Published online Nov 15, 2019. doi: 10.4239/wjd.v10.i11.534
Peer-review started: July 21, 2019
First decision: August 31, 2019
Revised: September 10, 2019
Accepted: October 7, 2019
Article in press: October 7, 2019
Published online: November 15, 2019
Processing time: 108 Days and 3.6 Hours
Type 1 diabetes (T1D) is a complex disease with a higher incidence in Europeans than other populations. The Colombians Living in Medellin (CLM) is admixed with ancestry contributions from Europeans, Native Americans (NAT) and Africans (AFR).
Our aim was to analyze the genetic admixture component at candidate T1D loci in Colombian individuals with the disease.
Seventy-four ancestry informative markers (AIMs), which tagged 41 T1D candidate loci/genes, were tested by studying a cohort of 200 Northwest Colombia diseased individuals. T1D status was classified by testing for glutamic acid decarboxylase (GAD-65 kDa) and protein tyrosine-like antigen-2 auto-antibodies in serum samples. Candidate loci/genes included HLA, INS, PTPN22, CTLA4, IL2RA, SUMO4, CLEC16A, IFIH1, EFR3B, IL7R, NRP1 and RNASEH1, amongst others. The 1,000 genome database was used to analyze data from 94 individuals corresponding to the reference CLM. As the data did not comply with a normal distribution, medians were compared between groups using the Mann-Whitney U-test.
Both T1D patients and individuals from CLM displayed mainly European ancestry (61.58 vs 62.06) followed by Native American (27.34 vs 27.46) and to a lesser extent the AFR ancestry (10.28 vs 10.65) components. However, compared to CLM, ancestry of T1D patients displayed a decrease of NAT ancestry at gene EFR3B (24.30 vs 37.10) and an increase at genes IFIH1 (32.07 vs 14.99) and IL7R (52.18 vs 39.18). Also, for gene NRP1 (36.67 vs 0.003), we observed a non-AFR contribution (attributed to NAT). Autoimmune patients (positive for any of two auto-antibodies) displayed lower NAT ancestry than idiopathic patients at the MHC region (20.36 vs 31.88). Also, late onset patients presented with greater AFR ancestry than early onset patients at gene IL7R (19.96 vs 6.17). An association analysis showed that, even after adjusting for admixture, an association exists for at least seven such AIMs, with the strongest findings on chromosomes 5 and 10 (gene IL7R, P = 5.56 × 10-6 and gene NRP1, P = 8.70 × 10-19, respectively).
Although Colombian T1D patients have globally presented with higher European admixture, specific T1D loci have displayed varying levels of Native American and AFR ancestries in diseased individuals.
Core tip: We have tested the effect of genetic admixture in a set of Colombian patients with Type 1 Diabetes (T1D). We show that, although no differences between T1Ds and Colombians living in Medellin arose globally, there appear to be ancestry differences when looking at specific T1D loci/genes (e.g., genes EFR3B, IFIH1, IL7R and NRP1). Also, when comparing patient ancestry according to the presence/absence of T1D-related auto-antibodies or age at onset of the disease, differences were also observed. The most striking differences in ancestry occurred outside the HLA region, which is considered the master risk locus in T1D and for autoimmune diseases overall. This in itself is a striking observation.
- Citation: Gomez-Lopera N, Alfaro JM, Leal SM, Pineda-Trujillo N. Type 1 diabetes loci display a variety of native American and African ancestries in diseased individuals from Northwest Colombia. World J Diabetes 2019; 10(11): 534-545
- URL: https://www.wjgnet.com/1948-9358/full/v10/i11/534.htm
- DOI: https://dx.doi.org/10.4239/wjd.v10.i11.534
Type 1 diabetes (T1D) is a heterogeneous disease with pathogenic processes and phenotypic characteristics that show marked variation. It is accepted that genetic effects are an important factor for this heterogeneity. HLA confers the major genetic susceptibility to T1D, contributing up to 50%; it is located on chromosome 6p21[1]. In addition, over 50 non-HLA genes (so far) increase susceptibility to T1D[2,3]. Recently, we have identified that RNASEH1 gene variants associate with T1D in Northwest Colombia[4]. This gene, which is located on chromosomal region 2p25, has not thus far been associated elsewhere with the disease. A wide geographical variation in the incidence of T1D both among and within countries has been reported[5]. Incidence of T1D is higher in Europeans[6-8] than in Latin American countries[7,8]. Genetic admixture is a factor that influences allelic frequencies in a population; this, in part, may contribute to explaining the differences observed in T1D epidemiology.
Three studies in Latin America have tested the admixture effect on T1D. Two of these were carried out in Brazil[9,10] and the third in Cuba[11]. These three studies found that T1D patients are mostly of European descendant and not necessarily different than controls. Thus, one Brazilian study and the one from Cuba reported that patients carried a greater European component than their controls; this observation was established as a risk factor[9,11].
In Colombia, the admixture process was produced differently in each region of the country. Populations in southern Colombia show higher values of Native American ancestry (NAT, average 60%), whilst African (AFR) ancestry is more observed in the region of Chocó (average 68%) and the Caribbean coast (average 30%)[12-14]. On the other hand, northwest Colombia, inhabited by the “paisa” population, exhibits the highest percentage of European ancestry, which ranges in studies from 47-79%[15-19]. In Colombia, the admixture effect has been examined for some complex diseases such as type 2 diabetes[20], asthma[21], cancer[22,23], dengue patients[24], Alzheimer’s disease[17], as well as for cardio-metabolic parameters[25].
Although much of the work on the admixture effect on several phenotypes has been done in Latin America and Colombia, none has tested this effect on T1D in Colombian patients. Our purpose was to analyze the genetic admixture composition of a set of Colombian T1D patients, by testing previously reported admixture informative markers (AIMs) in the vicinity of previously reported T1D candidate genes/loci. Besides, two chromosomal regions of high relevance to T1D in our population were tested more thoroughly. These loci were 6p21 (HLA), which is globally accepted as the T1D master risk locus, and 2p25 (RNASEH1), which has been reported solely in Colombia, so far. We inferred individual patient proportions of European, AFR and NAT ancestry components. Although the European component was higher than the two other parental contributions in a global analysis, some loci are clearly non-Europeans in cases vs the reference population, or between T1D categories. This study shed light on the genetics of T1D in a Colombian population, and reinforces the importance of including different approaches when looking for T1D genetic architecture. This is suggested by finding no admixture differences in strongly associated T1D loci, such as HLA (IDDM1) or IDDM2. In contrast, a strong genetic admixture effect was observed for other loci not described as high determinants for developing T1D. For instance, this was the case for chromosomal regions 5p13.2 and 10p11.22.
The study group consisted of 200 Colombian individuals with T1D. Their age at onset was < 15 years. Diagnostic criteria were according to the American Diabetes Association[26]. Patients were considered as “Paisas” according to a self-reported questionnaire asking for their geographical origin back until their great-grandparents. Other questions included gender, age at onset, and other family members with autoimmune diseases.
Patients were identified in the main pediatric endocrinology institutes from Antioquia: Program of Pediatric Endocrinology (Universidad de Antioquia and Hospital San Vicente Fundación), IPS Universitaria, Universidad Pontificia Bolivariana, Instituto Antioqueño de Diabetes and Clinica Integral de Diabetes. This study was approved by the ethics committee of the Faculty of Medicine at Universidad de Antioquia. Informed consent was obtained from patients and their parents before drawing blood samples.
Two diabetes-related autoantibodies (AABs) were tested in sera samples from the 200 patients. These AABs were glutamic acid decarboxylase (GAD-65 kDa) and protein tyrosine-like antigen-2 (IA-2), as reported previously[4]. They were measured using a commercial ELISA-based kit (AESKULISA and LifeSpan BioSciences, Inc) according to the manufacturer's instructions. If a patient presented with at least one of these AABs, he/she was classified as autoimmune (T1AD), or was otherwise classified as idiopathic (T1BD).
Genomic DNA was isolated from peripheral blood samples using either the phenol-chloroform or salting out protocols. A set of 75 AIMs was tested in 200 T1D patient samples using the Competitive genotyping Allele-Specific PCR technology (KASP™), which was undertaken by the Company LGC Genomics Ltd. Details of this method can be obtained from https://www.lgcgroup.com/genotyping/.
The AIMs used have a high discriminatory power (δ > 45%) among ancestral populations (Supplementary Table S1), which increases the statistical power for estimating individual ancestry. We selected these markers from Latino populations panels reported by Mao et al[27], Galanter et al[28] and Ruiz-Linares et al[29]. The AIMs were distributed throughout the genome, tagging previously reported T1D candidate loci. However, we chose a higher density of markers for chromosome 2 (23 AIMs) where the RNASEH1 gene is; and for chromosome 6 (18 AIMs) where the HLA region is.
| Chromosomal region | Ancestry | T1D, median (IQR) | CLM, median (IQR) | P value1 |
| Overall AIMs | EUR | 61.58 (52.84-69.85) | 62.06 (49.67-73.74) | 0.675 |
| NAT | 27.34 (21.36-34.05) | 25.46 (16.12- 32.90) | 0.106 | |
| AFR | 10.28 (4.0-16.83) | 10.65 (6.05-16.75) | 0.575 | |
| Chr2_EFR3B | EUR | 60.27 (34.05-80.79) | 47.88 (34.04-76.43) | 0.189 |
| NAT | 24.30 (0.01-51.24) | 37.10 (1.61-62.49) | 0.02 | |
| AFR | 7.17 (0.01-25.05) | 0.01 (0.01-15.67) | 0.06 | |
| Chr2_CTLA4 | EUR | 46.24 (16.82-69.52) | 58.26 (30.82-82.79) | 0.167 |
| NAT | 27.96 (0.04-41.31) | 25.01 (0.01-45.16) | 0.829 | |
| AFR | 20.54 (0.01-41.72) | 11.78 (0.01-36.59) | 0.183 | |
| Chr2_RNASEH1 | EUR | 56.91 (31.33-73.91) | 58.80 (32.75-78.43) | 0.482 |
| NAT | 27.41 (11.69-46.36) | 24.81 (0.01-48.84) | 0.241 | |
| AFR | 8.37 (0.01-28.24) | 13.04 (0.01-23.87) | 0.430 | |
| Chr2_IFIH1 | EUR | 42.42 (0.01-77.63) | 52.01 (16.33-82.16) | 0.05 |
| NAT | 32.07 (0.01-56.74) | 14.99 (0.01-41.35) | 0.246 | |
| AFR | 14.99 (0.01-32.31) | 17.83 (0.01-42.31) | 0.181 | |
| Chr5_IL7R | EUR | 11.56 (1.04-41.14) | 24.75 (7.20-46.98) | 7.0 × 10-3 |
| NAT | 52.18 (3.74-98.06) | 39.18 (0.04-52.80) | 1.0 × 10-4 | |
| AFR | 15.21 (3.0-35.75) | 33.89 (11.23-59.94) | 1.56 × 10-5 | |
| Chr6_MHC | EUR | 51.35 (32.92-70.32) | 55.86 (32.61-71.21) | 0.76 |
| NAT | 23.28 (8.92-40.0) | 21.61 (5.83-43.13) | 0.835 | |
| AFR | 18.87 (1.63-36.08) | 19.53 (0.3-34.11) | 0.660 | |
| Chr10_NRP1 | EUR | 63.32 (23.70-63.32) | 0.03 (0.001-20.34) | 2.2 × 10-16 |
| NAT | 36.67 (7.93-36.67) | 0.003 (0.001-7.93) | 2.2 × 10-16 | |
| AFR | 0.03 (0.001-30.91) | 94.23 (53.94-99.99) | 2.2 × 10-16 |
The 1,000 genome database was used to extract genetic information from 94 Colombians living in Medellin (CLM) for the 74 AIMs successfully typed (http://ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/). These population individuals are from the same geographical region as the patients. We calculated allele and genotypic frequencies, and Hardy-Weinberg equilibrium (HWE) using PLINK v. 1.07[30]. In addition, we considered markers that were not in linkage disequilibrium with each other. We used markers with a genotyping rate higher than 95%, without significant deviations from HWE after a Bonferroni correction.
We estimated individual European, NAT and AFR ancestry proportions using ADMIXTURE software[31]. The proportions of each component were estimated using a supervised-learning strategy, providing the genotypes of 74 AIMs from reference populations AFR, European and NAT (k = 3). We used 74/75 AIMs since one failed the PCR optimization.
To find the parental population allele frequencies, genotypes from 165 Europeans (Utah residents with ancestry from northern Europe and the West, named as CEU), and 165 AFR (Yoruba people in Ibadan, Nigeria, named as YRI) genotyped in the HapMap project were selected, which are deposited in the 1,000 genome database. Since we did not have access to NAT DNA samples or publicly available NAT genotype data on all 74 AIMs, we generated the genotypes of the 74 AIMs for 150 simulated individuals, according to the allele frequencies of NAT previously reported in the panels.
Comparison between groups for continuous variables that did not comply with normal distribution was performed using the Mann-Whitney U-test. Thus, comparisons of ancestry medians between T1D subtypes (T1AD and T1BD) according to AABs, and individuals with early/late age at onset (i.e. ≤ 5 years or > 5 years, respectively) were performed. In addition, these comparisons were also done to the CLM population. We performed these analyses for AIMs distributed across the set of candidate loci and independently for loci at different chromosomes. We ran all statistical analyses and graphs in the R package V3.3.3[32]. We also tested allelic association of these AIMs between T1D and CLM using PLINK 1.07[30].
One out of 75 AIMs did fail the PCR optimization. Therefore, we tested a total of 74 AIMs in 200 T1D patients from Antioquia, Colombia. AIMs characteristics are shown in Supplementary Table S1. Overall, the rate of genotyping was > 96% for every AIM, and there was no deviation from the HWE, after Bonferroni correction for multiple testing (P = 6.75 × 10-4). Also, as expected, none of the AIMs was in linkage disequilibrium with each other (data not shown).
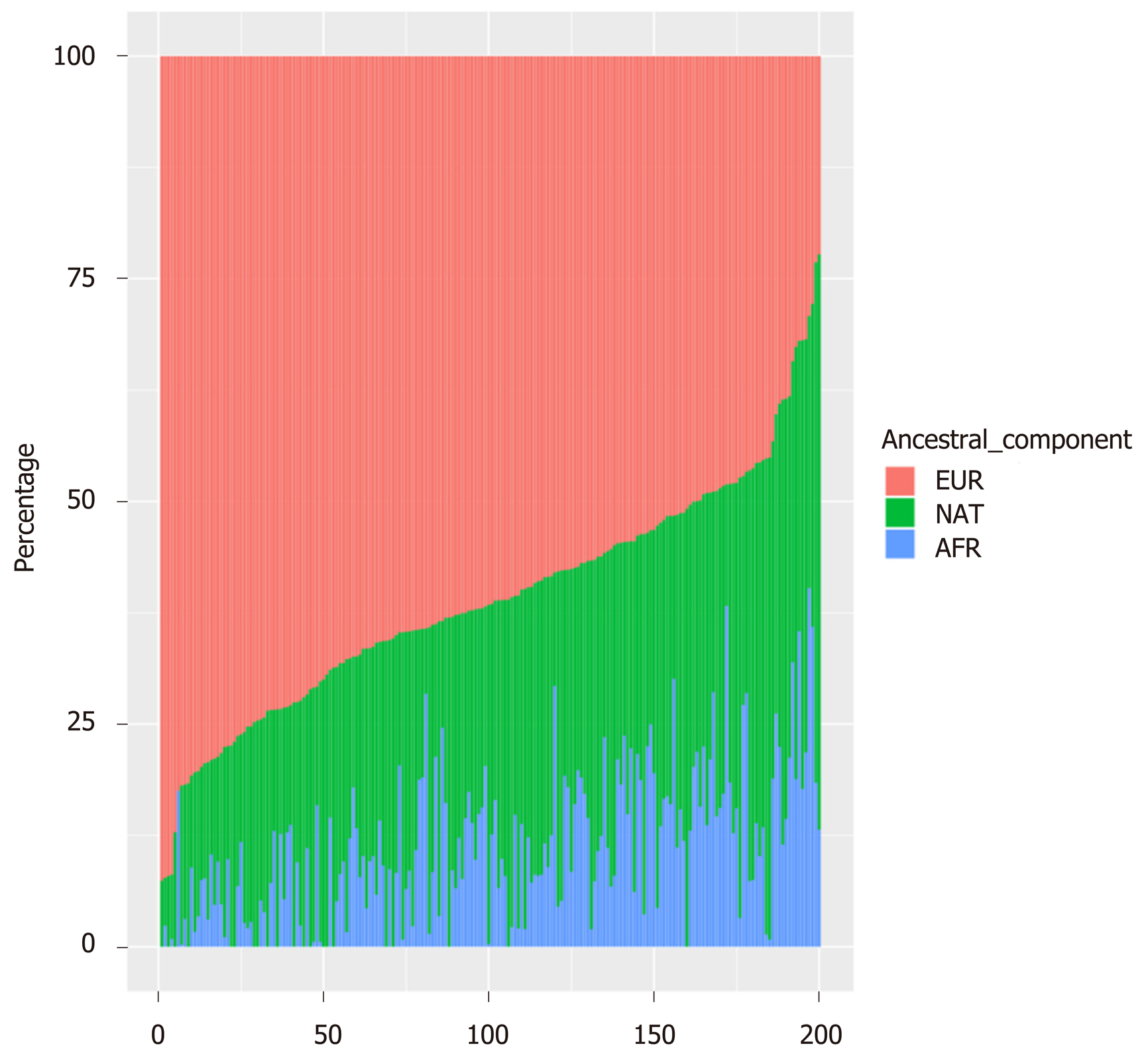
The overall ancestral genetic makeup of the 200 T1D children showed a predominant proportion of European ancestry (EUR, Median = 61.58) followed by NAT ancestry (Median = 27.34), and AFR ancestry was found at a lower proportion (Median = 10.28, Table 1 and Figure 1). Figure 1 presents the ancestry distribution for the 200 T1D children studied here. It can be noticed that the European component is the prominent one. European ancestry ranged from 22% to 93%; the NAT ancestry ranged from 0 to 65%, and the AFR ancestry ranged from 0 to 40%.
Looking at the overall set of AIMs, and also at their distribution in specific loci, it was observed that diseased individuals of EUR ancestry had a median from 61.58-11.56. The lowest value was found for chromosome 5 AIMs (Table 1). NAT ancestry ranged from 52.18-24.30 in the diseased subjects. The highest value was found for gene IL7R AIMs (chromosome 5), and the lowest value was found for gene EFR3B AIMs (chromosome 2). The AFR component (AFR) ranged from 20.58 to 0.01. the lowest AFR ancestry was found for gene IFIH1 AIMs (chromosome 2, Table 1). The wide ancestry variation across chromosomal regions is noticeable.
Overall, the CLM reference population displayed a very similar ancestry distribution compared to T1D cases. Nonetheless, specific T1D loci presented marked differences between the two groups; one such difference was observed for the gene EFR3B, which presented with higher NAT in the CLM population (P = 0.02), suggesting a protective role for developing T1D (Table 1). Also, at gene IFIH1, T1D patients presented with lower European ancestry (P = 0.05), at the expense of a higher NAT component than in CLM (Table 1). Other differences between T1D and CLM were observed for the IL7R and NRP1 genes (Chromosomes 5 and 10, respectively) as follows.
Chromosome 5 AIMs (gene IL7R) showed less European (P = 7.0 × 10-3) and less AFR ancestries (1.56 × 10-5) in diseased subjects than the CLM population; consequently, T1D patients had more NAT ancestry than CLM subjects at this chromosomal region (P = 1.0 × 10-4, Table 1). Regarding chromosome 10 AIMs (gene NRP1), it was observed that T1D patients presented a high European component compared to CLM (63.32 vs 0.03, Table 1). Conversely, patients presented an almost zero AFR component for this chromosomal region compared to CLM (0.03 vs 94.23, Table 1). Consequently, T1D patients displayed a predominance of NAT ancestry at this locus compared to CLM (36.67 vs 0.003, Table 1).
An exploratory association analysis showed that, after adjusting for admixture, seven markers were associated with T1D (Supplementary Table S2 and Table 2). The most significant findings were located on chromosomes 5 and 10 (P = 5.56 × 10-6 and 8.70 × 10-19, respectively). It is interesting that only one MHC marker (rs2395656) presented an association with the disease, and this happened with less strength in its association (P = 0.04) than markers at chromosomes 5 and 10 (Table 2).
| CHR | SNP | A1 | MAF | ORa | 95%CI | P value1 | EMP1 | |
| T1D | CLM | |||||||
| 2 | rs798364 | A | 0.18 | 0.27 | 0.62 | 0.40-0.96 | 0.034 | 0.035 |
| 2 | rs1606237 | T | 0.33 | 0.26 | 1.55 | 1.04-2.30 | 0.031 | 0.031 |
| 5 | rs700164 | T | 0.56 | 0.35 | 2.38 | 1.62-3.48 | 5.56 × 10-6 | 3.0 × 10-6 |
| 6 | rs9378428 | C | 0.38 | 0.48 | 0.61 | 0.42-0.89 | 0.010 | 0.018 |
| 6 | rs2523747 | G | 0.42 | 0.30 | 1.68 | 1.13-2.51 | 0.010 | 0.012 |
| 6 | rs2395656 | G | 0.23 | 0.28 | 0.63 | 0.41-0.98 | 0.040 | 0.041 |
| 10 | rs3123687 | G | 0.15 | 0.86 | 0.04 | 0.02-0.08 | 8.70 × 10-19 | 1.0 × 10-6 |
We stratified the T1D sample according to the presence (T1AD, autoimmune) or absence (T1BD, idiopathic) of diabetes-related AABs; we also stratified the patient group according to their age at onset, e.g., early (≤ 5 years) or late (> 5 years). We found that 78% (n = 156) of the patients had at least one T1D specific autoantibody (GAD-65 and IA-2), while the 22% remaining (n = 44) were negative for these two antibodies. T1AD average age at onset was 8.25 years, whilst for T1BD it was 7.22. We did not find significant differences between men and women within these two groups (data not shown).
Over thirty percent (n = 61, 30.5%) of T1D individuals developed the disease before the age of 5 years, with an average age at onset of 2.66 years. The remaining sample (69.5%, n = 139) presented with an age at onset after 5 years, with a mean for this category of 10.28 years. As in the stratification by AABs, we did not find significant differences between men and women within the age at onset categories (data not shown). Regarding the autoimmune category, comparisons among ancestral genetic composition led to the identification of no differences for the 74 AIMs taken together (Table 3). However, looking at individual loci, it was observed that MHC AIMs present with lower NAT ancestry in the autoimmune subgroup (P = 0.019, Table 3).
| Chromosomal region | Ancestry | T1AD, median (IQR) | T1BD, median (IQR) | P value1 |
| Overall AIMs | EUR | 61.92 (53.57-70.89) | 59.79 (49.70-68.03) | 0.333 |
| NAT | 27.07 (20.82-33.48) | 29.16 (21.57-37.06) | 0.312 | |
| AFR | 10.19 (2.88-16.01) | 11.54 (5.55-17.80) | 0.344 | |
| Chr2_EFR3B | EUR | 60.28 (36.24-83.66) | 59.79 (32.34-80.01) | 0.551 |
| NAT | 25.16 (0.01-51.24) | 21.92 (0.01-45.95) | 0.979 | |
| AFR | 4.53 (0.01-23.32 | 14.41 (0.01-27.36) | 0.119 | |
| Chr2_CTLA4 | EUR | 53.34 (16.82-70.53) | 41.16 (16.62-60.86) | 0.231 |
| NAT | 26.28 (4.55-41.31) | 32.69 (9.35-50.25) | 0.343 | |
| AFR | 20.54 (0.01-41.73) | 19.47 (0.01-42.52) | 0.764 | |
| Chr2_RNASEH1 | EUR | 58.96 (37.72-73.87) | 47.50 (26.38-78.10) | 0.506 |
| NAT | 26.92 (14.19-45.66) | 31.59 (0.01-51.98) | 0.885 | |
| AFR | 6.85 (0.01-28.24) | 11.71 (0.01-27.86) | 0.657 | |
| Chr2_IFIH1 | EUR | 41.90 (0.01-76.72) | 42.42 (0.01-67.93) | 0.708 |
| NAT | 32.56 (0.01-55.56) | 27.39 (2.01-56.89) | 0.985 | |
| AFR | 14.99 (0.01-42.31) | 0.01 (0.01-36.48) | 0.654 | |
| Chr5_IL7R | EUR | 13.27 (1.02-43.73) | 9.19 (1.77-33.67) | 0.555 |
| NAT | 50.94 (38.08-98.60) | 53.12 (31.82-95.13) | 0.984 | |
| AFR | 14.68 (3.17-33.94) | 17.98 (2.75-41.01) | 0.634 | |
| Chr6_MHC | EUR | 52.31 (37.26-72.43) | 45.73 (19.25-67.03) | 0.087 |
| NAT | 20.32 (7.12-37.06) | 31.88 (17.65-44.62) | 0.019 | |
| AFR | 18.50 (3.21-36.04) | 21.13 (0.53-36.56) | 0.905 | |
| Chr10_NRP1 | EUR | 63.32 (28.61-69.09) | 63.31 (6.89-63.32) | 0.092 |
| NAT | 36.67 (7.93-36.67) | 36.67 (7.93-59.79) | 0.282 | |
| AFR | 0.25 (1e-05-26.0) | 0.52 (1e-05-37.80) | 0.848 |
In addition, when comparing diseased individuals in the autoimmune categories to CLM population, it was observed that gene EFR3B AIMs present differences in their ancestral components (Supplementary Table S3). Thus, autoimmune patients presented with less NAT ancestry (P = 0.032), whilst T1D idiopathic category presented with higher AFR ancestry (P = 0.016). Regarding the age at onset categories, it was observed that the AFR ancestry is significantly higher in the late onset subgroup at gene IL7R AIMs (P = 0.023, Table 4). Comparing these two categories to the CLM population showed no significant differences for either the overall set of AIMs nor specific loci (Supplementary Table S3).
| Chromosomal region | Ancestry | Early age at onset, ≤ 5 yr | Late age at onset, > 5 yr | P value1 |
| Overall AIMs | EUR | 62.06 (54.50-71.38) | 61.12 (51.66-68.68) | 0.420 |
| NAT | 25.40 (18.14-33.55) | 27.75 (21.86-34.35) | 0.345 | |
| AFR | 11.09 (3.90-17.25) | 10.19 (3.88-16.91) | 0.927 | |
| Chr2_EFR3B | EUR | 57.65 (34.05-82.17) | 62.45 (42.04-80.67) | 0.419 |
| NAT | 25.16 (1.27-52.11) | 24.30 (0.01-51.73) | 0.607 | |
| AFR | 5.02 (0.01-27.29) | 7.37 (0.01 23.32) | 0.941 | |
| Chr2_CTLA4 | EUR | 58.49 (24.63-78.80) | 46.09 (12.90-66.30) | 0.180 |
| NAT | 26.28 (3.98-39.82) | 30.19 (4.55-42.10) | 0.362 | |
| AFR | 18.05 (0.01-37.80) | 20.68 (0.01-41.72) | 0.701 | |
| Chr2_RNASEH1 | EUR | 56.84 (44.50-72.18) | 56.91 (28.02-74.15) | 0.672 |
| NAT | 27.22 (0.01-31.22) | 28.03 (8.15-48.58) | 0.472 | |
| AFR | 3.38 (0.01-31.22) | 8.62 (0.01-27.20) | 0.917 | |
| Chr2_IFIH1 | EUR | 41.35 (0.01-66.94) | 42.42 (4.44-76.73) | 0.126 |
| NAT | 33.05 (0.67-57.49) | 29.05 (0.01-53.09) | 0.339 | |
| AFR | 14.57 (0.01-42.31) | 1.01 (0.01-42.31) | 0.796 | |
| Chr5_IL7R | EUR | 9.38 (0.33-41.70) | 12.86 (1.73-43.11) | 0.338 |
| NAT | 53.83 (39.76-99.60) | 50.94 (30.21-88.75) | 0.197 | |
| AFR | 6.17 (1.01-29.03) | 19.96 (9.6-37.36) | 0.023 | |
| Chr6_MHC | EUR | 51.67 (32.23-63.29) | 52.07 (34.26-71.84) | 0.596 |
| NAT | 30.57 (13.47-43.21) | 26.71 (7.69-37.39) | 0.480 | |
| AFR | 15.03 (4.46-36.05) | 20.52 (1.08-36.73) | 0.118 | |
| Chr10_NRP1 | EUR | 63.32 (23.70-63) | 63.32(43.07-69.08) | 0.357 |
| NAT | 36.68 (7.93-36.67) | 36.67 (7.93-36.68) | 0.797 | |
| AFR | 0.16 (0.001-39-1) | 0.38 (0.001-63.3) | 0.498 |
T1D incidence differences among countries, mainly related to European versus non-Europeans, led us to assess whether our T1D patients had a predominantly European ancestral component or other. Our analyses were based on 74 AIMs located on previously reported T1D loci/genes. AIM deltas (δs) between the NAT, European and AFR populations indicated that they were appropriate discriminators. We found that T1D patients from northwest Colombia are predominantly of European ancestry, followed by NAT and AFR components. Proportion estimates of the three parental populations for this sample were consistent with those reported in previous studies for Colombians, but using different sets of markers[13,16,19,20,29].
We also compared T1D children to CLM. Analyzing the overall set of AIMs found no statistically significant differences in the ancestral genetic component between the two groups. Comparable results were obtained by Gomes et al[10] in Sao Paulo-Brazil; they noted that the European component predominated in both T1D patients and controls, followed by AFR and NAT ancestry; however, no significant differences between cases and controls were observed. For the contrary, a study conducted in ten Brazilian cities showed that T1D patients presented a higher percentage of European component than the healthy population[9]. Similarly, a study by Diaz-Horta et al[11] found a higher proportion of European component in cases than in controls. Even more, they found a risk association with the European ancestry.
Further analysis disaggregating the candidate loci tested led us to find a different ancestry composition for MHC AIMs. Lower NAT ancestry was observed in T1AD compared to T1BD patients (Table 3). Ancestry variation at the HLA region has been reported for Latin American populations. However, such variation has shown an excess of the AFR component in these populations, including CLM[16,33,34]. It has been suggested that the excess of the AFR component in the HLA region in Latin America is due to a positive selection orchestrated by the presence of infectious agents during the process of the conquest. The European conquerors brought to America, African and European diseases such as smallpox, measles, and influenza, which caused massive epidemics and were responsible for the extinction of many native populations[34]. Given this historical background, these AFR fragments could obtain a selective advantage, since the AFR populations have the most diverse repertoire in HLA[35,36]. However, the ancestry variation observed here shows that the European component is higher in autoimmune (T1AD) than T1BD, in combination with lower NAT in T1AD than T1BD (Table 3).
Another gene with remarkable findings is IFIH1. This observation is of particular interest to our population, since we had found in the past that SNP rs10930046, which is located at IFIH1, associates with T1D in our population[37]. This SNP has been reported as a rare variant in European populations (MAF = 0.02) related to Psoriasis[38]. Interestingly, we found in our previous study that this variant MAF = 0.3[37]. Therefore, such an allele frequency difference could have been speculatively explained by random genetic drift, involving over-representation of European chromosomes with such variants at the time of conquering Colombia. However, in the present study, evidence suggests that this allele frequency difference between populations might be a NAT contribution.
It is worth mentioning that IFIH1 AIMs presented wide values for the AFR component comparing autoimmune to idiopathic patients (14.99 vs 0.01, Table 3), without reaching statistical significance. This was the case since the interquartile range overlapped between these two autoimmune categories. Neither gene CTLA4 nor RNASEH1 AIMs revealed significant contributions to T1D, either looking to the overall set of AIMs or in any of the loci/genes analyzed. Regarding CTLA4, this observation makes sense when related to our previous finding of no association of this gene variant with T1D[37]. However, a different situation holds for the RNASEH1 gene.
RNASEH1 gene variants have thus far been associated with T1D only in the northwest Colombia population and not elsewhere in the world[4]. It has not even been reported in GWA studies using large sample sizes, albeit mostly of European origin[3]. Analyzing a larger sample size of T1D patients from this region in Colombia will allow us to conclude whether there really is an ancestry effect related to RNASEH1 gene variants in T1D.
Unexpectedly, we found that ancestry for chromosomes 5 and 10 were sharply different between T1D patients and the CLM population (Tables 1 and 3). The former involves chromosomal region 5p13.2 (IL7R)[39]. This region was assessed with only one AIM, which clearly discriminates between NAT and non-NAT (Supplementary Table S1). As shown in Table 1, the T1D ancestry observed for this locus is confidently greater for NAT, at the expense of the two other ancestries. It is also apparent that AFR ancestry at this locus contributes to late onset of the disease (Table 4). Such results, in turn, should be taken with caution since this AIM does not clearly discriminate between EUR and AFR (Supplementary Table S1). Therefore, we cannot rule out the possibility that this effect is of European origin.
The second striking finding involves chromosomal region 10p11.22 (gene NRP1)[40]. Although the opposite ancestry contributions between T1D and CLM are evident (Table 1), it is worth keeping in mind that the only AIM (rs3123687) used for this locus is highly informative for AFR and non-AFR ancestries (i.e., either EUR or NAT). Given this information, we are aware that the conclusion regarding greater NAT contribution in our study could eventually go towards greater EUR ancestry. Therefore we can only tell that the difference observed is non-AFR, but are not able to define whether it is European or NAT.
The actual SNPs reported as associated with disease in these two genes (IL7R and NRP1) have not yet been tested in the sample presented here. However, a test of association using the AIMs analyzed here, after adjusting for the admixture effect, revealed that AIM rs700164 associates with affected status (5.56 × 10-6, Supplementary Table S2 and Table 2) and that similarly rs3123687 strongly associates with the disease (P = 8.07 × 10-19, Supplementary Table S2 and Table 2) for IL7R and NRP1 genes, respectively. A verification of this finding should be performed using the transmission disequilibrium test (TDT). The TDT is not susceptible to population structure issues, such as admixture. This analysis is to be done for the actual SNPs, as the parents for the patients presented here are available. Such association analyses should include choosing gene variants from the genetic variability in this set of patients, and should also consider the LD blocks observed in this population.
No ancestry differences were found overall when comparing T1AD to idiopathic (T1BD) (Table 3). T1AD, whose etiology and pathology are better characterized, has a higher incidence in Europe[6]; on the contrary, T1BD is reported mainly in AFR and Asian countries[26]. Our results are different from those by Piñero-Piloña et al[41], who reported a high incidence of T1BD in Mexican patients, whose predominant ancestral component was NAT. Our cohort presents a majority of autoimmune cases (78%) and, as described here, their predominant ancestry is of European contribution.
However, looking at chromosomal regions along the analysis stratified by age at onset of T1D, we found that patients with a late onset of the disease have a greater AFR component, which was more marked on chromosome 5 (Table 4). This suggests that AFR ancestry could be a risk factor for developing the disease at a late age in our population (over 2/3 of the sample had age at onset > 5 years), which can modify the metabolic phenotype of patients, and influence the risk of late complications of diabetes[42].
Our study has an important limitation regarding the number and location of the AIMs. Thus, chromosomes 5 and 10 were tested with just a few such markers. It will be worth testing more AIMs nearby these two loci to further examine the differences revealed. Also, the reference population we used (CLM from the 1,000 genome database), although supposedly unaffected and older than our patients, were typed by a different method from the one we used to type our T1D cases. Nonetheless, both groups share comparable genetic ancestries.
Our study’s strength is its population choice. As described, the northwest Colombia population is the one with a greater European component in the country[15-19]. Thus, our results make much more sense regarding the overall European contribution, together with the apparent unexplored NAT input to T1D, in addition to certain contributions of the AFR ancestry for late age at onset.
In conclusion, this study describes the ancestral genetic composition of 200 T1D patients from an admixed population from northwest Colombia. Consistently, we found a predominant proportion of European followed by NAT ancestry. No statistical difference was observed in the distribution of the proportions of ancestral genetic components between T1D patients and the CLM reference population. A variation in chromosomal segments derived from the parental populations was observed when comparing individuals with T1AD versus T1BD, and those who had an early (≤ 5 years) or late (> 5 years) age at onset of the disease. These results demonstrate that the study of the genetic admixture provides new perspectives in the delineation of the genetic architecture underlying autoimmune diseases. Finally, performing a novel study in this sample, including unbiased distribution of AIMs through the whole genome, could help find undetected loci in previous studies, which would contribute to complete the T1D genetic architecture for our population. This will also contribute to making approaches, such as the polygenic risk score, become more accurate for these types of populations.
Type 1 diabetes (T1D) is described as a disease predominantly in white populations. Subtypes of the disease are also more frequent in different ethnicities. Thus, the autoimmune form of the disease is observed more frequently in Caucasian countries, whilst the idiopathic form is more frequently observed in African and Asian countries. The patients included in this study are from Northwest Colombia. This is an admixed population originated by a three ethnic contribution. This population has been described as the most European in the country, followed by the Native American ancestry, and with its least significant component being African contribution.
In this study, we looked at the genetic ancestry of a set of 200 diseased subjects from Northwest Colombia. We were interested in describing whether their global ancestry, as well as some specific genomic regions, were of which particular ancestry. Only a few of these types of studies have been reported in Latin American populations, and none have occurred in Colombia.
We aimed at describing the ancestry composition of a cohort of Colombian patients with T1D. This description included both global analysis as well as specific tests on loci/genes previously related to the disease.
We studied 200 diseased subjects from Northwest Colombia. We tested 75 admixture informative markers (AIMs) distributed through a set of previously reported genes (or chromosomal regions) associated with T1D. The disease was classified as either autoimmune or idiopathic in the study subjects. This was done by testing two disease-related auto-antibodies (AABs). If at least one such AAB was present, then the disease was classified as autoimmune. We also classified the age at onset of the disease as early (≤ 5 years) or late (> 5 years). The reference population of Colombians living in Medellin (CLM) was compared to the set of patients presented here. We applied appropriate statistical tests given the non-normality of the data obtained.
Seventy eight percent of the patients presented at least one AAB. Over two thirds (69.5%) of the subjects developed the disease after 5-years-old. There were no significant differences between genders among the affected individuals. Seventy four AIMs were successfully tested (one failed the PCR optimization). It was observed that both the diseased and CLM groups were predominantly of European ancestry (61.58 vs 62.06), followed by Native American (24.30 vs 37.10) and African ancestries (10.28 vs 10.65). In addition, specific genes such as EFR3B, IFIH1, IL7R and NRP1 displayed differential Native American or African rather than European contributions. In addition, we found that autoimmune patients displayed lower Native American ancestry than idiopathic cases.
Our study shows that diseased individuals from Northwest Colombia are predominantly of European ancestry, followed by native American and African ancestries. Also, other European contributions were found for specific genes in our study.
MHC is expected to play the strongest role in T1D susceptibility. However, this was not the observation in our study. Our results suggest that different loci effect sizes might be at play in our admix population. This is inferred from the observation of the significance strength observed for MHC ancestry compared to other loci. Therefore, it would be worth testing AIMs in this sample (expanded with extra individuals from the same region in Colombia) throughout the whole genome. This way, it would be feasible to reveal differences in local ancestry either for known or unknown loci associated with T1D in our population. This would help complete the genetic architecture of the disease, particularly for our population. In turn, this would contribute to the knowledge of the disease biology, and would also make this sample population appropriate for applying approaches such as the polygenic risk score.
We are very grateful to the patients that participated in this study. We are also very grateful to Doctors Martin Toro, Maria Victoria Lopera, Jorge García and Alejandra Velez for contributing patients to this study.
Manuscript source: Unsolicited manuscript
Specialty type: Endocrinology and metabolism
Country of origin: Colombia
Peer-review report classification
Grade A (Excellent):
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Dabla PK, Neri V S-Editor: Yan JP L-Editor: Filipodia E-Editor: Zhang YL
| 1. | Morran MP, Vonberg A, Khadra A, Pietropaolo M. Immunogenetics of type 1 diabetes mellitus. Mol Aspects Med. 2015;42:42-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 92] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 2. | Onengut-Gumuscu S, Chen WM, Burren O, Cooper NJ, Quinlan AR, Mychaleckyj JC, Farber E, Bonnie JK, Szpak M, Schofield E, Achuthan P, Guo H, Fortune MD, Stevens H, Walker NM, Ward LD, Kundaje A, Kellis M, Daly MJ, Barrett JC, Cooper JD, Deloukas P; Type 1 Diabetes Genetics Consortium, Todd JA, Wallace C, Concannon P, Rich SS. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381-386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 562] [Cited by in RCA: 520] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 3. | Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS; Type 1 Diabetes Genetics Consortium. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703-707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1427] [Cited by in RCA: 1360] [Article Influence: 85.0] [Reference Citation Analysis (0)] |
| 4. | Pineda-Trujillo N, Rodríguez-Acevedo A, Rodríguez A, Ruíz-Linares A, Bedoya G, Rivera A, Alfaro JM. RNASEH1 gene variants are associated with autoimmune type 1 diabetes in Colombia. J Endocrinol Invest. 2018;41:755-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 5. | Patterson C, Guariguata L, Dahlquist G, Soltész G, Ogle G, Silink M. Diabetes in the young - a global view and worldwide estimates of numbers of children with type 1 diabetes. Diabetes Res Clin Pract. 2014;103:161-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 273] [Article Influence: 24.8] [Reference Citation Analysis (2)] |
| 6. | Diaz-Valencia PA, Bougnères P, Valleron AJ. Covariation of the incidence of type 1 diabetes with country characteristics available in public databases. PLoS One. 2015;10:e0118298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 7. | Aschner P. Diabetes trends in Latin America. Diabetes Metab Res Rev. 2002;18 Suppl 3:S27-S31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 46] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Gómez-Díaz RA, Garibay-Nieto N, Wacher-Rodarte N, Aguilar-Salinas CA. Epidemiology of type 1 diabetes in Latin America. Curr Diabetes Rev. 2014;10:75-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 9. | Gomes MB, Gabrielli AB, Santos DC, Pizarro MH, Barros BSV, Negrato CA, Dib SA, Porto LC, Silva DA. Self-reported color-race and genomic ancestry in an admixed population: A contribution of a nationwide survey in patients with type 1 diabetes in Brazil. Diabetes Res Clin Pract. 2018;140:245-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 10. | Gomes KF, Santos AS, Semzezem C, Correia MR, Brito LA, Ruiz MO, Fukui RT, Matioli SR, Passos-Bueno MR, Silva ME. The influence of population stratification on genetic markers associated with type 1 diabetes. Sci Rep. 2017;7:43513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Diaz-Horta O, Cintado A, Fernandez-De-Cossio ME, Nazabal M, Ferrer A, Roca J, Camacho H, Benítez J, Ale M, Villarreal A, Molina G, Vera M, Cabrera-Rode E, Novoa L. Relationship of type 1 diabetes to ancestral proportions and HLA DR/DQ alleles in a sample of the admixed Cuban population. Ann Hum Biol. 2010;37:778-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 12. | Ossa H, Aquino J, Pereira R, Ibarra A, Ossa RH, Pérez LA, Granda JD, Lattig MC, Groot H, Fagundes de Carvalho E, Gusmão L. Outlining the Ancestry Landscape of Colombian Admixed Populations. PLoS One. 2016;11:e0164414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 13. | Rojas W, Parra MV, Campo O, Caro MA, Lopera JG, Arias W, Duque C, Naranjo A, García J, Vergara C, Lopera J, Hernandez E, Valencia A, Caicedo Y, Cuartas M, Gutiérrez J, López S, Ruiz-Linares A, Bedoya G. Genetic make up and structure of Colombian populations by means of uniparental and biparental DNA markers. Am J Phys Anthropol. 2010;143:13-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 14. | Ibarra A, Freire-Aradas A, Martínez M, Fondevila M, Burgos G, Camacho M, Ostos H, Suarez Z, Carracedo A, Santos S, Gusmão L. Comparison of the genetic background of different Colombian populations using the SNPforID 52plex identification panel. Int J Legal Med. 2014;128:19-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 15. | Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, Tandon A, Schirmer C, Neubauer J, Bedoya G, Duque C, Villegas A, Bortolini MC, Salzano FM, Gallo C, Mazzotti G, Tello-Ruiz M, Riba L, Aguilar-Salinas CA, Canizales-Quinteros S, Menjivar M, Klitz W, Henderson B, Haiman CA, Winkler C, Tusie-Luna T, Ruiz-Linares A, Reich D. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80:1024-1036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 217] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 16. | Rishishwar L, Conley AB, Wigington CH, Wang L, Valderrama-Aguirre A, Jordan IK. Ancestry, admixture and fitness in Colombian genomes. Sci Rep. 2015;5:12376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 77] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 17. | Moreno DJ, Pino S, Ríos Á, Lopera F, Ostos H, Via M, Bedoya G. Genetic Ancestry and Susceptibility to Late-Onset Alzheimer Disease (LOAD) in the Admixed Colombian Population. Alzheimer Dis Assoc Disord. 2017;31:225-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Wang S, Ray N, Rojas W, Parra MV, Bedoya G, Gallo C, Poletti G, Mazzotti G, Hill K, Hurtado AM, Camrena B, Nicolini H, Klitz W, Barrantes R, Molina JA, Freimer NB, Bortolini MC, Salzano FM, Petzl-Erler ML, Tsuneto LT, Dipierri JE, Alfaro EL, Bailliet G, Bianchi NO, Llop E, Rothhammer F, Excoffier L, Ruiz-Linares A. Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet. 2008;4:e1000037. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 316] [Cited by in RCA: 314] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 19. | Bedoya G, Montoya P, García J, Soto I, Bourgeois S, Carvajal L, Labuda D, Alvarez V, Ospina J, Hedrick PW, Ruiz-Linares A. Admixture dynamics in Hispanics: a shift in the nuclear genetic ancestry of a South American population isolate. Proc Natl Acad Sci USA. 2006;103:7234-7239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 161] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 20. | Campbell DD, Parra MV, Duque C, Gallego N, Franco L, Tandon A, Hünemeier T, Bortolini C, Villegas A, Bedoya G, McCarthy MI, Price A, Reich D, Ruiz-Linares A. Amerind ancestry, socioeconomic status and the genetics of type 2 diabetes in a Colombian population. PLoS One. 2012;7:e33570. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 21. | Vergara C, Murray T, Rafaels N, Lewis R, Campbell M, Foster C, Gao L, Faruque M, Oliveira RR, Carvalho E, Araujo MI, Cruz AA, Watson H, Mercado D, Knight-Madden J, Ruczinski I, Dunston G, Ford J, Caraballo L, Beaty TH, Mathias RA, Barnes KC. African ancestry is a risk factor for asthma and high total IgE levels in African admixed populations. Genet Epidemiol. 2013;37:393-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 22. | Lopera EA, Baena A, Florez V, Montiel J, Duque C, Ramirez T, Borrero M, Cordoba CM, Rojas F, Pareja R, Bedoya AM, Bedoya G, Sanchez GI. Unexpected inverse correlation between Native American ancestry and Asian American variants of HPV16 in admixed Colombian cervical cancer cases. Infect Genet Evol. 2014;28:339-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 23. | Hernandez-Suarez G, Sanabria MC, Serrano M, Herran OF, Perez J, Plata JL, Zabaleta J, Tenesa A. Genetic ancestry is associated with colorectal adenomas and adenocarcinomas in Latino populations. Eur J Hum Genet. 2014;22:1208-1216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 24. | Chacón-Duque JC, Adhikari K, Avendaño E, Campo O, Ramirez R, Rojas W, Ruiz-Linares A, Restrepo BN, Bedoya G. African genetic ancestry is associated with a protective effect on Dengue severity in colombian populations. Infect Genet Evol. 2014;27:89-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 25. | Muñoz AM, Velásquez CM, Bedoya G. Cardio-metabolic parameters are associated with genetic admixture estimates in a pediatric population from Colombia. BMC Genet. 2016;17:93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | American Diabetes Association. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2019. Diabetes Care. 2019;42:S13-S28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1621] [Cited by in RCA: 1964] [Article Influence: 327.3] [Reference Citation Analysis (0)] |
| 27. | Mao X, Bigham AW, Mei R, Gutierrez G, Weiss KM, Brutsaert TD, Leon-Velarde F, Moore LG, Vargas E, McKeigue PM, Shriver MD, Parra EJ. A genomewide admixture mapping panel for Hispanic/Latino populations. Am J Hum Genet. 2007;80:1171-1178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 177] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 28. | Galanter JM, Fernandez-Lopez JC, Gignoux CR, Barnholtz-Sloan J, Fernandez-Rozadilla C, Via M, Hidalgo-Miranda A, Contreras AV, Figueroa LU, Raska P, Jimenez-Sanchez G, Zolezzi IS, Torres M, Ponte CR, Ruiz Y, Salas A, Nguyen E, Eng C, Borjas L, Zabala W, Barreto G, González FR, Ibarra A, Taboada P, Porras L, Moreno F, Bigham A, Gutierrez G, Brutsaert T, León-Velarde F, Moore LG, Vargas E, Cruz M, Escobedo J, Rodriguez-Santana J, Rodriguez-Cintrón W, Chapela R, Ford JG, Bustamante C, Seminara D, Shriver M, Ziv E, Burchard EG, Haile R, Parra E, Carracedo A; LACE Consortium. Development of a panel of genome-wide ancestry informative markers to study admixture throughout the Americas. PLoS Genet. 2012;8:e1002554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 183] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 29. | Ruiz-Linares A, Adhikari K, Acuña-Alonzo V, Quinto-Sanchez M, Jaramillo C, Arias W, Fuentes M, Pizarro M, Everardo P, de Avila F, Gómez-Valdés J, León-Mimila P, Hunemeier T, Ramallo V, Silva de Cerqueira CC, Burley MW, Konca E, de Oliveira MZ, Veronez MR, Rubio-Codina M, Attanasio O, Gibbon S, Ray N, Gallo C, Poletti G, Rosique J, Schuler-Faccini L, Salzano FM, Bortolini MC, Canizales-Quinteros S, Rothhammer F, Bedoya G, Balding D, Gonzalez-José R. Admixture in Latin America: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet. 2014;10:e1004572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 280] [Cited by in RCA: 318] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 30. | Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24221] [Cited by in RCA: 24217] [Article Influence: 1345.4] [Reference Citation Analysis (0)] |
| 31. | Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655-1664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4449] [Cited by in RCA: 5741] [Article Influence: 358.8] [Reference Citation Analysis (0)] |
| 32. | R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria; 2014. Available from: https://www.R-project.org/. |
| 33. | Meyer D, C Aguiar VR, Bitarello BD, C Brandt DY, Nunes K. A genomic perspective on HLA evolution. Immunogenetics. 2018;70:5-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 128] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 34. | Deng L, Ruiz-Linares A, Xu S, Wang S. Ancestry variation and footprints of natural selection along the genome in Latin American populations. Sci Rep. 2016;6:21766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 35. | Sanchez-Mazas A, Lemaître JF, Currat M. Distinct evolutionary strategies of human leucocyte antigen loci in pathogen-rich environments. Philos Trans R Soc Lond B Biol Sci. 2012;367:830-839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 36. | Ramos PS, Shedlock AM, Langefeld CD. Genetics of autoimmune diseases: insights from population genetics. J Hum Genet. 2015;60:657-664. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 117] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 37. | Rodríguez A, Alfaro JM, Balthazar V, Pineda Trujillo N. Association analysis of PTPN22, CTLA4 and IFIH1 genes with type 1 diabetes in Colombian families. J Diabetes. 2015;7:402-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 38. | Li Y, Liao W, Cargill M, Chang M, Matsunami N, Feng BJ, Poon A, Callis-Duffin KP, Catanese JJ, Bowcock AM, Leppert MF, Kwok PY, Krueger GG, Begovich AB. Carriers of rare missense variants in IFIH1 are protected from psoriasis. J Invest Dermatol. 2010;130:2768-2772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 39. | Santiago JL, Alizadeh BZ, Martínez A, Espino L, de la Calle H, Fernández-Arquero M, Figueredo MA, de la Concha EG, Roep BO, Koeleman BP, Urcelay E. Study of the association between the CAPSL-IL7R locus and type 1 diabetes. Diabetologia. 2008;51:1653-1658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 40. | Hasan NM, Kendrick MA, Druckenbrod NR, Huelsmeyer MK, Warner TF, MacDonald MJ. Genetic association of the neuropilin-1 gene with type 1 diabetes in children: Neuropilin-1 expression in pancreatic islets. Diabetes Res Clin Pract. 2010;87:e29-e32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 41. | Piñero-Piloña A, Raskin P. Idiopathic Type 1 diabetes. J Diabetes Complications. 2001;15:328-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 42. | Valdes AM, Erlich HA, Carlson J, Varney M, Moonsamy PV, Noble JA. Use of class I and class II HLA loci for predicting age at onset of type 1 diabetes in multiple populations. Diabetologia. 2012;55:2394-2401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |