C, Majoor-Krakauer D, Angrist M, Chakravarti A, Meijers C, Buys CH. A homozygous mutation in the endothelin-3 gene associated with a combined Waardenburg type 2 and Hirschsprung phenotype (Shah-Waardenburg syndrome). Nat Genet. 1996;12:445-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 214] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 28. | Baynash AG, Hosoda K, Giaid A, Richardson JA, Emoto N, Hammer RE, Yanagisawa M. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 720] [Cited by in RCA: 686] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 29. | Wu CH, Liu IJ, Lu RM, Wu HC. Advancement and applications of peptide phage display technology in biomedical science. J Biomed Sci. 2016;23:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 229] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 30. | Day LA, Wiseman L. A Comparison of DNA Packaging in the Virions of fd, Xf, and Pf1. Cold Spring Harbor: New York, 1978: 605-625. [RCA] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 31. | Jain A, Mehrotra V, Yong H, Hiremath K, Jain A, Johnson M, Jha I. Creating a Soluble Binder to Endothelin-1 based on the natural ligand binding domains of the endothelin-1 (G-protein-coupled) receptor. Int J Pept Res Ther. 2019;25:107-114. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 32. | Smith GP, Petrenko VA. Phage Display. Chem Rev. 1997;97:391-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1233] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 33. | Zeiher AM, Goebel H, Schächinger V, Ihling C. Tissue endothelin-1 immunoreactivity in the active coronary atherosclerotic plaque. A clue to the mechanism of increased vasoreactivity of the culprit lesion in unstable angina. Circulation. 1995;91:941-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 154] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 34. | Haufschild T, Shaw SG, Kesselring J, Flammer J. Increased endothelin-1 plasma levels in patients with multiple sclerosis. J Neuroophthalmol. 2001;21:37-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 70] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 35. | Pache M, Kaiser HJ, Akhalbedashvili N, Lienert C, Dubler B, Kappos L, Flammer J. Extraocular blood flow and endothelin-1 plasma levels in patients with multiple sclerosis. Eur Neurol. 2003;49:164-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 36. | Sethi AS, Lees DM, Douthwaite JA, Dawnay AB, Corder R. Homocysteine-induced endothelin-1 release is dependent on hyperglycaemia and reactive oxygen species production in bovine aortic endothelial cells. J Vasc Res. 2006;43:175-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 37. | Chang I, Bramall AN, Baynash AG, Rattner A, Rakheja D, Post M, Joza S, McKerlie C, Stewart DJ, McInnes RR, Yanagisawa M. Endothelin-2 deficiency causes growth retardation, hypothermia, and emphysema in mice. J Clin Invest. 2013;123:2643-2653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 38. | Liu Q, Cheng J, Lu Y, Zhou J, Wang L, Yang C, Yang G, Yang H, Cao J, Zhang Z, Sun Y. The clinical and genetic research of Waardenburg syndrome type I and II in Chinese families. Int J Pediatr Otorhinolaryngol. 2020;130:109806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 39. | Sánchez-Mejías A, Fernández RM, López-Alonso M, Antiñolo G, Borrego S. New roles of EDNRB and EDN3 in the pathogenesis of Hirschsprung disease. Genet Med. 2010;12:39-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |