Copyright
©2011 Baishideng Publishing Group Co.
World J Diabetes. Oct 15, 2011; 2(10): 164-175
Published online Oct 15, 2011. doi: 10.4239/wjd.v2.i10.164
Published online Oct 15, 2011. doi: 10.4239/wjd.v2.i10.164
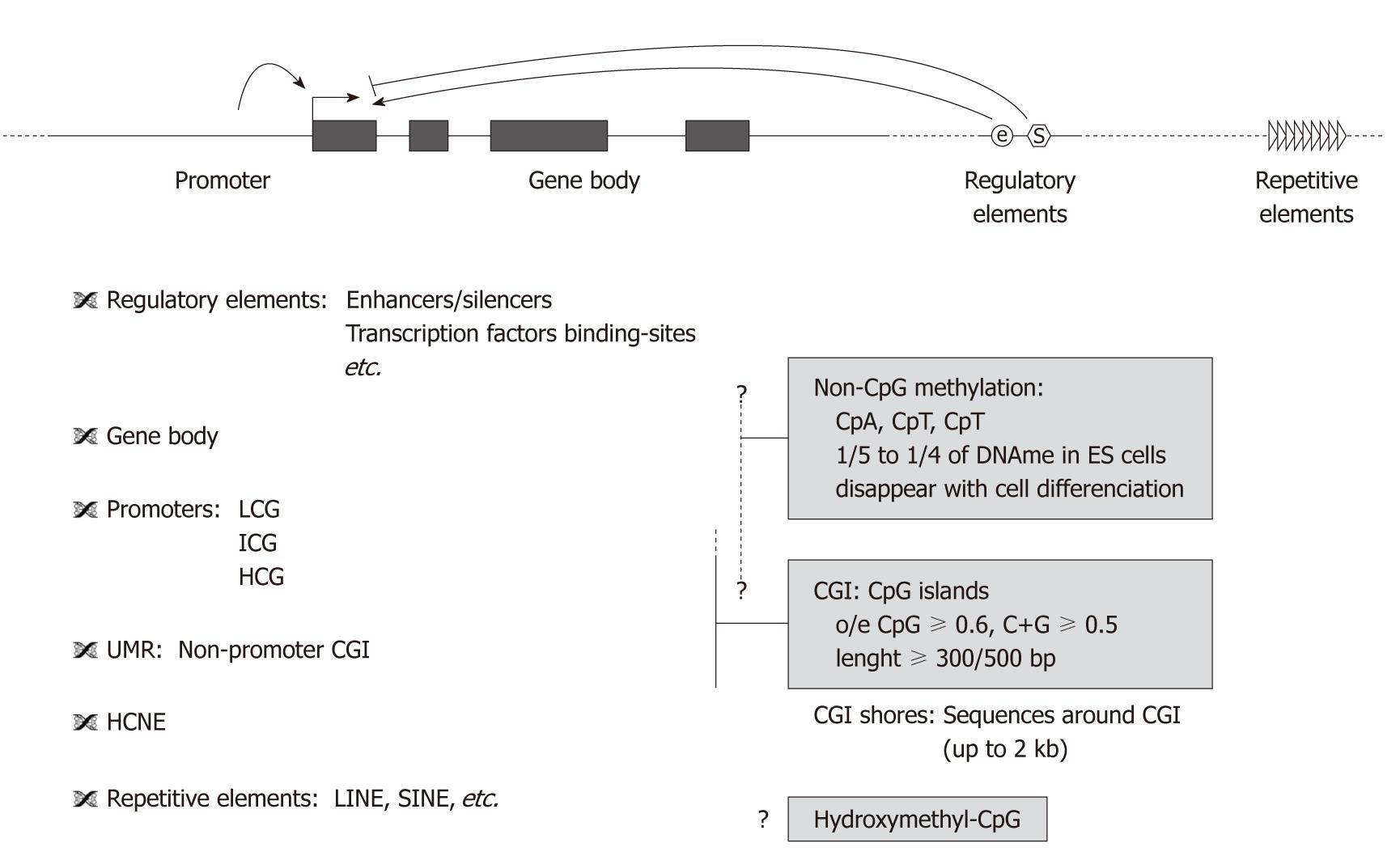
Figure 1 Target sequences for DNA methylation studies.
The majority of DNA methylation concerns methylcytosine on CpG dinucleotides but recently hydroxymethylcytosine and methylation on non-CpG sites were identified. Non-CpG methylation was reported in gene body, promoters and repetitive elements; its expanse needs to be further investigated. CpG islands (CGI) and gene promoters are preferred targets in many studies as they correspond to a tractable fraction of the genome with obvious regulatory potential. CGIs are defined algorithmically, as sequences with an observed-to-expected ratio of CpG greater than 0.6, a G+C content greater than 0.5 and, in most cases, a length of more than 500 bp. Three classes of promoters were defined according to their CpG content: LCP have the highest probability to be methylated but methylation correlates poorly to transcription, HCP have low probability to be methylated but this correlates with gene expression. However, transcriptional regulation of genes depends also on distal regulatory elements such as enhancers, insulators, locus control regions and silencing elements. In addition, recent studies show that gene-bodies in active transcription sites are enriched in DNA methylation. Moreover, non-promoter CGIs unmethylated regions (UMR) were recently identified, initially unmethylated they become methylated during development in a tissue-specific manner. “CGI shores” sequences were described around CGI, their methylation in normal tissues is highly conserved, tissue-specific and strongly related to gene expression and were highly sensitive to DNA alterations in colon cancer, as opposed to promoters or CGIs. Highly methylated repetitive elements and highly conserved non-coding elements can also be interesting targets for DNA methylation studies. LCG: Low-CpG promoters; ICG: Intermediate-CpG promoters; HCG: High-CpG promoters; HCNE: Highly conserved non-coding elements.
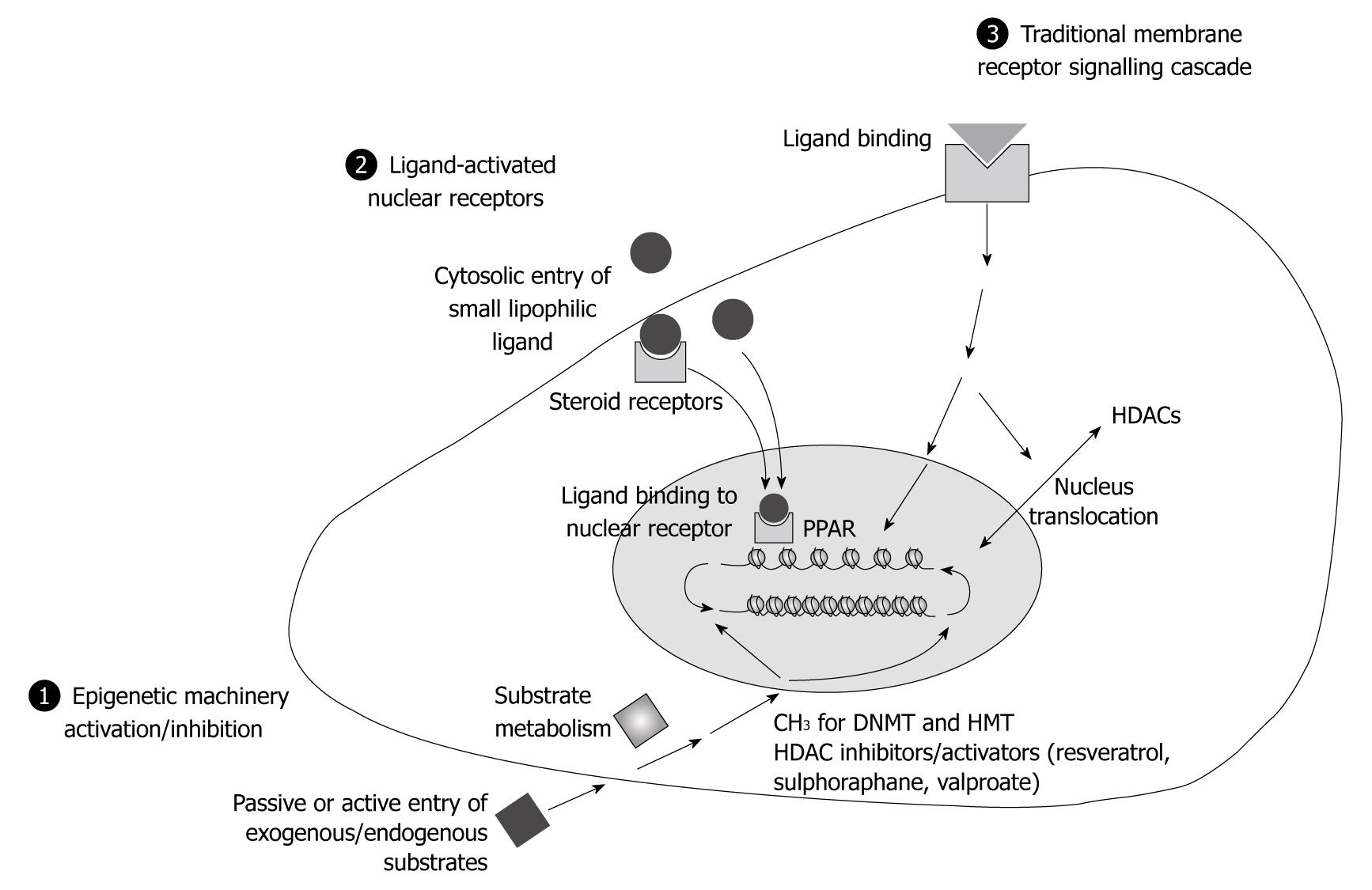
Figure 2 Mechanistic pathways for environmental factors involved in epigenetic reprogramming.
There are three ways to link environmental factors such as nutrients or drugs from the cell membrane to the chromatin structure: (1) Some environmental factors, aging and gender may target chromatin modifying enzymes or their substrate availability. Exogenous/endogenous substrates [methyl donors such as folates, histone deacetylase (HDAC) inhibitors such as TSA, etc.], after passive or active entry through the cell membrane, undergo cell specific metabolism. Thus, endogenous or exogenous compounds may lead to the alteration of a critical balance of chromatin remodelling enzymes, at the whole genome level, or to specific regions targeted by specific enzymes, e.g. HDACs; (2) Some other compounds (like endocrine disruptors) specifically bind to nuclear receptors, like steroid receptors, may be present in the cytoplasm, bind to their ligand, undergo several modifications and be subsequently translocated to the nucleus where they bind to their responsive elements (RE). The binding of other nuclear receptors, like PPARs and retinoid X receptor (RXR), with their natural polyunsaturated fatty acids ligands or drugs like fibrates lead to the recruitment of co-activators and chromatin remodelling factors. The appropriate modifications of the epigenetic marks at PPAR/RXR RE in target gene promoters modulate the expression of a particular set of genes, in a tissue-specific manner depending on the presence of appropriate co-factors; and (3) Traditional membrane receptor-signalling cascades may be involved. It is possible, depending on the type of ligand, that different pathways could be used. The maintenance of epigenetic patterns is dependent on the preservation of the balance of factors such as DNMTs, Histone acetyl transferases/HDACs or histone methyltransferases/histone demethylases and on the translocation of these enzymes into the nucleus. Extra- or intracellular signalling pathways could trigger activation of one of these factors and result in loci-specific modifications. PPAR: Peroxisome proliferator-activated receptor; DNMT: DNA methyltransferase.
- Citation: Gabory A, Attig L, Junien C. Epigenetic mechanisms involved in developmental nutritional programming. World J Diabetes 2011; 2(10): 164-175
- URL: https://www.wjgnet.com/1948-9358/full/v2/i10/164.htm
- DOI: https://dx.doi.org/10.4239/wjd.v2.i10.164