Published online Nov 15, 2021. doi: 10.4239/wjd.v12.i11.1957
Peer-review started: May 18, 2021
First decision: July 4, 2021
Revised: July 14, 2021
Accepted: October 27, 2021
Article in press: October 27, 2021
Published online: November 15, 2021
Processing time: 180 Days and 22.8 Hours
Monogenic forms of diabetes (MFD) are single gene disorders. Their diagnosis is challenging, and symptoms overlap with type 1 and type 2 diabetes.
To identify the genetic variants responsible for MFD in the Pakistani population and their frequencies.
A total of 184 patients suspected of having MFD were enrolled. The inclusion criterion was diabetes with onset below 25 years of age. Brief demographic and clinical information were taken from the participants. The maturity-onset diabetes of the young (MODY) probability score was calculated, and glutamate decarboxylase ELISA was performed. Antibody negative patients and features resembling MODY were selected (n = 28) for exome sequencing to identify the pathogenic variants.
A total of eight missense novel or very low-frequency variants were identified in 7 patients. Three variants were found in genes for MODY, i.e. HNF1A (c.169C>A, p.Leu57Met), KLF11 (c.401G>C, p.Gly134Ala), and HNF1B (c.1058C>T, p.Ser353Leu). Five variants were found in genes other than the 14 known MODY genes, i.e. RFX6 (c.919G>A, p.Glu307Lys), WFS1 (c.478G>A, p.Glu160Lys) and WFS1 (c.517G>A, p.Glu173Lys), RFX6 (c.1212T>A, p.His404Gln) and ZBTB20 (c.1049G>A, p.Arg350His).
The study showed wide spectrum of genetic variants potentially causing MFD in the Pakistani population. The MODY genes prevalent in European population (GCK, HNF1A, and HNF4a) were not found to be common in our population. Identification of novel variants will further help to understand the role of different genes causing the pathogenicity in MODY patient and their proper management and diagnosis.
Core Tip: There was a lack of data on monogenic forms of diabetes (MFD) from Pakistan, therefore this study was designed to determine the genetic variants responsible for MFD in the country. The study identified wide spectrum of genetic variants potentially causing MFD. The identification of novel variants paved the way for better understanding of genetic landscape and risk factors of MFD in the country.
- Citation: Rafique I, Mir A, Siddiqui S, Saqib MAN, Fawwad A, Marchand L, Adnan M, Naeem M, Basit A, Polychronakos C. Comprehensive genetic screening reveals wide spectrum of genetic variants in monogenic forms of diabetes among Pakistani population. World J Diabetes 2021; 12(11): 1957-1966
- URL: https://www.wjgnet.com/1948-9358/full/v12/i11/1957.htm
- DOI: https://dx.doi.org/10.4239/wjd.v12.i11.1957
Monogenic forms of diabetes (MFD) result from changes in single gene. Maturity-onset diabetes of the young (MODY) is a monogenic form of diabetes. It is inherited in an autosomal dominant pattern[1] and is often misdiagnosed as type 1 or type 2 diabetes[2]. It is estimated that MODY accounts for 1%-2% of all the diabetic cases[3]. There are fourteen sub-types of MODY listed in OMIM(On-Line Mendelian Inheritance in Man) (#606391). Most common of them are GCK, HNF1A, and HNF4A.The other listed genes are PDX1, HNF1B, NEUROD1, KLF11, CEL, PAX4, INS, BLK, ABCC8, KCNJ11, and APPL1[1]. Mutations in a number of additional genes are also known to cause diabetes.
The subtypes of MODY differ with respect to hyperglycemia, age of disease onset, treatment pattern, and complications reported[4].Therefore timely diagnosis is of vital importance in MFD. Previously, most common genes, i.e. GCK, HNF1A, and HNF4A were generally sequenced for suspected cases, but n ow with the advent of latest technologies, targeted panel sequencing or exome sequencing are standard[5,6].
Pakistan has a huge burden of diabetes. The recent surveys showed that one out of every four people in the general population is suffering from diabetes in the country[7,8]. Being a resource poor country, advanced diagnostic facilities are not available to the public. The literature from Pakistan is scarce on MFD[9,10]. Therefore, this study was designed to enroll suspected MFD patients and assess the causal variants involved. To our knowledge, this was the first comprehensive study to be conducted in Pakistan on MFD.
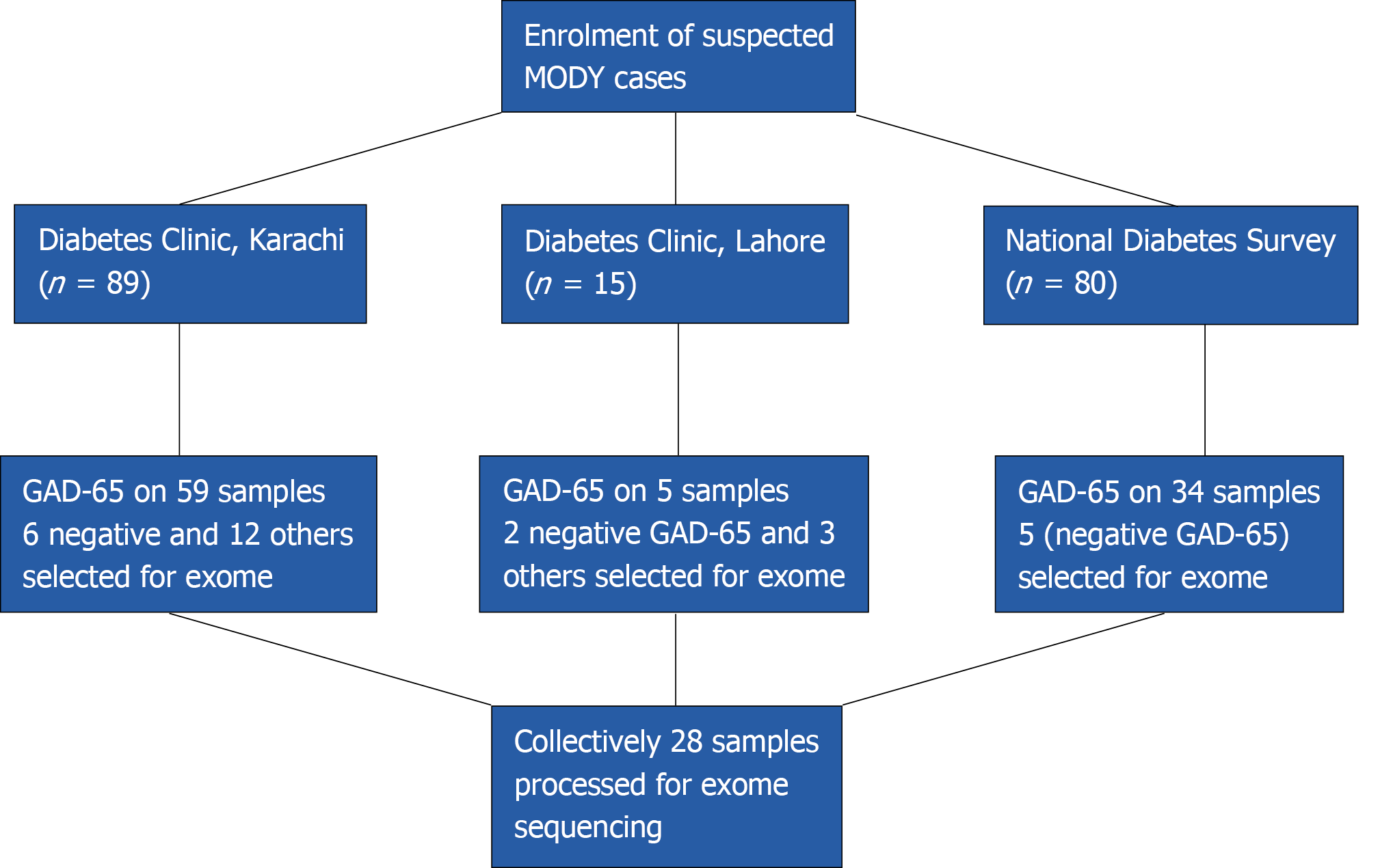
A total of 184 patients with diabetes onset before 25 years of age were considered for participation in the study. The participants were chosen from the sources indicated below:
The record of patients who had an onset of disease before 25 years of age was obtained from National Diabetes Survey data. This was a population-based survey carried out all over the four provinces of Pakistan. The detailed methodology is described elsewhere[7]. According to World Health Organization, the patients were diagnosed as diabetic if having fasting glucose level > 126 mg/dL (7.0 mmol/L) or HbA1c > 6.5% (48 mmol/mol). Clinical information in 34 cases suggested MFD [young onset, low body mass index (BMI), mild hyperglycemia]. The glutamate decarboxylase 65 (GAD-65) ELISA testing was negative in 5 cases, which were selected for exome sequencing.
A total of 15 patients were enrolled from The Diabetes Clinic at PHRC Research Centre, Fatima Jinnah Medical University, Lahore. The inclusion criteria were onset of diabetes before 25 years of age with preferential first-degree family history of diabetes. The demographic information along with history of disease (onset, complications, treatment details and family history) was collected on proforma. On the basis of BMI and preserved fasting C-peptide, GAD-65 ELISA was performed on three samples where serum was available. The two that were found negative for GAD-65 and, along with another three patients having clinical features like low BMI but for whom serum was not available, were finally selected for exome sequencing. All the selected subjects had normal c-peptide values (range: 0.8-3.8 ng/mL)
The patients coming to the diabetes clinic for type-1 and type 2 diabetes treatments were enrolled if onset of the disease was below 25 years of age, with family history of diabetes as preference. Information on demography, treatment, and diagnosis was taken on pre-designed proforma. Height and weight were recorded for BMI. The GAD-65 ELISA was performed on 59 patients. Patients that were GAD-65 negative and additional patients with low BMI but no available serum for testing were selected for exome sequencing (n = 18) (Figure 1).
A total of 28 patients were selected for exome sequencing. Their median age at diagnosis was 18 years and median BMI was 22. Among them, 17 were taking treatment (14 insulin and 3 were taking OHA agent) and 5 were not taking any treatment.
The DNA was extracted using a QIAamp DNA mini kit (QIAGEN). The GAD-65 autoantibody test was done by using a KRONUS Elisa kit. C-peptide test was commercially tested in a diagnostic laboratory. Exome sequencing was performed by Genome Quebec, Canada. Exome sequencing was carried out with 50 Mb Agilent Sure select array and sequencing on Illumina Hi-seq at 50 × depth.
Written informed consent was taken from participants/parents/guardians prior to enrollment. The Ethical Clearance was taken from Institutional Bioethics Committee of Pakistan Health Research Council and Ethical Review Board of Pakistan Institute of Medical Sciences, Shaheed Zulfiqar Ali Bhutto Medical University, Islamabad.
For bioinformatics analysis, the FastQ files were processed using the best practices recommendations of the genome analysis tool kit (GATK). The reads were mapped to human reference genome GRCh37 using Burrow-Wheeler alignment (BWA-MEM). The Picard tool was used to mark and remove duplicate alignments and then indel realignment and base quality score recalibration was done. The gVCFs were generated with GATK HaplotypeCaller and joint variant calling was done. The variants with low map reads (DP < 20) and low genotype quality (GQ < 20) were discarded. Annovar was used to annotate the variants. UTR, synonymous, and intronic (unless splicing) variants were discarded by standard procedure to focus on protein altering ones. These variants were filtered for frequency in three public databases (Exome Aggregation Consortium, 1000 genomes, and gnomAD version v2.1.1). For dominant genes, maximal allele frequency cutoff was 0.0001 in any population while it was 0.005 for recessive genes (ACMG PM2 criterion). The missense variants were selected only if predicted disease-causing by the majority of 10 algorithms used (see legend to Table 1) which satisfies PP3 by ACMG/AMP criteria). The computational evidence supports deleterious effects on the gene[11]. Analysis of the exome focused on the genes listed in the University of Chicago monogenic diabetes panel (https://dnatesting.uchicago.edu/tests/monogenic-diabetes-panel). Variant coordinates were searched for functional domains in uniport.org (results shown in Table 1).
| Case ID | Chromosome position | Gene symbol | cDNA change | Protein change | Maxfreq. | In silico prediction | Protein region byUniport (uniport.org) |
| 612 | 12; 1416740 | HNF1A | c. 169C>A | p.Leu57Met | 0 | T, NA, D, D, N, L, D, D, D, D | DNA-interacting |
| 705 | 2; 10187916 | KLF11 | c. 401G>C | p.Gly134Ala | 0 | T, N, N, T, N, N, T, T, T, N | NA |
| 830 | 6;117237424 | RFX6 | c. 919G>A | p.Glu307Lys | 0.0001 | T, D, D, T, N, N, T, T, D, D | NA |
| P-9 | 17; 6070581 | HNF1B | c. 1136C>T | p.Ser379Leu | 0.00003 | NA, D, D, NA, NA, L, D, D, D, D | NA |
| P-17 | 4; 6292941 | WFS1 | c. 478G>A | p.Glu160Lys | 0 | T, N, N, D, N, M, D, D, D, D | NA |
| 4; 6292980 | c. 517G>A | p.Glu173Lys | 0 | D, D, D, D, D, M, T, T, D, D | |||
| P-68 | 6; 117241502 | RFX6 | c. 1212T>A | p.His404Gln | 0.0001 | T, D, D, T, N, L, T, T, T, D | NA |
| P-87 | 3; 114069876 | ZBTB20 | c. 1049G>A | p.Arg350His | 0 | D, D, D, T, N, N, T, T, T, D | NA |
A total of 80 cases selected from the National Diabetes Survey with the criteria of having diabetes and onset of disease before 25 years of age. The median age was 24 years. Most of them (62%) were females and average BMI was 28 kg/m2 (Table 2). Of them, based on clinical criteria and negativity for antibody, five were selected for exome sequencing. The results revealed three novel missense variants identified in three patients. Two of the variants belong to the OMIM-listed MODY genes, i.e. HNF1A (c. 169C>A, pLeu57Met) and KLF11 (c. 401G>C, p.Gly134Ala) and one in RFX6 (c. 919G>A, p.Glu307Lys), a gene recently described as mutated in dominant, MODY-like diabetes.
In the prospective enrollment from Lahore, 15 patients were enrolled. The mean age for onset of disease in patients was 23 years and mean BMI was 21.5 kg/m2. The fasting blood glucose on average was 239.26 mg/dL. The HbA1c ranged from 6% to 11% (Supplementary Table 1). The exome analysis revealed a variant in HNF1B (c. 1058C>T, p.Ser353Leu) for one patient.
In the prospective enrollment from Karachi, 89 diabetic patients were enrolled. The mean age of onset of disease was 16.5 years and the current age was 26.2 years. Mean BMI was 24.3 kg/m2. Most of them (95%) had family history of diabetes. Among them, 93% were taking treatment for diabetes and 68% were taking insulin. About 25% of the patients had a MODY probability score more than 75%. The serum was available for 59 patients. The GAD-65 autoantibody test was conducted on all patients and 15 of them were negative. The patients whose serum was not available were shortlisted on the basis of low BMI, family history, and HbA1c levels.
Among them, 18 were selected for exome sequencing, which revealed potentially causal variants in three patients (Table 1). One had a variant in RFX6 (c. 1212T>A, p.His404Gln) and a second one was a compound heterozygote (Supplementary Figure 1) for WFS1 (c. 478G>A, p.Glu160Lys and c. 517G>A, p.Glu173Lys). Recessive WFS1 mutations cause Wolfram syndrome, but non-syndromic diabetes alone is also seen[12]. A third patient had a variant in ZBTB20 (c. 1049G>A, p.Arg350His).
The mutation in this gene is responsible for causing primrose syndrome[13]. The patient did not have the other manifestations of this syndrome.
All variants reported here were missense, all satisfied the PP3 and PM2 criteria but, being novel and not having previously been reported, all classified as VUS. Nevertheless, our extremely low cut-off for allele frequency of 0.0001 (more than two orders of magnitude lower than the ACMG/AMP cut-off for a VUS), minimizes the probability of spurious variants. The MODY probability score was calculated for all the participants. More than half of the patients in National Diabetes Survey, all patients from Lahore and one fourth of patients from Karachi had probability scores more than 75%. The probability score was calculated by MODY probability calculator developed for Caucasian population.
Exome sequencing of 28 suspected patients for MFD identified missense novel variants in 7 patients (with the caveat that KLF11, BLK, and PAX4 are not universally accepted as genes whose mutation causes diabetes). Previous studies from Pakistan have discussed the importance of diagnosing MFD in Pakistan[9,10]. However, to the best of our knowledge, this is the first comprehensive study from the country to enroll suspected MFD patients for exome sequencing.
We enrolled diabetic patients with early onset of disease below 25 years of age with clinical features suggestive of MFD and negative (or unknown) for GAD65 auto antibodies. The probability score was calculated by using the MODY probability calculator developed for the Caucasian population[14]. However there is a need to validate this with South Asian populations[15].
In determining the type of MFD, ethnic differences play an important role. There is wide variation of prevalence of different MFD types in different areas. The HNF1A MODY type was reported to be more prevalent in northern Europe while GCK MODY types in southern Europe[16]. There were similar findings from United States regarding the three major prevalent types of MODY in their population[17]. The study on Russian children with non-type 1 diabetes showed that the most prevalent MODY type was GCK, with only 18% of variants in other than known MODY genes. The studies from China reported that HNF4A MODY types were relatively less common as compared to Europeans[18,19]. A study from Korea also has a similar finding that common MODY types prevalent in Europe were not common there, but instead they found variants in three new genes, including the WFS1 gene[20]. The MODY landscape in India is also complex, reporting MODY types other than common genes known in European population[21,22]. A study from Oman reported that variants were not found in three common MODY genes[23]. Similar findings were also reported from Tunisia that common MODY types were not found there and concluded that other genes might be responsible for young onset diabetes in their population[24,25]. These discrepant results may be only partially explained by different methodologies and different selection criteria for testing.
We found in our study in a total 28 screened patients that 3 of the patients tested had variants in OMIM-listed MODY genes while 4 had variants were in other genes also known to be mutated in MFD. Similar findings were also reported from Norway[26], France[27] and Sweden[28]. Two novel missense variants were found in RFX6 (c.G919A, p.E307K). In addition to Europeans[29], variants from this gene was also reported in studies from India[22] and Japan[30]. One patient was compound heterozygous at WFS1, the gene mutated in Wolfram syndrome and also responsible for non-syndromic diabetes. WFS1variants have been reported from India[22], China[12,31], Korea[20,32,33], Russia[6], and European ancestry[34]. Finally, one variant was identified in ZBTB20, a transcription factor that regulates the function of beta cells and glucose homeostasis[35-37]. In humans, dominant ZBTB20 mutations cause Primrose syndrome.
The three variants in OMIM-listed MODY genes were found in HNF1A, KLF11, and HNF1B. The HNF1A (MODY 3) is most common type prevalent in some European and Asian countries[38] and variants in this gene have been reported from various countries all around the world[5,39,40]. The patients with HNF1A variants respond well to sulfonylurea therapy[41]. The other variant was found in the KLF11 (MODY 7)[42]; variants from this gene have been reported from France[43] and Japan[44], although recent literature disputes this gene as true MODY gene. One variant was found in HNF1B (MODY 5) as reported to account for 2%-6% of all the diagnosed MODY cases[45]. This type was generally found to be associated with kidney dysfunction[46]. Variants in this gene have been reported from different countries[47-49]. Although it has been considered that KLF11, BLK, and PAX4 are not MODY causing genes, the OMIM and recent literature reported it as involved in MODY[3,6].
Pakistan, being a developing country, is facing a huge burden of diabetes as evident from the recent survey findings that 26% of adults in the general population were suffering from diabetes[7]. There is a need to identify the genetic basis of the diabetes in Pakistan, with large-scale efforts of screening. As Pakistan is a limited-resource society, it is important to develop sensitive and population-specific criteria. We propose our paper contributes as the first step in this direction.
A study reported that 56% of the marriages were consanguineous, and among them, over 49% were first cousin marriages[50,51], suggesting unknown recessive genes, such as seen in non-syndromic WFS1 cases. Studies from Pakistan on MFD were very scarce. According to American Diabetes Association, the diagnosis of MFD should be considered when there is a family history of diabetes with atypical features of diabetes, such as lacking obesity[52]. There is a need to conduct large scale genetic studies on young onset diabetes to understand the genetic aspects from our country.
A wide spectrum of genetic variants involved in MFD was identified from this study. The common genes prevalent in European countries were not found common in this study. The genes other than commonly known MODY genes were identified. There is need for large scale genetic studies on early onset of diabetes in the country.
The data on monogenic forms of diabetes (MFD) was lacking from Pakistan.
The identification of MFD from Pakistan will paved the way for better diagnostics and treatment for patients
To identify the genetic variants for MFD.
Exome sequencing was used.
The wide spectrum of genetic variants was identified.
The MODY genes prevalent in other countries, like those in Europe, were not found common in our population
More studies are required, keeping in view the consanguinity rate in Pakistan
The authors thank all the participants and their guardians for participating in the study. Thanks to Ms. Raheela and Bilal Tahir from Baqai Institute of Diabetology and Endocrinology for assistance in patient enrollment and all National Diabetes Survey of Pakistan members.
Provenance and peer review: Unsolicited article; Externally peer reviewed
Specialty type: Genetics and heredity
Country/Territory of origin: Canada
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D, D, D
Grade E (Poor): 0
P-Reviewer: Sun XD, Wen XL, Xiao X S-Editor: Liu M L-Editor: Filipodia P-Editor: Liu M
| 1. | Urakami T. Maturity-onset diabetes of the young (MODY): current perspectives on diagnosis and treatment. Diabetes Metab Syndr Obes. 2019;12:1047-1056. [PubMed] [DOI] [Full Text] |
| 2. | Thanabalasingham G, Pal A, Selwood MP, Dudley C, Fisher K, Bingley PJ, Ellard S, Farmer AJ, McCarthy MI, Owen KR. Systematic assessment of etiology in adults with a clinical diagnosis of young-onset type 2 diabetes is a successful strategy for identifying maturity-onset diabetes of the young. Diabetes Care. 2012;35:1206-1212. [PubMed] [DOI] [Full Text] |
| 3. | Firdous P, Nissar K, Ali S, Ganai BA, Shabir U, Hassan T, Masoodi SR. Genetic Testing of Maturity-Onset Diabetes of the Young Current Status and Future Perspectives. Front Endocrinol (Lausanne). 2018;9:253. [PubMed] [DOI] [Full Text] |
| 4. | Murphy R, Ellard S, Hattersley AT. Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat Clin Pract Endocrinol Metab. 2008;4:200-213. [PubMed] [DOI] [Full Text] |
| 5. | Ağladıoğlu SY, Aycan Z, Çetinkaya S, Baş VN, Önder A, Peltek Kendirci HN, Doğan H, Ceylaner S. Maturity onset diabetes of youth (MODY) in Turkish children: sequence analysis of 11 causative genes by next generation sequencing. J Pediatr Endocrinol Metab. 2016;29:487-496. [PubMed] [DOI] [Full Text] |
| 6. | Glotov OS, Serebryakova EA, Turkunova ME, Efimova OA, Glotov AS, Barbitoff YA, Nasykhova YA, Predeus AV, Polev DE, Fedyakov MA, Polyakova IV, Ivashchenko TE, Shved NY, Shabanova ES, Tiselko AV, Romanova OV, Sarana AM, Pendina AA, Scherbak SG, Musina EV, Petrovskaia-Kaminskaia AV, Lonishin LR, Ditkovskaya LV, Zhelenina LА, Tyrtova LV, Berseneva OS, Skitchenko RK, Suspitsin EN, Bashnina EB, Baranov VS. Wholeexome sequencing in Russian children with nontype 1 diabetes mellitus reveals a wide spectrum of genetic variants in MODYrelated and unrelated genes. Mol Med Rep. 2019;20:4905-4914. [PubMed] [DOI] [Full Text] |
| 7. | Basit A, Fawwad A, Qureshi H, Shera AS; NDSP Members. Prevalence of diabetes, pre-diabetes and associated risk factors: second National Diabetes Survey of Pakistan (NDSP), 2016-2017. BMJ Open. 2018;8:e020961. [PubMed] [DOI] [Full Text] |
| 8. | Aamir AH, Ul-Haq Z, Mahar SA, Qureshi FM, Ahmad I, Jawa A, Sheikh A, Raza A, Fazid S, Jadoon Z, Ishtiaq O, Safdar N, Afridi H, Heald AH. Diabetes Prevalence Survey of Pakistan (DPS-PAK): prevalence of type 2 diabetes mellitus and prediabetes using HbA1c: a population-based survey from Pakistan. BMJ Open. 2019;9:e025300. [PubMed] [DOI] [Full Text] |
| 9. | Kanwal A FS, Asghar S, Naeem M. Mody genes; linkage analysis and subgroup discovery from text documents. Professional Med J. 2013;20:623-633. |
| 10. | Ali FB, Sohail S, Majid Z. MODY (maturity onset diabetes of the young). J Pak Med Assoc. 2013;63:1327. [PubMed] |
| 11. | Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405-424. [PubMed] [DOI] [Full Text] |
| 12. | Li M, Wang S, Xu K, Chen Y, Fu Q, Gu Y, Shi Y, Zhang M, Sun M, Chen H, Han X, Li Y, Tang Z, Cai L, Li Z, Yang T, Polychronakos C. High Prevalence of a Monogenic Cause in Han Chinese Diagnosed With Type 1 Diabetes, Partly Driven by Nonsyndromic Recessive WFS1 Mutations. Diabetes. 2020;69:121-126. [PubMed] [DOI] [Full Text] |
| 13. | Cleaver R, Berg J, Craft E, Foster A, Gibbons RJ, Hobson E, Lachlan K, Naik S, Sampson JR, Sharif S, Smithson S; Deciphering Developmental Disorders Study, Parker MJ, Tatton-Brown K. Refining the Primrose syndrome phenotype: A study of five patients with ZBTB20 de novo variants and a review of the literature. Am J Med Genet A. 2019;179:344-349. [PubMed] [DOI] [Full Text] |
| 14. | Shields BM, McDonald TJ, Ellard S, Campbell MJ, Hyde C, Hattersley AT. The development and validation of a clinical prediction model to determine the probability of MODY in patients with young-onset diabetes. Diabetologia. 2012;55:1265-1272. [PubMed] [DOI] [Full Text] |
| 15. | Ma RC, Chan JC. Type 2 diabetes in East Asians: similarities and differences with populations in Europe and the United States. Ann N Y Acad Sci. 2013;1281:64-91. [PubMed] [DOI] [Full Text] |
| 16. | Kleinberger JW, Pollin TI. Undiagnosed MODY: Time for Action. Curr Diab Rep. 2015;15:110. [PubMed] [DOI] [Full Text] |
| 17. | Bennett JT, Vasta V, Zhang M, Narayanan J, Gerrits P, Hahn SH. Molecular genetic testing of patients with monogenic diabetes and hyperinsulinism. Mol Genet Metab. 2015;114:451-458. [PubMed] [DOI] [Full Text] |
| 18. | Liang H, Zhang Y, Li M, Yan J, Yang D, Luo S, Zheng X, Yang G, Li Z, Xu W, Groop L, Weng J. Recognition of maturity-onset diabetes of the young in China. J Diabetes Investig. 2021;12:501-509. [PubMed] [DOI] [Full Text] |
| 19. | Xu A, Lin Y, Sheng H, Cheng J, Mei H, Ting TH, Zeng C, Liang C, Zhang W, Li C, Li X, Liu L. Molecular diagnosis of maturity-onset diabetes of the young in a cohort of Chinese children. Pediatr Diabetes. 2020;21:431-440. [PubMed] [DOI] [Full Text] |
| 20. | Shim YJ, Kim JE, Hwang SK, Choi BS, Choi BH, Cho EM, Jang KM, Ko CW. Identification of Candidate Gene Variants in Korean MODY Families by Whole-Exome Sequencing. Horm Res Paediatr. 2015;83:242-251. [PubMed] [DOI] [Full Text] |
| 21. | Chapla A, Mruthyunjaya MD, Asha HS, Varghese D, Varshney M, Vasan SK, Venkatesan P, Nair V, Mathai S, Paul TV, Thomas N. Maturity onset diabetes of the young in India - a distinctive mutation pattern identified through targeted next-generation sequencing. Clin Endocrinol (Oxf). 2015;82:533-542. [PubMed] [DOI] [Full Text] |
| 22. | Mohan V, Radha V, Nguyen TT, Stawiski EW, Pahuja KB, Goldstein LD, Tom J, Anjana RM, Kong-Beltran M, Bhangale T, Jahnavi S, Chandni R, Gayathri V, George P, Zhang N, Murugan S, Phalke S, Chaudhuri S, Gupta R, Zhang J, Santhosh S, Stinson J, Modrusan Z, Ramprasad VL, Seshagiri S, Peterson AS. Comprehensive genomic analysis identifies pathogenic variants in maturity-onset diabetes of the young (MODY) patients in South India. BMC Med Genet. 2018;19:22. [PubMed] [DOI] [Full Text] |
| 23. | Woodhouse NJ, Elshafie OT, Al-Mamari AS, Mohammed NH, Al-Riyami F, Raeburn S. Clinically-Defined Maturity Onset Diabetes of the Young in Omanis: Absence of the common Caucasian gene mutations. Sultan Qaboos Univ Med J. 2010;10:80-83. [PubMed] |
| 24. | Ben Khelifa S, Martinez R, Dandana A, Khochtali I, Ferchichi S, Castaño L. Maturity Onset Diabetes of the Young (MODY) in Tunisia: Low frequencies of GCK and HNF1A mutations. Gene. 2018;651:44-48. [PubMed] [DOI] [Full Text] |
| 25. | Dallali H, Pezzilli S, Hechmi M, Sallem OK, Elouej S, Jmel H, Ben Halima Y, Chargui M, Gharbi M, Mercuri L, Alberico F, Mazza T, Bahlous A, Ben Ahmed M, Jamoussi H, Abid A, Trischitta V, Abdelhak S, Prudente S, Kefi R. Genetic characterization of suspected MODY patients in Tunisia by targeted next-generation sequencing. Acta Diabetol. 2019;56:515-523. [PubMed] [DOI] [Full Text] |
| 26. | Johansson BB, Irgens HU, Molnes J, Sztromwasser P, Aukrust I, Juliusson PB, Søvik O, Levy S, Skrivarhaug T, Joner G, Molven A, Johansson S, Njølstad PR. Targeted next-generation sequencing reveals MODY in up to 6.5% of antibody-negative diabetes cases listed in the Norwegian Childhood Diabetes Registry. Diabetologia. 2017;60:625-635. [PubMed] [DOI] [Full Text] |
| 27. | Donath X, Saint-Martin C, Dubois-Laforgue D, Rajasingham R, Mifsud F, Ciangura C, Timsit J, Bellanné-Chantelot C; Monogenic Diabetes Study Group of the Société Francophone du Diabète. Next-generation sequencing identifies monogenic diabetes in 16% of patients with late adolescence/adult-onset diabetes selected on a clinical basis: a cross-sectional analysis. BMC Med. 2019;17:132. [PubMed] [DOI] [Full Text] |
| 28. | Carlsson A, Shepherd M, Ellard S, Weedon M, Lernmark Å, Forsander G, Colclough K, Brahimi Q, Valtonen-Andre C, Ivarsson SA, Elding Larsson H, Samuelsson U, Örtqvist E, Groop L, Ludvigsson J, Marcus C, Hattersley AT. Absence of Islet Autoantibodies and Modestly Raised Glucose Values at Diabetes Diagnosis Should Lead to Testing for MODY: Lessons From a 5-Year Pediatric Swedish National Cohort Study. Diabetes Care. 2020;43:82-89. [PubMed] [DOI] [Full Text] |
| 29. | Patel KA, Kettunen J, Laakso M, Stančáková A, Laver TW, Colclough K, Johnson MB, Abramowicz M, Groop L, Miettinen PJ, Shepherd MH, Flanagan SE, Ellard S, Inagaki N, Hattersley AT, Tuomi T, Cnop M, Weedon MN. Heterozygous RFX6 protein truncating variants are associated with MODY with reduced penetrance. Nat Commun. 2017;8:888. [PubMed] [DOI] [Full Text] |
| 30. | Akiba K, Ushijima K, Fukami M, Hasegawa Y. A heterozygous protein-truncating RFX6 variant in a family with childhood-onset, pregnancy-associated and adult-onset diabetes. Diabet Med. 2020;37:1772-1776. [PubMed] [DOI] [Full Text] |
| 31. | Wang Y, Zhang J, Zhao Y, Wang S, Han Q, Zhang R, Guo R, Li H, Li L, Wang T, Tang X, He C, Teng G, Gu W, Liu F. COL4A3 Gene Variants and Diabetic Kidney Disease in MODY. Clin J Am Soc Nephrol. 2018;13:1162-1171. [PubMed] [DOI] [Full Text] |
| 32. | Kwak SH, Jung CH, Ahn CH, Park J, Chae J, Jung HS, Cho YM, Lee DH, Kim JI, Park KS. Clinical whole exome sequencing in early onset diabetes patients. Diabetes Res Clin Pract. 2016;122:71-77. [PubMed] [DOI] [Full Text] |
| 33. | Park SS, Jang SS, Ahn CH, Kim JH, Jung HS, Cho YM, Lee YA, Shin CH, Chae JH, Choi SH, Jang HC, Bae JC, Won JC, Kim SH, Kim JI, Kwak SH, Park KS. Identifying Pathogenic Variants of Monogenic Diabetes Using Targeted Panel Sequencing in an East Asian Population. J Clin Endocrinol Metab. 2019;. [PubMed] [DOI] [Full Text] |
| 34. | Marchand L, Li M, Leblicq C, Rafique I, Alarcon-Martinez T, Lange C, Rendon L, Tam E, Courville-Le Bouyonnec A, Polychronakos C. Monogenic Causes in the Type 1 Diabetes Genetics Consortium Cohort: Low Genetic Risk for Autoimmunity in Case Selection. J Clin Endocrinol Metab. 2021;106:1804-1810. [PubMed] [DOI] [Full Text] |
| 35. | Hattersley AT, Greeley SAW, Polak M, Rubio-Cabezas O, Njølstad PR, Mlynarski W, Castano L, Carlsson A, Raile K, Chi DV, Ellard S, Craig ME. ISPAD Clinical Practice Consensus Guidelines 2018: The diagnosis and management of monogenic diabetes in children and adolescents. Pediatr Diabetes. 2018;19 Suppl 27:47-63. [PubMed] [DOI] [Full Text] |
| 36. | Stellacci E, Steindl K, Joset P, Mercurio L, Anselmi M, Cecchetti S, Gogoll L, Zweier M, Hackenberg A, Bocchinfuso G, Stella L, Tartaglia M, Rauch A. Clinical and functional characterization of two novel ZBTB20 mutations causing Primrose syndrome. Hum Mutat. 2018;39:959-964. [PubMed] [DOI] [Full Text] |
| 37. | Cordeddu V, Redeker B, Stellacci E, Jongejan A, Fragale A, Bradley TE, Anselmi M, Ciolfi A, Cecchetti S, Muto V, Bernardini L, Azage M, Carvalho DR, Espay AJ, Male A, Molin AM, Posmyk R, Battisti C, Casertano A, Melis D, van Kampen A, Baas F, Mannens MM, Bocchinfuso G, Stella L, Tartaglia M, Hennekam RC. Mutations in ZBTB20 cause Primrose syndrome. Nat Genet. 2014;46:815-817. [PubMed] [DOI] [Full Text] |
| 38. | Rafique I, Saqib MAN, Mir A, Naeem M. Maturity onset diabetes of the young–an overview of common types. a review. Romanian J Diabetes Nutr Metab Dis. 2018;25:209-213. [DOI] [Full Text] |
| 39. | Kyithar MP, Bacon S, Pannu KK, Rizvi SR, Colclough K, Ellard S, Byrne MM. Identification of HNF1A-MODY and HNF4A-MODY in Irish families: phenotypic characteristics and therapeutic implications. Diabetes Metab. 2011;37:512-519. [PubMed] [DOI] [Full Text] |
| 40. | Pavić T, Juszczak A, Pape Medvidović E, Burrows C, Šekerija M, Bennett AJ, Ćuća Knežević J, Gloyn AL, Lauc G, McCarthy MI, Gornik O, Owen KR. Maturity onset diabetes of the young due to HNF1A variants in Croatia. Biochem Med (Zagreb). 2018;28:020703. [PubMed] [DOI] [Full Text] |
| 41. | Bacon S, Kyithar MP, Rizvi SR, Donnelly E, McCarthy A, Burke M, Colclough K, Ellard S, Byrne MM. Successful maintenance on sulphonylurea therapy and low diabetes complication rates in a HNF1A-MODY cohort. Diabet Med. 2016;33:976-984. [PubMed] [DOI] [Full Text] |
| 42. | Neve B, Fernandez-Zapico ME, Ashkenazi-Katalan V, Dina C, Hamid YH, Joly E, Vaillant E, Benmezroua Y, Durand E, Bakaher N, Delannoy V, Vaxillaire M, Cook T, Dallinga-Thie GM, Jansen H, Charles MA, Clément K, Galan P, Hercberg S, Helbecque N, Charpentier G, Prentki M, Hansen T, Pedersen O, Urrutia R, Melloul D, Froguel P. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proc Natl Acad Sci U S A. 2005;102:4807-4812. [PubMed] [DOI] [Full Text] |
| 43. | Lomberk G, Grzenda A, Mathison A, Escande C, Zhang JS, Calvo E, Miller LJ, Iovanna J, Chini EN, Fernandez-Zapico ME, Urrutia R. Krüppel-like factor 11 regulates the expression of metabolic genes via an evolutionarily conserved protein interaction domain functionally disrupted in maturity onset diabetes of the young. J Biol Chem. 2013;288:17745-17758. [PubMed] [DOI] [Full Text] |
| 44. | Ushijima K, Narumi S, Ogata T, Yokota I, Sugihara S, Kaname T, Horikawa Y, Matsubara Y, Fukami M, Kawamura T; Japanese Study Group of Insulin Therapy for Childhood and Adolescent Diabetes. KLF11 variant in a family clinically diagnosed with early childhood-onset type 1B diabetes. Pediatr Diabetes. 2019;20:712-719. [PubMed] [DOI] [Full Text] |
| 45. | Horikawa Y, Enya M, Fushimi N, Fushimi Y, Takeda J. Screening of diabetes of youth for hepatocyte nuclear factor 1 mutations: clinical phenotype of HNF1β-related maturity-onset diabetes of the young and HNF1α-related maturity-onset diabetes of the young in Japanese. Diabet Med. 2014;31:721-727. [PubMed] [DOI] [Full Text] |
| 46. | Omura Y, Yagi K, Honoki H, Iwata M, Enkaku A, Takikawa A, Kuwano T, Watanabe Y, Nishimura A, Liu J, Chujo D, Fujisaka S, Enya M, Horikawa Y, Tobe K. Clinical manifestations of a sporadic maturity-onset diabetes of the young (MODY) 5 with a whole deletion of HNF1B based on 17q12 microdeletion. Endocr J. 2019;66:1113-1116. [PubMed] [DOI] [Full Text] |
| 47. | Dotto RP, Santana LS, Lindsey SC, Caetano LA, Franco LF, Moisés RCMS, Sa JR, Nishiura JL, Teles MG, Heilberg IP, Dias-da-Silva MR, Giuffrida FMA, Reis AF. Searching for mutations in the HNF1B gene in a Brazilian cohort with renal cysts and hyperglycemia. Arch Endocrinol Metab. 2019;63:250-257. [PubMed] [DOI] [Full Text] |
| 48. | Kim EK, Lee JS, Cheong HI, Chung SS, Kwak SH, Park KS. Identification and Functional Characterization of P159L Mutation in HNF1B in a Family with Maturity-Onset Diabetes of the Young 5 (MODY5). Genomics Inform. 2014;12:240-246. [PubMed] [DOI] [Full Text] |
| 49. | Ozsu E, Cizmecioglu FM, Yesiltepe Mutlu G, Yuksel AB, Calıskan M, Yesilyurt A, Hatun S. Maturity Onset Diabetes of the Young due to Glucokinase, HNF1-A, HNF1-B, and HNF4-A Mutations in a Cohort of Turkish Children Diagnosed as Type 1 Diabetes Mellitus. Horm Res Paediatr. 2018;90:257-265. [PubMed] [DOI] [Full Text] |
| 50. | Hina S, Malik S. Pattern of consanguinity and inbreeding coefficient in sargodha district, punjab, pakistan. J Biosoc Sci. 2015;47:803-811. [PubMed] [DOI] [Full Text] |
| 51. | Pervaiz R, Faisal F, Serakinci N. Practice of consanguinity and attitudes towards risk in the pashtun population of khyber pakhtunkhwa, pakistan. J Biosoc Sci. 2018;50:414-420. [PubMed] [DOI] [Full Text] |
| 52. | American Diabetes Association. Classification and diagnosis of diabetes. Diabetes Care. 2015;38 Suppl:S8-S16. [PubMed] [DOI] [Full Text] |