Copyright
©The Author(s) 2025.
World J Diabetes. Apr 15, 2025; 16(4): 97201
Published online Apr 15, 2025. doi: 10.4239/wjd.v16.i4.97201
Published online Apr 15, 2025. doi: 10.4239/wjd.v16.i4.97201
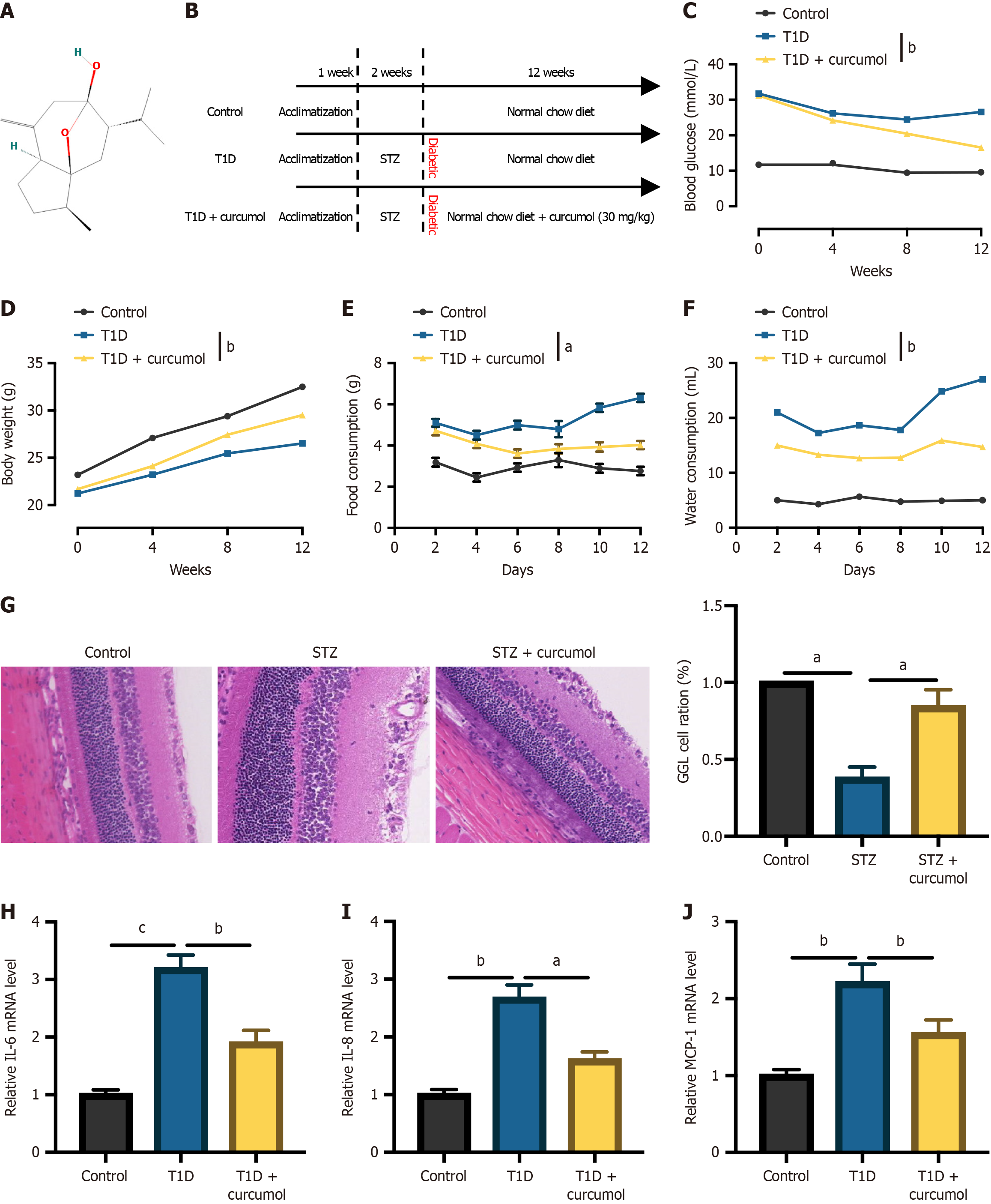
Figure 1 Curcumol attenuates diabetic retinopathy progression in mouse models.
A: The molecular configuration of curcumol; B: A schematic illustration of the animal model assembly; C: Systematic monitoring of blood glucose levels of all mice; D: Body weight measurements; E: Food intake; F: Water intake; G: Representative retinal morphology images obtained through hematoxylin and eosin staining; H-J: Quantitative real-time polymerase chain reaction analysis of mRNA expression levels of pro-inflammatory factors, including interleukin-6, interleukin-8, and monocyte chemoattractant protein-1. Data represent the mean ± SD, n = 5-7 per group. aP < 0.05, bP < 0.01, cP < 0.001. T1D: Type 1 diabetes; STZ: Streptozotocin; IL: Interleukin; MCP-1: Chemoattractant protein-1.
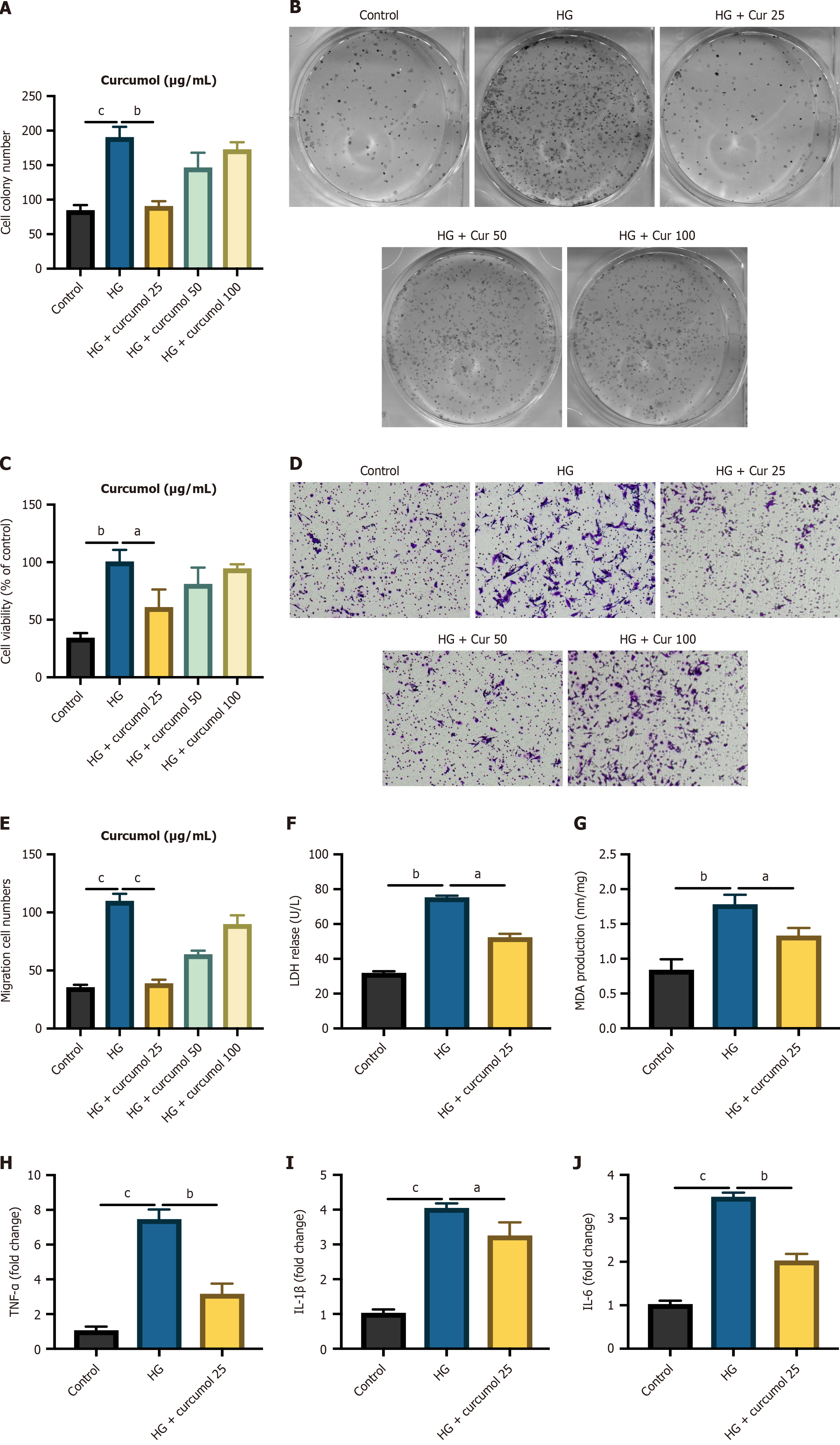
Figure 2 Curcumol inhibits high glucose-induced proliferation, migration, and inflammation in human retinal vascular endothelial cells.
A: Methyl tetrazolium assay was used to measure the proliferation of high glucose (HG)-treated human retinal vascular endothelial cells under different doses of curcumol; B and C: Cell colony assay to detect cell viability; D and E: Transwell migration assay to evaluate cell migration level; F and G: Levels of lactate dehydrogenase and malondialdehyde, both indicative of HG-induced cellular injury, were detected upon treatment with curcumol; H-J: The expressions of HG-induced transcription of inflammatory cytokines upon curcumol treatment were detected by quantitative real-time polymerase chain reaction. Data represent the mean ± SD. aP < 0.05, bP < 0.01, cP < 0.001. HG: High glucose; Cur: Curcumol; LDH: Lactate dehydrogenase; MDA: Malondialdehyde; TNF: Tumor necrosis factor; IL: Interleukin.
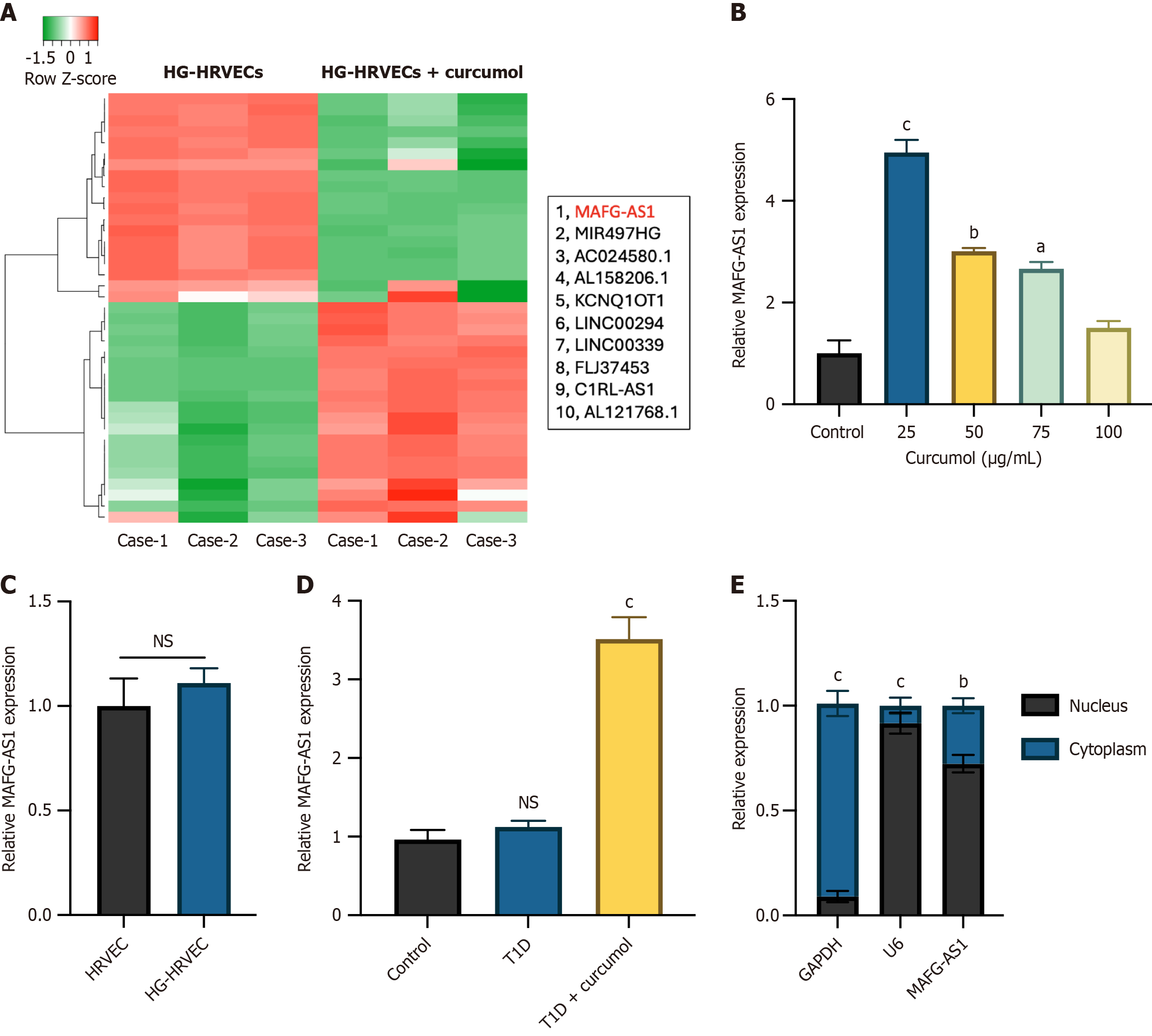
Figure 3 Identification of dysregulated long non-coding RNAs upon curcumol treatment.
A: Long non-coding RNA (lncRNA) microarray analysis was utilized to identify significantly dysregulated lncRNAs expression in high glucose-human retinal vascular endothelial cells (HG-HRVECs) and curcumol-treated HG-HRVECs. The top 10 upregulated lncRNAs are shown; B: LncRNA MAF transcription factor G antisense RNA 1 (MAFG-AS1) expression in HG-HRVECs and HG-HRVECs treated with different doses of curcumol was measured by quantitative real-time polymerase chain reaction (qRT-PCR); C: MAFG-AS1 expression in HRVECs and HG-HRVECs was detected by qRT-PCR; D: MAFG-AS1 expression the retinal tissues of control mice, type 1 diabetes mice, and type 1 diabetes mice treated with curcumol was detected by qRT-PCR; E: Cellular distribution of MAFG-AS1 in HRVECs. Data represent the mean ± SD. aP < 0.05, bP < 0.01, cP < 0.001, NS: No significance. HG-HRVECs: High glucose-human retinal vascular endothelial cells; MAFG-AS1: MAF transcription factor G antisense RNA 1; T1D: Type 1 diabetes; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
Figure 4 MAF transcription factor G antisense RNA 1 mediates the inhibitory effects of curcumol on high glucose-induced human retinal vascular endothelial cells.
A: MAF transcription factor G antisense RNA 1 expression in high glucose (HG)-human retinal vascular endothelial cells models (control, curcumol, and curcumol + si-MAF transcription factor G antisense RNA 1) was measured by quantitative real-time polymerase chain reaction; B: Methyl tetrazolium assay to measure cell proliferation; C and D: Cell colony assay to detect cell viability; E and F: Transwell migration assay to evaluate cell migration; G and H: Levels of lactate dehydrogenase and malondialdehyde, both indicative of HG-induced cellular injury, were detected upon treatment with curcumol; I-K: The expression of HG-induced transcription of inflammatory cytokines upon curcumol treatment was detected by quantitative real-time polymerase chain reaction. Data represent the mean ± SD. aP < 0.05, bP < 0.01, cP < 0.001. HG-HRVECs: High glucose-human retinal vascular endothelial cells; MAFG-AS1: MAF transcription factor G antisense RNA 1; LDH: Lactate dehydrogenase; MDA: Malondialdehyde; TNF: Tumor necrosis factor; IL: Interleukin.
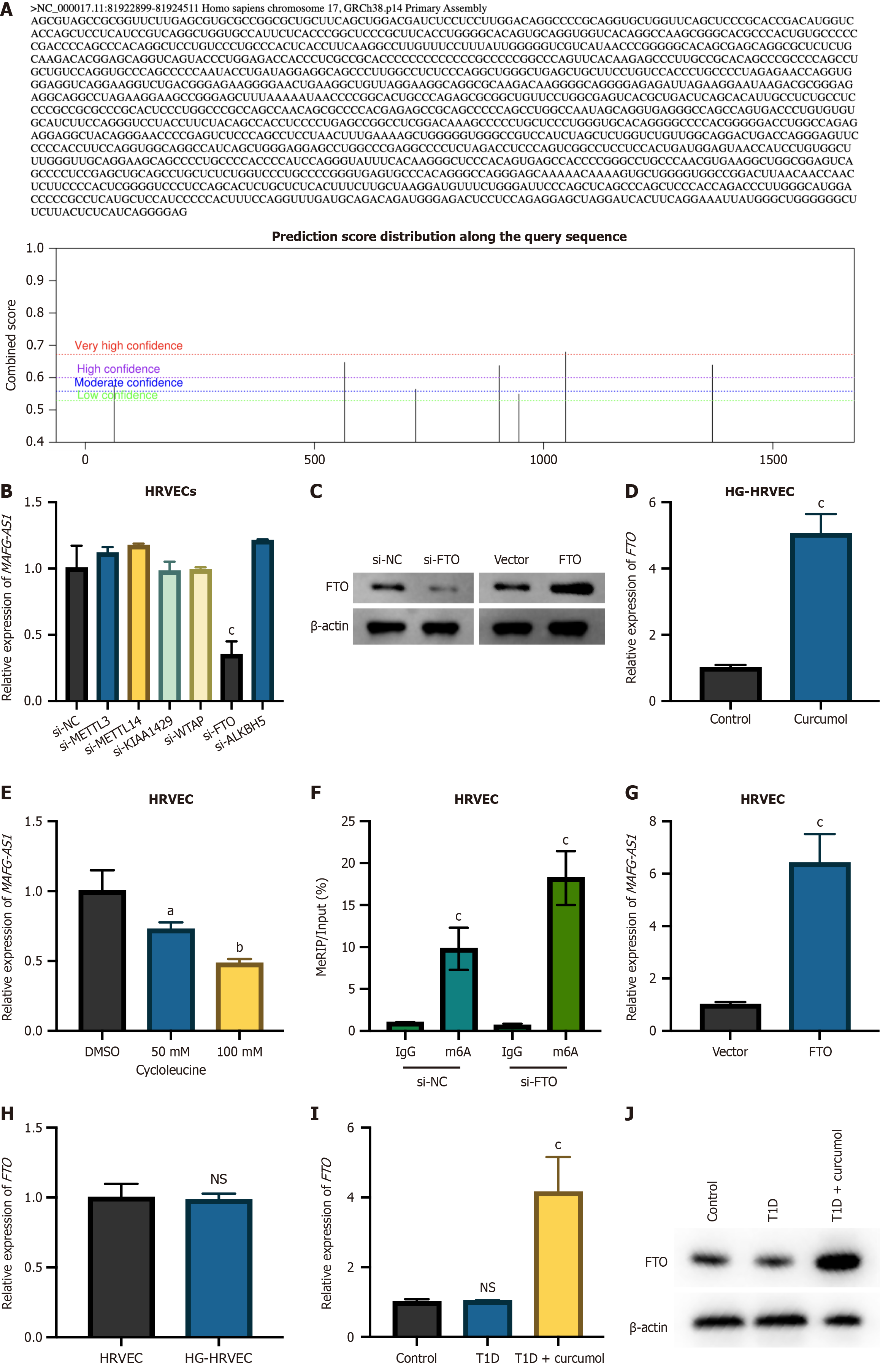
Figure 5 MAF transcription factor G antisense RNA 1 expression is demethylated by fat mass and obesity-associated in human retinal vascular endothelial cells.
A: Predicted m6A modification sites on MAF transcription factor G antisense RNA 1 (MAFG-AS1) using the SRAMP tool; B: Expression of MAFG-AS1 after knockdown of common m6A writers; C: Fat mass and obesity-associated protein (FTO) knockdown or overexpression in human retinal vascular endothelial cells (HRVECs) confirmed by Western blot; D: MAFG-AS1 expression in FTO overexpressed HRVECs was measured by quantitative real-time polymerase chain reaction (qRT-PCR); E: MAFG-AS1 expression in cycloleucine-treated HRVECs analyzed by qRT-PCR; F: Methylated RNA immunoprecipitation-qPCR analysis of the m6A modification level of MAFG-AS1 in HRVECs after FTO knockdown; G: qRT-PCR analysis of FTO expression in high glucose (HG)-HRVECs and HG-HRVECs treated with curcumol; H: qRT-PCR analysis of FTO expression in HRVECs and HG-HRVECs; I and J: FTO expression in the retinal tissue of control mice, type 1 diabetes mice, and type 1 diabetes mice treated with curcumol detected by qRT-PCR and Western blot. Data represent the mean ± SD. aP < 0.05, bP < 0.01, cP < 0.001, NS: No significance. HRVECs: Human retinal vascular endothelial cells; MAFG-AS1: MAF transcription factor G antisense RNA 1; METTL: Methyltransferase-like; WTAP: Wilms’ tumor 1-associating protein; FTO: Fat mass and obesity-associated protein; ALKBH5: Anaplastic lymphoma kinase B homolog 5; HG: High glucose; MeRIP: Methylated RNA immunoprecipitation; T1D: Type 1 diabetes.
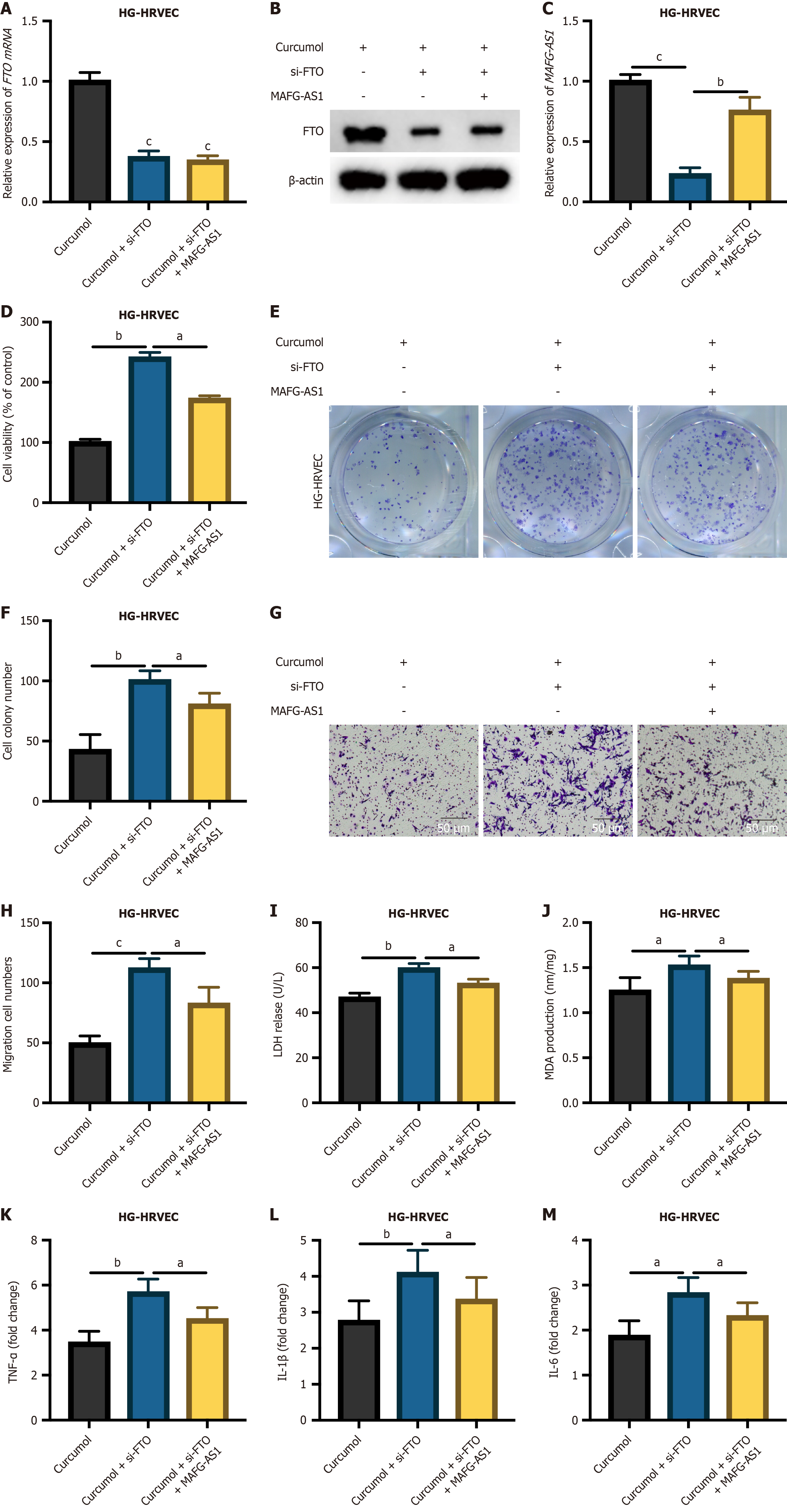
Figure 6 Curcumol exerts its protective effects via the fat mass and obesity-associated/MAF transcription factor G antisense RNA 1 axis.
A and B: Fat mass and obesity-associated protein (FTO) expression in high glucose-human retinal vascular endothelial cells models (curcumol, curcumol + si-FTO, curcumol + si-FTO + MAF transcription factor G antisense RNA 1) detected by quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot assays; C: MAF transcription factor G antisense RNA 1 expression in cell models measured by qRT-PCR; D: Methyl tetrazolium assay to measure cell proliferation; E and F: Cell colony assay to detect cell viability; G and H: Transwell migration assay to evaluate cell migration; I and J: Levels of lactate dehydrogenase and malondialdehyde, both indicative of HG-induced cellular injury, detected after treatment with curcumol; K-M: qRT-PCR analysis of high glucose-induced transcription of inflammatory cytokines upon curcumol treatment. Data represent the mean ± SD. aP < 0.05, bP < 0.01, cP < 0.001. HG-HRVECs: High glucose-human retinal vascular endothelial cells; FTO: Fat mass and obesity-associated protein; MAFG-AS1: MAF transcription factor G antisense RNA 1; LDH: Lactate dehydrogenase; MDA: Malondialdehyde; TNF: Tumor necrosis factor; IL: Interleukin.
- Citation: Rong H, Hu Y, Wei W. Curcumol ameliorates diabetic retinopathy via modulating fat mass and obesity-associated protein-demethylated MAF transcription factor G antisense RNA 1. World J Diabetes 2025; 16(4): 97201
- URL: https://www.wjgnet.com/1948-9358/full/v16/i4/97201.htm
- DOI: https://dx.doi.org/10.4239/wjd.v16.i4.97201