Copyright
©The Author(s) 2025.
World J Diabetes. Jun 15, 2025; 16(6): 102727
Published online Jun 15, 2025. doi: 10.4239/wjd.v16.i6.102727
Published online Jun 15, 2025. doi: 10.4239/wjd.v16.i6.102727
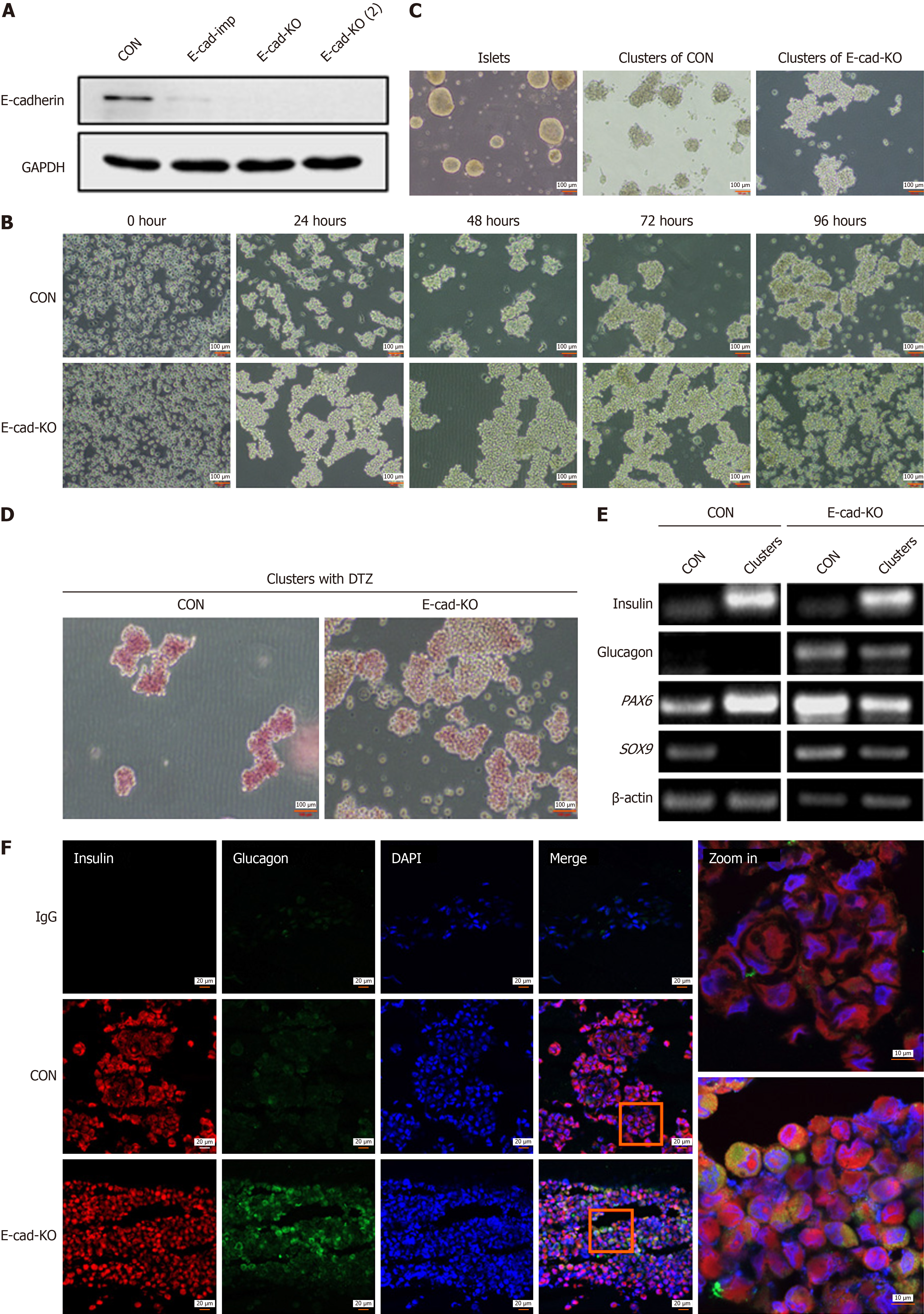
Figure 1 E-cadherin contributes to aggregation of human pancreatic cancer cell line-1 into islet-like clusters induced by trypsin.
A: Expression of E-cadherin protein was detected by western blotting, reflecting knockout (KO) efficiency of E-cadherin 1 gene (CDH1), E-cad-imp group: Stable transgenic cells with CDH1 gene KO by clustered regularly interspaced short palindromic repeats (CRISPR)- -associated 9 (Cas9) system impure, E-cad-KO and E-cad-KO (2): Monoclonal cells screening from stable KO cells; B: Representative images of KO cells at five time points after digestion with trypsin before and after CDH1 KO, magnification 10 ×; C: Morphology of islet-like cell clusters differentiated in vitro was compared with that of natural islets; D: Representative image of diphenylterazine (DTZ) staining of cell clusters formed at 96 hours before and after CDH1 KO, magnification 10 ×; E: Polymerase chain reaction detection reflected the changes in expression of the transcripts for mature islet-related genes insulin, glucagon, paired box 6 (PAX6), SRY-box transcription factor 9 (SOX9) before and after CDH1 KO; F: Representative immunofluorescence images of insulin, glucagon, and 4’,6-diamidino-2-phenylindole (DAPI) signal in differentiated cell clusters before and after CDH1 KO, magnification 20 ×, increased to 63 ×. CON: Control; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
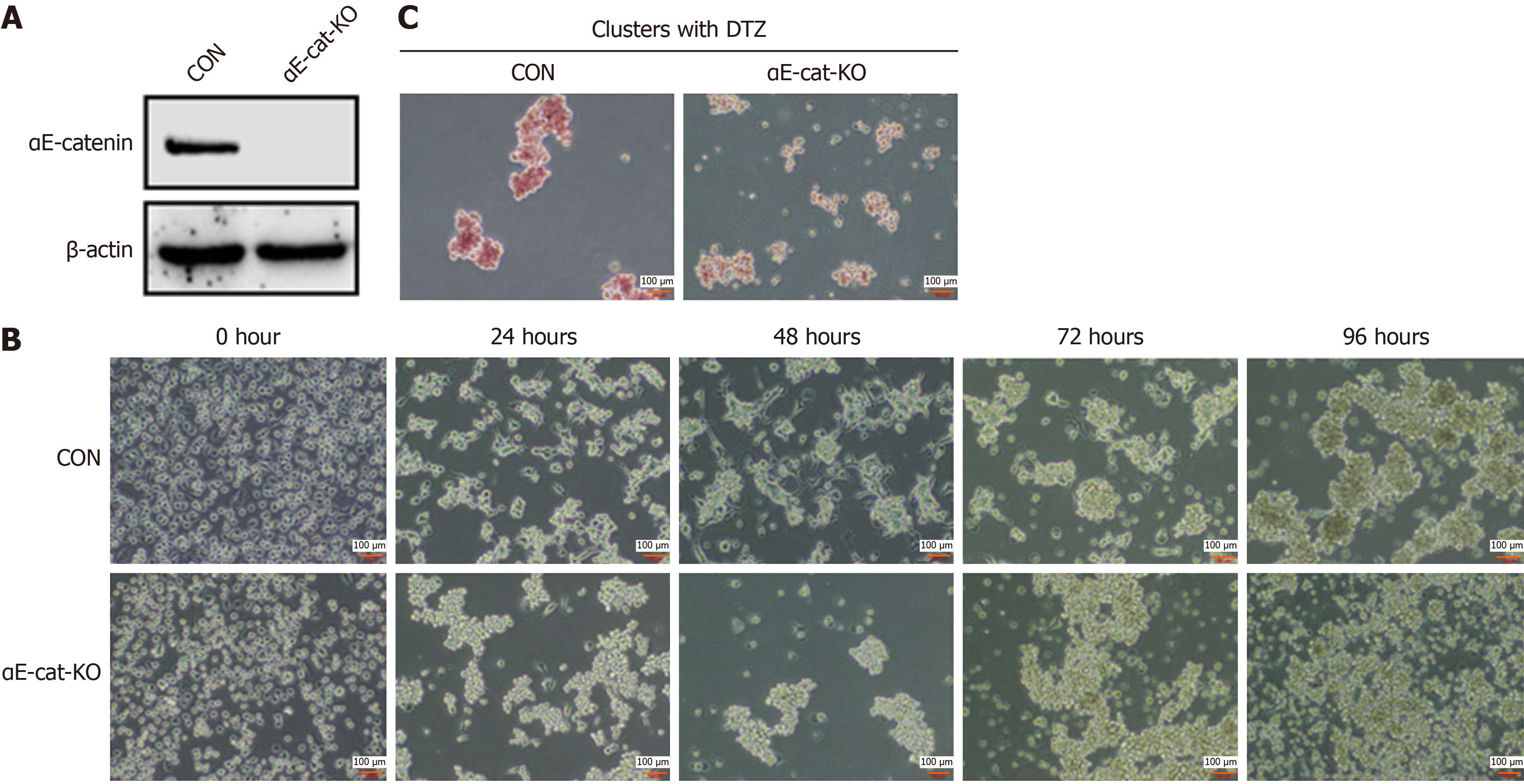
Figure 2 αE-catenin is related to trypsin-mediated human pancreatic cancer cell line-1 aggregation and differentiation.
A: Expression of αE-catenin protein was detected by western blotting, reflecting knockout (KO) efficiency of catenin alpha 1 (CTNNA1); B: Representative images of KO cells at five time points after digestion with trypsin before and after CTNNA1 KO, magnification 10 ×; C: Representative images of diphenylterazine (DTZ) staining of cell clusters formed at 96 hours before and after KO of CTNNA1, magnification 10 ×. CON: Control.
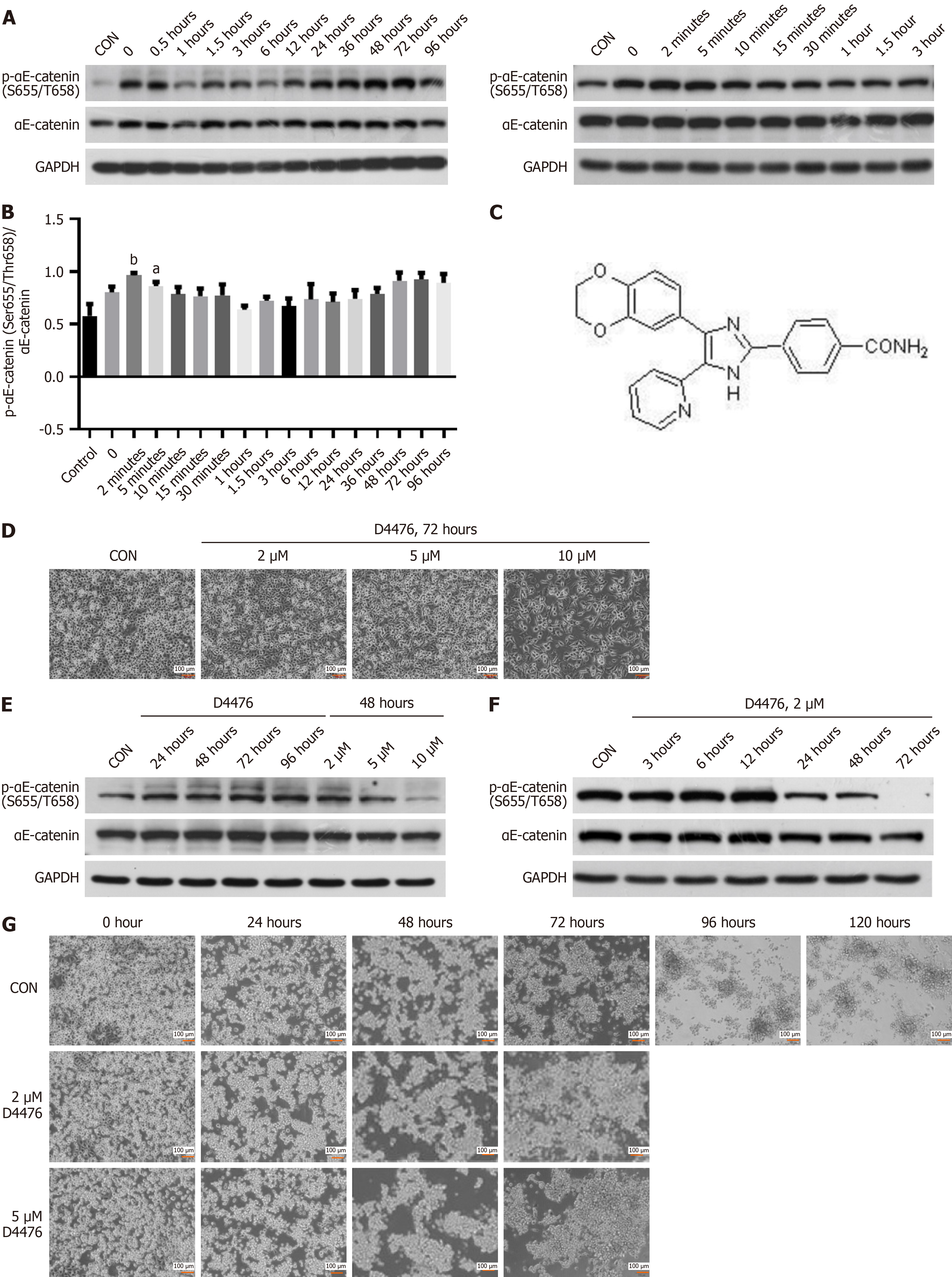
Figure 3 Phosphorylation of αE-catenin at Ser655/Thr658 during human pancreatic cancer cell line-1 differentiation and effects of casein kinase-1 inhibitor D4476 on human pancreatic cancer cell line-1.
A: Representative western blot images reflecting protein expression of phospho-αE-catenin (p-αE-catenin) (Ser655/Thr658) at different time points during human pancreatic cancer cell line-1 (PANC-1) differentiation; B: Statistical analyses of the gray value of western blot images using ImageJ and GraphPad Prism, reflecting the differences in expression of p-αE-catenin (Ser655/Thr658); C: Chemical structure of D4476, inhibitor of casein kinase-1; D: Representative images of PANC-1 cells treated with 2, 5, or 10 μM D4476 for 72 hours, magnification 10 ×; E: Western blot images showed expression of p-αE-catenin (Ser655/Thr658) of PANC-1 cells treated with 2 μM D4476; F: Western blot images showed expression of p-αE-catenin (Ser655/Thr658) of PANC-1 cells treated with 2 μM D4476 at different times; G: Representative images of PANC-1 after 72 hours treatment with 2 μM D4476 at different time points after digestion with trypsin, magnification 10 ×. aP < 0.05. bP < 0.01. P calculated vs control (CON). GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
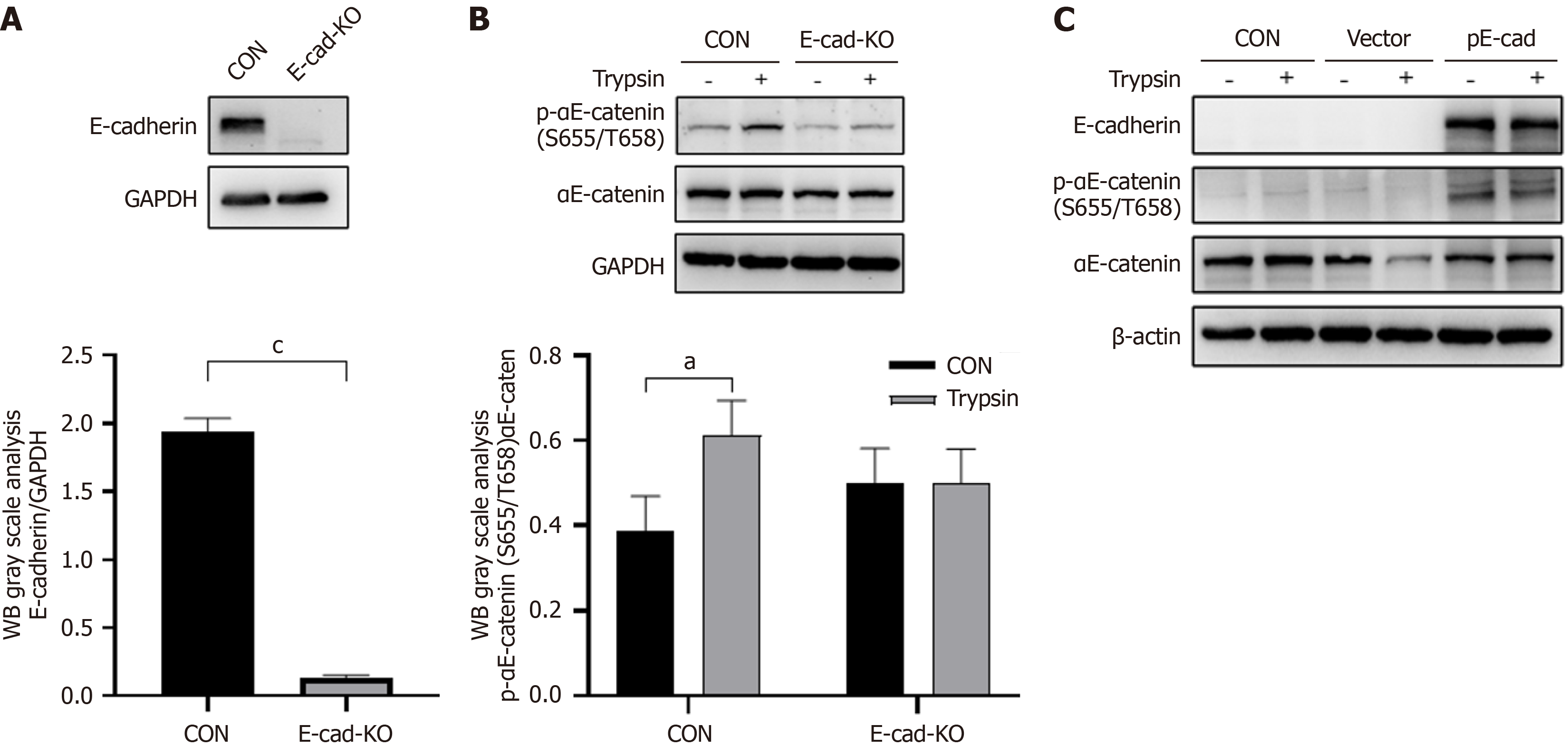
Figure 4 E-cadherin effects on phosphorylation of αE-catenin (Ser655/T658) mediated by trypsin.
A: Representative western blot images reflecting protein expression of E-cadherin of cells with knockout (KO) of E-cadherin gene (CDH1); B: Representative western blot images reflecting protein expression of phospho-αE-catenin (p-αE-catenin) (Ser655/Thr658) in CDH1 KO cells and difference in trypsin-mediated αE-catenin (Ser655/Thr658) phosphorylation before and after KO; C: Representative western blot images reflecting protein expression of p-αE-catenin (Ser655/Thr658) of restoration of E-cadherin in CDH1 KO cells and difference in trypsin-mediated αE-catenin (Ser655/Thr658) phosphorylation before and after restoration (n = 2). aP < 0.05. cP < 0.001. P calculated vs control (CON). GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
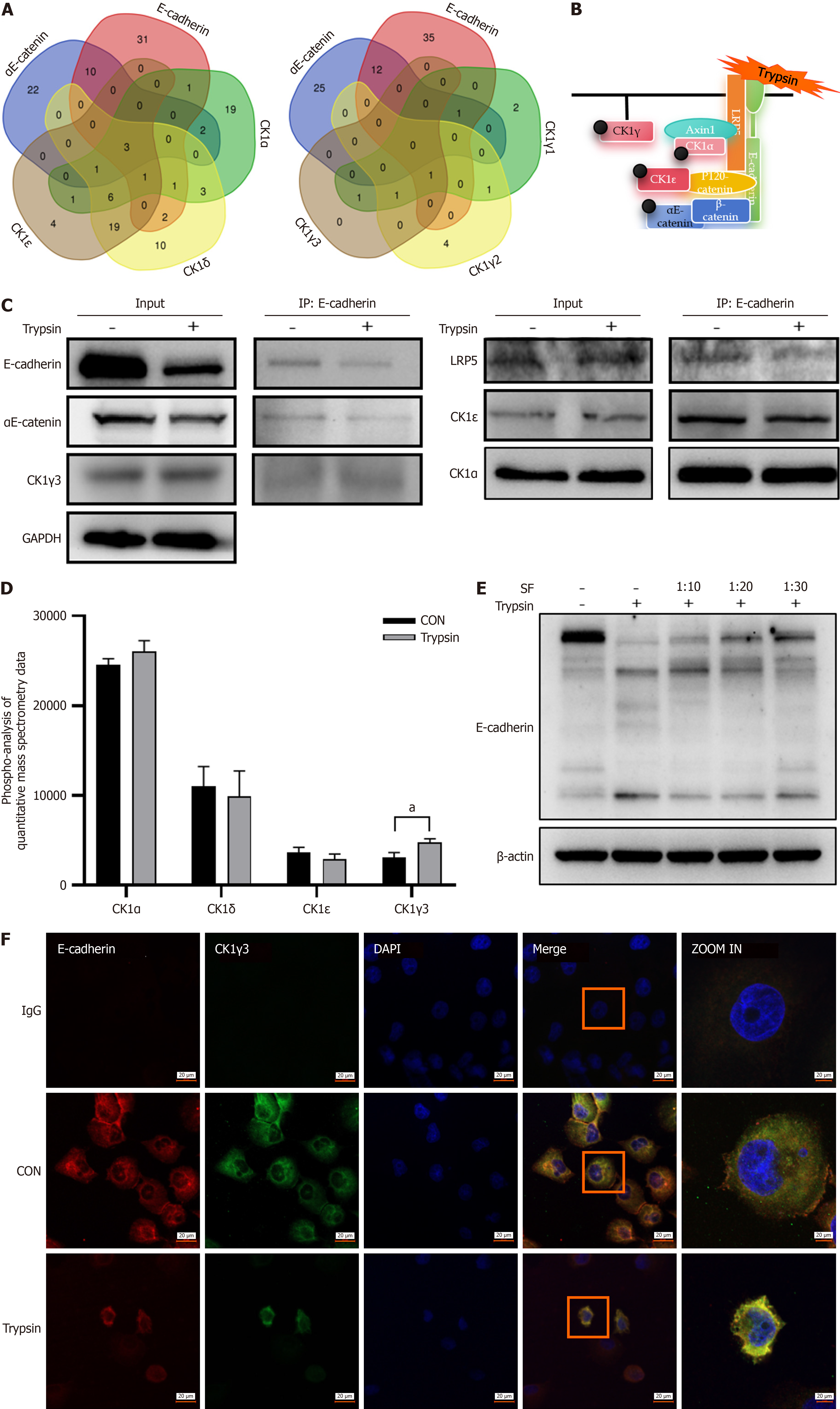
Figure 5 Trypsin promotes binding of casein kinase-1γ3 to the E-cadherin complex.
A: Wayne diagram, showing intersection of E-cadherin, αE-catenin, and casein kinase-1 (CK1) subtypes interacting proteins; B: Hypothetical model of the interaction among CK1, E-cadherin, and αE-catenin; C: Immunoprecipitation (IP) reflecting the protein composition of the E-cadherin complex; D: Quantitative mass spectrometric data analysis of phosphorylated protein, control group and trypsin representing the samples before and after trypsin treatment, and the ordinate reflects the level of phosphorylation; E: Western blot images reflect the hydrolysis of E-cadherin by trypsin and the blocking effect of different concentrations of trypsin inhibitor soybean flour (SF); F: Representative confocal images show colocalization changes of E-cadherin and CK1γ3 before and after trypsin treatment, Cy3 labeling E-cadherin (red), fluorescein isothiocyanate isomer labeling CK1γ3 (green), 4’,6-diamidino-2-phenylindole (DAPI) labeling nucleus (blue), magnification 20 ×, increased to 63 ×. aP < 0.05. P calculated vs control (CON). GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
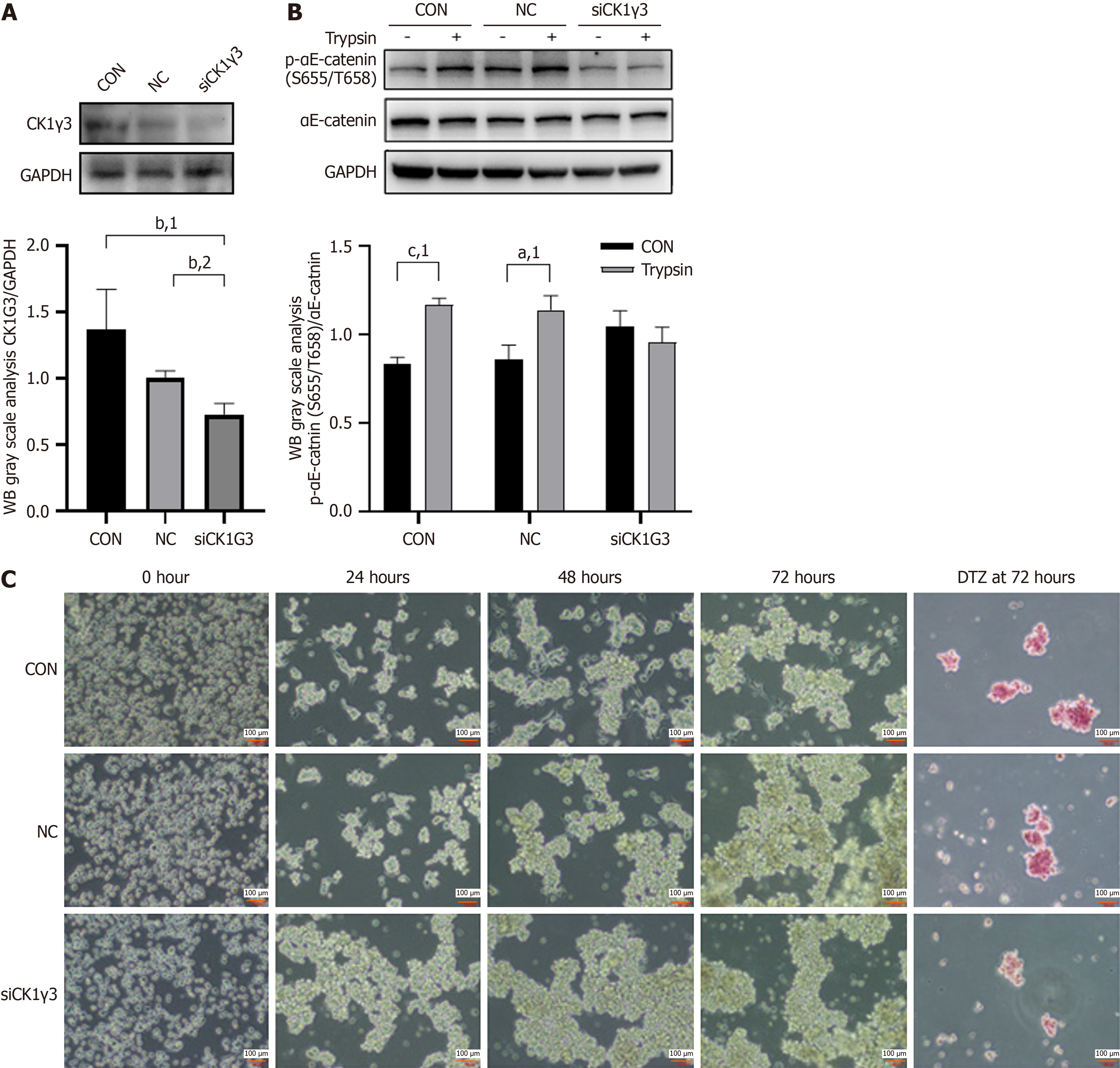
Figure 6 Casein kinase-1γ3 participated in the αE-catenin (Ser655/Thr658) phosphorylation mediated by trypsin.
A: Representative western blot (WB) images reflecting the efficiency of small interfering RNA knockdown of casein kinase-1γ3 (CK1γ3); B: Representative WB images reflecting protein expression of phospho-αE-catenin (Ser655/Thr658) in CK1γ3 knockdown cells and differences in trypsin-mediated αE-catenin (Ser655/Thr658) phosphorylation before and after knockdown; C: Representative images of knockout (KO) cells at five time points after digestion with trypsin and diphenylterazine (DTZ) staining of cell clusters formed at 72 hours before and after CK1γ3 knockdown, magnification 10 ×. aP < 0.05. bP < 0.01. cP < 0.001. 1P calculated vs control. 2P calculated vs negative control (NC). GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; CON: Control.
- Citation: Gao L, Lai JS, Chen H, Qian LX, Hong WJ, Li LC. Mechanism of trypsin-mediated differentiation of pancreatic progenitor cells into functional islet-like clusters. World J Diabetes 2025; 16(6): 102727
- URL: https://www.wjgnet.com/1948-9358/full/v16/i6/102727.htm
- DOI: https://dx.doi.org/10.4239/wjd.v16.i6.102727