Published online Mar 15, 2019. doi: 10.4239/wjd.v10.i3.181
Peer-review started: February 10, 2019
First decision: February 19, 2019
Revised: March 8, 2019
Accepted: March 11, 2019
Article in press: March 11, 2019
Published online: March 15, 2019
Processing time: 37 Days and 20.3 Hours
The regulatory factor X6 (RFX6), a member of regulatory factor X family, is known to play a key role in the development and differentiation of pancreatic beta cells as well as insulin production and secretion. However, the potential role of RFX6 in type 2 diabetes (T2D) is still unclear.
Recent studies have indicated that RFX6 binding to DNA could be disrupted in diabetes. Therefore, in this study we investigated whether genetic mutations are present in the DNA binding domain of RFX6 gene that could abrogate its function in T2D.
A cohort of T2D patients was enrolled in this study, and the gene encoding the DNA binding domain of RFX6 was amplified by polymerase chain reaction and then analysed by direct DNA sequencing.
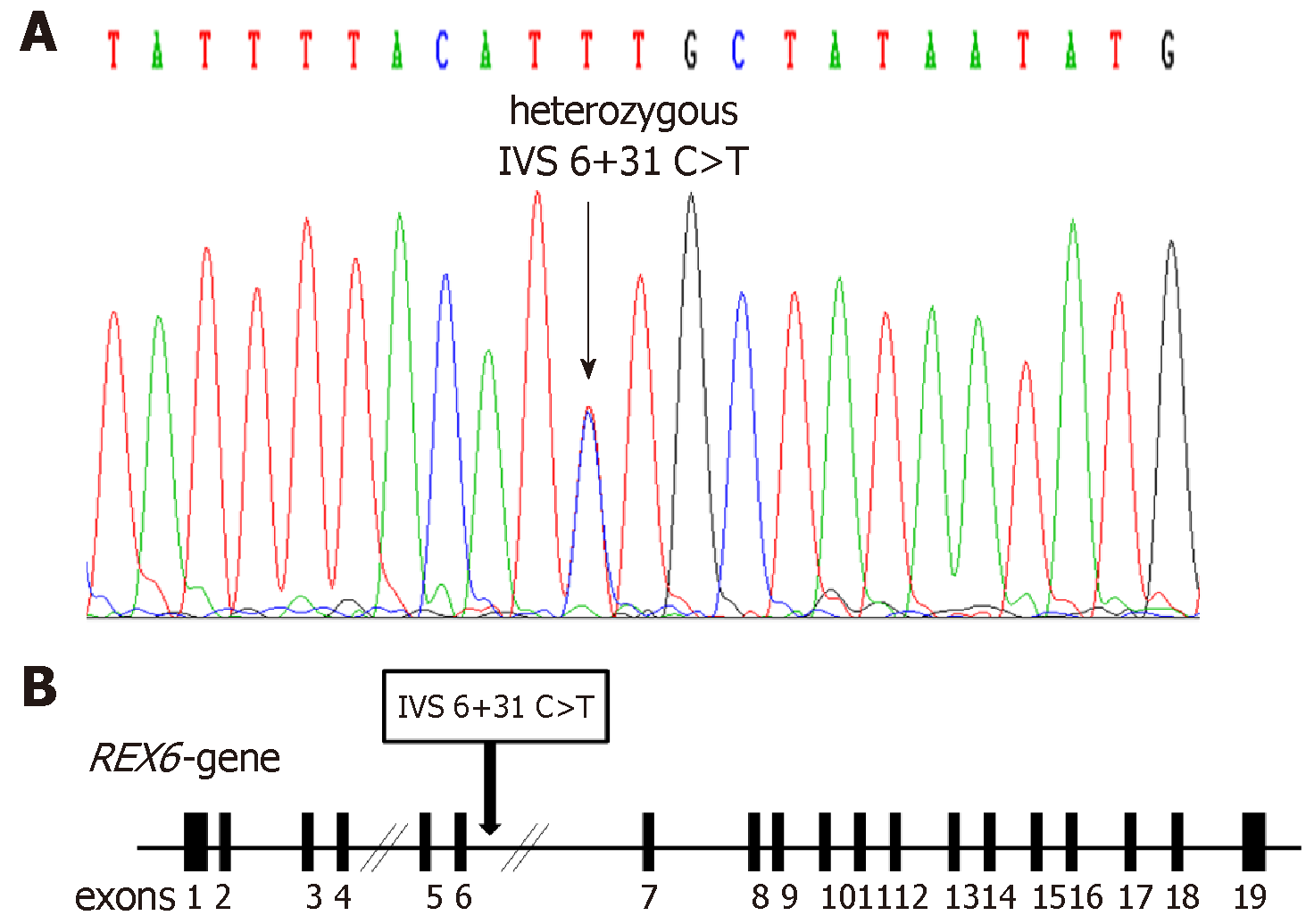
The DNA sequence analysis revealed the absence of any exonic mutation. However, we have identified a new heterozygous single nucleotide polymorphism (IVS6+31 C>T) in the intronic region of DNA binding domain gene that is present in 9.2% and 8.5% of diabetic and control people, respectively (P = 0.97).
We report the absence of any significant genetic variant that could affect the function of RFX6-DNA binding domain in T2D.
Core tip: Regulatory factor X6 (RFX6) protein plays a key role in the differentiation of pancreatic beta cells as well as insulin production and secretion. Several lines of evidence have indicated that RFX6 binding to DNA could be disrupted in diabetes; however, the mechanism underlying this process is still unknown. In this case-control study, we analysed the genotype of RFX6-DNA binding domain in diabetes patients in comparison to healthy controls. Our results indicate the absence of any significant genetic variant in the DNA binding domain that could affect the function of RFX6 in type 2 diabetes.
- Citation: Mahmoud IS, Homsi A, Al-Ameer HJ, Alzyoud J, Darras M, Shhab MA, Zihlif M, Hatmal MM, Alshaer W. Screening the RFX6-DNA binding domain for potential genetic variants in patients with type 2 diabetes. World J Diabetes 2019; 10(3): 181-188
- URL: https://www.wjgnet.com/1948-9358/full/v10/i3/181.htm
- DOI: https://dx.doi.org/10.4239/wjd.v10.i3.181
Diabetes mellitus is a group of glucose metabolism disorders characterized by high levels of blood glucose. The disease affects millions of people worldwide, which is usually associated with serious complications that affect various systems of human body[1]. Type 2 diabetes (T2D) is mainly manifested by low insulin production by pancreatic cells and/or the produced insulin does not function effectively[2]. Alterations in both beta cells average mass and function have been observed and reported in people with diabetes[3-5].
Regulatory factor X (RFX) proteins constitute a family of DNA binding proteins that is conserved in the eukaryotic kingdom[6]. In humans, RFXs act as regulatory transcription factors that bind to a conserved cis-regulatory element called the X-box motif, which is typically 14-mer DNA sequences located in specific promoter regions of the genome[7]. The function of mammalian RFX proteins has only recently started to emerge, and has shown to play an important role in regulating growth and development, immune response and endocrine secretions[7]. The RFX family has seven members of RFX1-7 in mammals, which have wide expression in various tissues and organs. RFX6 is a main member of RFX family that is predominantly expressed in pancreatic islets and encoded by a gene on chromosome 6[7]. RFX6 possesses a highly conserved DNA binding domain which is critical for binding of RFX6 to X-box promoter motifs and thus regulating their function. Recent studies conducted in mice demonstrated that RFX6 is specifically required for pancreatic beta cells differentiation during embryonic development[8]. Moreover, RFX6 was shown to be an important factor to maintain key features of functionality of mature beta cells, and RFX6 gene deletion in adult mice beta cells was shown to disrupt glucose homeostasis and caused glucose intolerance, impaired beta cell glucose sensing and defective insulin secretion[9].
In 2017, Varshney et al[10] published an interesting study in the Proceedings of the National Academy of Sciences, where they performed an integrated analysis of molecular profiling data of the genomic DNA, epigenome and transcriptome in diabetic pancreatic beta islets, to understand the potential connections between genetic variants, chromatin landscape, and gene expression in T2D. The study showed that most of the reported genetic variants in T2D are enriched in regions of the DNA where RFX transcription factors are predicted to bind. The study also concluded that these genetic variants that increased the risk of T2D are predicted to disrupt mainly the binding of RFX6 to genomic DNA[10], indicating that RFX6 binding to X-box promoter motifs could be disrupted in T2D.
In this study, we sought to investigate if any structural genetic defects could be present in the RFX6-DNA binding domain in T2D patients that could potentially inhibit its function in diabetes.
Initially, a total of 98 blood samples (49 samples from T2D patients, 49 from healthy volunteers (control group)) were collected from Jordanian population (Table 1). The study was then extended to investigate the association between the identified intronic variant (IVS6+31 C>T) and diabetes. A total of 283 blood samples (141 from T2D patients, 142 from healthy volunteers) were included in the extended study (Table 1). Diabetic participants who enrolled in this study were Jordanian adults (age ≥ 20 years), including both females and males, with known history of diabetes and recruited from Jordanian medical centres during the time period between Dec 2015 and July 2017. Controls were unrelated to diabetic patients and had no history of diabetes, as determined by history and lab examination. All blood samples were collected according to protocols approved by the Institutional Review Board, and informed consents were obtained from participants included in the study.
| Control group | Diabetic patients | P-value | |
| Primary screening, (n = 98) | |||
| Male, n (%) | 32 (65%) | 24 (49%) | 0.285 |
| Age (mean ± SD) | 50.1 ± 12.3 | 54.7 ± 10.7 | 0.149 |
| Female, n (%) | 17 (35%) | 25 (51%) | 0.217 |
| Age (mean ± SD) | 47.7 ± 15.5 | 56.2 ± 13.2 | 0.065 |
| FBG (mg/dL) | 97± 10 | 192± 94 | < 0.001 |
| HbA1c (%) | 5.4± 0.4 | 8.2± 2.2 | < 0.001 |
| Screening the IVS6+31 C>T, (n = 283) | |||
| Male, n (%) | 78 (55%) | 59 (42%) | 0.105 |
| Age (mean ± SD) | 48.9 ± 15.2 | 57.4 ± 11.3 | 0.001 |
| Female, n (%) | 64 (45%) | 82 (58%) | 0.136 |
| Age (mean ± SD) | 51.8 ± 15.2 | 57.1 ± 11.5 | 0.024 |
| FBG (mg/dL) | 98± 10.4 | 177± 80 | < 0.001 |
| HbA1c (%) | 5.4± 0.5 | 7.8± 1.9 | < 0.001 |
The levels of blood glucose and glycosylated Hb (HbA1c) were evaluated in the participants of this study. Blood glucose level was measured by the glucose oxidase method using Cobas c111 analyzer (Switzerland). The percentage of HbA1c in blood was determined using ion-exchange high-performance liquid chromatography (D-10™ Bio Rad, United States).
For molecular assays, DNA was extracted from venous whole-blood samples, and then collected in EDTA containing tubes, using Wizard genomic DNA purification kit (Promega, United States). Extracted DNA was stored at −20°C. Polymerase chain reaction (PCR) was used to amplify genomic DNA encompassing the coding sequences and intronic borders of exons 3, 4, 5 and 6 of the RFX6 gene (NCBI: NG_027699.1). The primers used for PCR assay were as described in (Table 2). The PCR amplification reactions were performed in a total volume of 25 μL in 0.2 mL PCR tubes containing 200 ng of genomic DNA, 5 μL of 5× FIREPol® Master Mix (Solis BioDyne) with 7.5 mmol MgCl2 and 10 pmol of each primer (Gene Link, United States). All PCR reactions were performed using C1000 Touch™ Thermal Cycler (Bio-Rad; United Kingdom) and the reaction conditions were as follows: initial denaturation of 5 min at 95°C, followed by 29 cycles of 30 s at 95°C, 30 s at specific annealing temperature (Table 2), 1 min at 72°C and a final extension of 6 min at 72°C. All PCR products were checked by gel electrophoresis to verify correct product size, then purified and sequenced using the same forward primer used for the gene amplification. DNA sequencing reactions were performed at Macrogen Inc., South Korea, using BigDye(R) Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, United States) and the ABI PRISM 3730XL analyzer (Applied Biosystem, United States).
| Primer | Sequence (5’-3’) | Ta (°C) | Size (bp) |
| RFX6-3 | F: 5- CTT ATG TCT ACT CAT TAC CTC -3 | 50 | 306 |
| R: 5- TCA TGC TAT CTG CCT GAC -3 | |||
| RFX6-4 | F: 5- CAG TTC ATT CAG AGT TCA AC -3 | 56 | 216 |
| R: 5- CTT CAT GCA CAA GAG CAG -3 | |||
| RFX6-5 | F: 5- GTC ATC AGG GTT TGC AGT TC -3 | 50 | 258 |
| R: 5- ATT CAA TAG GTA TCA TGC -3 | |||
| RFX6-6 | F: 5- GTA AGT TGA GAA AGA TGC -3 | 56 | 258 |
| R: 5- CAT GTA TTG CTC AGC TTG -3 |
Data analyses were performed using IBM Statistical Package for the Social Sciences software (SPSS, version 19). Pearson’s chi-squared or Fisher’s exact tests were used to test an association between categorical variables (i.e., gender and genetic variant) and case-control status. Student t-test was used to assess the significance of difference of means of age, blood glucose and glycosylated Hb between case and control groups. A 95% confidence interval for the odds ratio was calculated and used to describe the results. For sample size calculation, the Cochran’s formula was used[11].
In this study, the screening for structural genetic variants in specific regions of RFX6 gene was carried out in T2D patients in attempt to discover new potential mutations that could mediate the pathogenesis of T2D. Evaluation of levels of blood glucose and HbA1c of enrolled subjects was performed to discriminate between diabetic and non-diabetic (Table 1). The DNA sequence analysis of exons 3, 4, 5 and 6 of RFX6 gene in 49 diabetic patients revealed the absence of any genetic mutation. However, the DNA sequencing of introns borders revealed the presence of a heterozygous genetic variant (IVS6+31 C>T) within the intron 6 of RFX6 gene, where C is substituted by T, 31 nucleotides downstream of the end of exon 6 (Figure 1). To investigate the significance of the identified intronic variant in T2D, the study was extended and the IVS6+31 C>T was screened in 283 samples (141 diabetic and 142 healthy controls). DNA sequence analysis of the samples revealed the identification of the heterozygous IVS6+31 C>T in 9.2% and 8.4% of diabetic and control groups respectively, with no significant association in genotype or allele frequency between diabetic and control groups (Table 3).
| Control, n (%) | T2D, n (%) | P-value | OR | 95%CI | |
| genotype | |||||
| C \ C | 130 (91.5) | 128 (90.8) | 0.97 | 0.9 | 0.39-2.06 |
| C \ T | 12 (8.5) | 13 (9.2) | 1.1 | 0.48-2.50 | |
| T \ T | 0 | 0 | 1 | 0.02-51.1 | |
| Allele | |||||
| C | 272 (95.7) | 269 ( 95.3) | 0.82 | 0.9 | 0.41-2.03 |
| T | 12 (4.3) | 13 (4.7) | 1.1 | 0.49-2.44 | |
The demographic characteristics of the study population (age and gender) are shown in Table 1. In primary screening study, there was no statistically significant difference in male/female proportion (P = 0.285 and P = 0.217, respectively) or age of gender (P = 0.149 and P = 0.065, respectively) between case and control groups. As for the extended screening study of IVS6+31 C>T variant, no statistically significant difference in male/female proportion was found between case and control groups (P = 0.105 and P = 0.136, respectively). However, there was a significant difference in age of male/female of case–control status (P = 0.001 and P = 0.024, respectively).
Recently, it has been proposed that a loss of pancreatic beta-cells mass, differentiation and function is a hallmark of T2D[1,2]. Concurrently, the regulation of beta cells growth and differentiation has been under intensive investigation, and several lines of evidences have indicated the role of key transcription factors in controlling the function state of beta cells. For example, evidences coming from loss-of-function studies in adult mice beta cells have revealed that transcription factors such as NeuroD1[12], Nkx6.1[13] and Pdx1[14] are important in maintaining the differentiation and function state of pancreatic beta cells. Thus, it appears that loss of function of key beta cell transcription factors results in the loss of both beta cell identity and function. More recently, RFX6 transcription factor has been shown to play a key role in regulating the state of pancreatic beta cells differentiation and function[9].
RFX6 contains a highly conserved DNA binding domain that facilitates their binding to X-box promoter motif of certain genes, which is essential to regulate the transcription of RFX6-target genes[7]. It has been shown that genetic alterations in the RFX6-DNA binding domain could be associated with neonatal diabetes. In fact, mutations in the RFX6-DNA binding domain are assumed to be the cause of neonatal diabetes in Mitchell-Riley syndrome, through the production of a defective RFX6 protein[15]. In this project, we sought to detect if any genetic mutation could be present in the RFX6-DNA binding domain in T2D. Based on our findings we conclude that structural mutations in the DNA binding domain of RFX6 are unlikely to exist in T2D. However, another large-scale study could increase the statistical power of our results. In addition, it is noteworthy to mention that RFX6 proteins contain other conserved regions including B, C, and D domains[6]. These domains are thought to be involved in RFX6 oligomerization which are required for DNA binding and activation[7]. Therefore, screening the other functional domains of RFX6 may provide more insights into the potential mechanism by which RFX6 binding to DNA is abrogated in diabetes.
Diabetes mellitus is a global health challenge, which is usually associated with the loss/dysfunction of insulin-producing pancreatic beta cells. Hence, understanding the molecular mechanisms that control beta cells differentiation and function represents a major interest in the medical field. Regulatory factor X6 (RFX6) is DNA binding protein that is predominantly expressed in pancreatic islets of human and plays a key role in regulating pancreatic beta cells differentiation and insulin production, and it has been recently. RFX6 contains a highly conserved DNA binding domain which is critical for binding of RFX6 to DNA and consequently regulates the amount of messenger RNA produced by the gene. Several lines of evidence have indicated that RFX6 binding to DNA could be disrupted in diabetes. However, the mechanism by which this could happen is still unknown.
The presence of genetic mutations in the gene coding for the RFX6-DNA binding domain could result in inhibition of binding of RFX6 to DNA and consequently loss of function. Defining such genetic mutations will provide valuable information to diagnose, treat, prevent and cure type 2 diabetes (T2D).
In this study, we sought to investigate if any structural genetic mutations could be present in the RFX6-DNA binding domain in T2D patients and whether they are associated with diabetes.
A case-control study was conducted in T2D patients and healthy volunteers. The DNA was extracted from all subjects and polymerase chain reaction (PCR) was used to amplify genomic DNA encompassing the coding sequences and intronic borders of exons 3, 4, 5 and 6 of the RFX6 gene, then PCR samples were analysed by DNA sequencing.
Our data showed the absence of any mutation in the exons coding for the RFX6-DNA binding domain. However, we have identified a new heterozygous single nucleotide polymorphism (IVS6+31 C>T) in the intronic region of DNA binding domain gene that is present in 9.2% and 8.5% of diabetic and control people, respectively (P = 0.97).
We conclude that genetic mutations in the DNA binding domain of RFX6 are unlikely to exist in T2D.
RFX6 binding to DNA is mediated by multiple of domains. Indeed, RFX6 proteins contain other conserved regions, including B, C, and D domains, which play a critical role in oligomerization of the protein and are required for DNA binding and activation. Thus, testing the other functional domains of RFX6 in future will provide more insights into the role of RFX6 in diabetes.
We would like to thank Dr Hussam Alhawari and the laboratory staff of the Molecular Biology Research Lab (MBRL) at the University of Jordan for technical support.
STROBE Statement: The manuscript was prepared and revised according to the STROBE Statement-checklist of items.
Manuscript source: Unsolicited manuscript
Specialty type: Endocrinology and metabolism
Country of origin: Jordan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Hosseinpour-Niazi S, Hamad ARA, Avtanski D S- Editor: Dou Y L- Editor: A E- Editor: Wu YXJ
| 1. | Cheng SK, Park EY, Pehar A, Rooney AC, Gallicano GI. Current progress of human trials using stem cell therapy as a treatment for diabetes mellitus. Am J Stem Cells. 2016;5:74-86. [PubMed] |
| 2. | Patil PD, Mahajan UB, Patil KR, Chaudhari S, Patil CR, Agrawal YO, Ojha S, Goyal SN. Past and current perspective on new therapeutic targets for Type-II diabetes. Drug Des Devel Ther. 2017;11:1567-1583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 3. | Butler AE, Janson J, Soeller WC, Butler PC. Increased beta-cell apoptosis prevents adaptive increase in beta-cell mass in mouse model of type 2 diabetes: evidence for role of islet amyloid formation rather than direct action of amyloid. Diabetes. 2003;52:2304-2314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3031] [Cited by in RCA: 3041] [Article Influence: 138.2] [Reference Citation Analysis (0)] |
| 4. | Rahier J, Guiot Y, Goebbels RM, Sempoux C, Henquin JC. Pancreatic beta-cell mass in European subjects with type 2 diabetes. Diabetes Obes Metab. 2008;10 Suppl 4:32-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 583] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 5. | Rosengren AH, Braun M, Mahdi T, Andersson SA, Travers ME, Shigeto M, Zhang E, Almgren P, Ladenvall C, Axelsson AS, Edlund A, Pedersen MG, Jonsson A, Ramracheya R, Tang Y, Walker JN, Barrett A, Johnson PR, Lyssenko V, McCarthy MI, Groop L, Salehi A, Gloyn AL, Renström E, Rorsman P, Eliasson L. Reduced insulin exocytosis in human pancreatic β-cells with gene variants linked to type 2 diabetes. Diabetes. 2012;61:1726-1733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 184] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 6. | Emery P, Durand B, Mach B, Reith W. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 1996;24:803-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 164] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 7. | Aftab S, Semenec L, Chu JS, Chen N. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol Biol. 2008;8:226. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 105] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 8. | Smith SB, Qu HQ, Taleb N, Kishimoto NY, Scheel DW, Lu Y, Patch AM, Grabs R, Wang J, Lynn FC, Miyatsuka T, Mitchell J, Seerke R, Désir J, Vanden Eijnden S, Abramowicz M, Kacet N, Weill J, Renard ME, Gentile M, Hansen I, Dewar K, Hattersley AT, Wang R, Wilson ME, Johnson JD, Polychronakos C, German MS. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010;463:775-780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 296] [Cited by in RCA: 255] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 9. | Piccand J, Strasser P, Hodson DJ, Meunier A, Ye T, Keime C, Birling MC, Rutter GA, Gradwohl G. Rfx6 maintains the functional identity of adult pancreatic β cells. Cell Rep. 2014;9:2219-2232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 10. | Varshney A, Scott LJ, Welch RP, Erdos MR, Chines PS, Narisu N, Albanus RD, Orchard P, Wolford BN, Kursawe R, Vadlamudi S, Cannon ME, Didion JP, Hensley J, Kirilusha A; NISC Comparative Sequencing Program, Bonnycastle LL, Taylor DL, Watanabe R, Mohlke KL, Boehnke M, Collins FS, Parker SC, Stitzel ML. Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proc Natl Acad Sci U S A. 2017;114:2301-2306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 139] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 11. | Bartlett JE, Kotrlik JW, Higgins CC. Organizational Research: Determining Appropriate Sample Size in Survey Research. ITLPG. 2001;19:43-50. |
| 12. | Gu C, Stein GH, Pan N, Goebbels S, Hörnberg H, Nave KA, Herrera P, White P, Kaestner KH, Sussel L, Lee JE. Pancreatic beta cells require NeuroD to achieve and maintain functional maturity. Cell Metab. 2010;11:298-310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 225] [Cited by in RCA: 216] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 13. | Taylor BL, Liu FF, Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep. 2013;4:1262-1275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 254] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 14. | Gao T, McKenna B, Li C, Reichert M, Nguyen J, Singh T, Yang C, Pannikar A, Doliba N, Zhang T, Stoffers DA, Edlund H, Matschinsky F, Stein R, Stanger BZ. Pdx1 maintains β cell identity and function by repressing an α cell program. Cell Metab. 2014;19:259-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 324] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 15. | Zegre Amorim M, Houghton JA, Carmo S, Salva I, Pita A, Pereira-da-Silva L. Mitchell-Riley Syndrome: A Novel Mutation in RFX6 Gene. Case Rep Genet. 2015;2015:937201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |