Copyright
©The Author(s) 2025.
World J Diabetes. Apr 15, 2025; 16(4): 102970
Published online Apr 15, 2025. doi: 10.4239/wjd.v16.i4.102970
Published online Apr 15, 2025. doi: 10.4239/wjd.v16.i4.102970
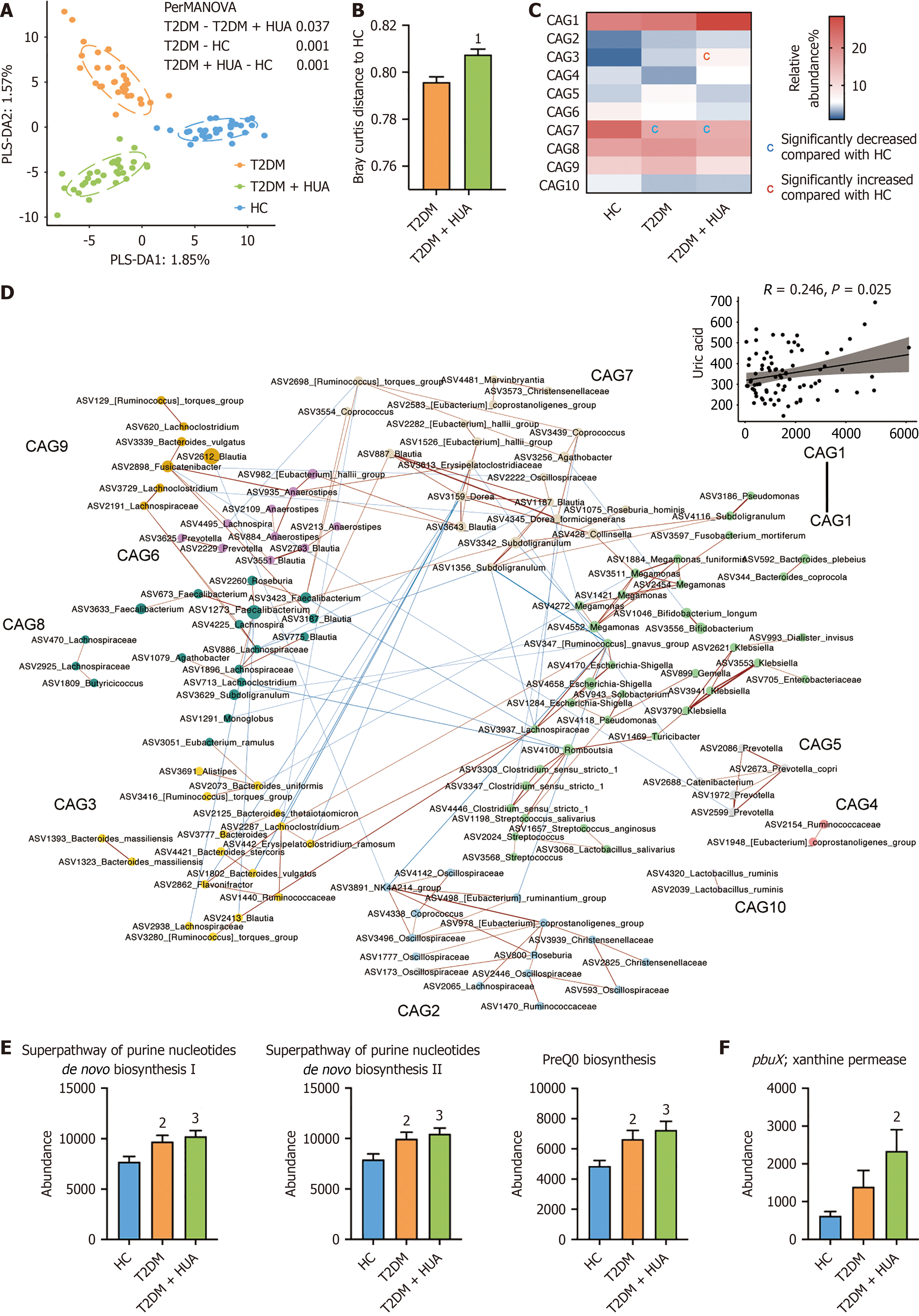
Figure 1 Characteristics of the microbiome in the cross-sectional study.
A: Partial least squares discrimination analysis was used to show the difference in microbiome structures, and the permutational multivariate analysis of variance test results based on the Bray Curtis distance were labeled in the figure; B: The Bray Curtis distance between the microbiome of the two patient groups and the healthy control group. The difference between groups was tested by the student’s t-test; C: Differences of microbiome co-abundance between groups were tested by ordinary one-way analysis of variance test, combined with Tukey’s multiple comparisons and the threshold of significance was P < 0.05; D: Network diagrams of the co-abundance groups. Lines between nodes represent correlations; only correlations with magnitudes > 0.3 are drawn. Red lines mean positive correlations, and blue lines mean negative correlations. The significant linear correlations between two co-abundance groups and uric acid are shown on the right; E: Metabolic pathways related to the uric acid metabolism which showed differences among the three groups; F: Kos related to the uric acid metabolism which showed differences among the three groups. Differences were tested by ordinary one-way analysis of variance test combined with Tukey’s multiple comparisons. aP < 0.05 vs type 2 diabetes mellitus group. bP < 0.05 vs healthy control group. cP < 0.01 vs healthy control group. PLS-DA: Partial least squares discrimination analysis; PERMANOVA: Permutational multivariate analysis of variance; HC: Healthy control; T2DM: Type 2 diabetes mellitus; HUA: Hyperuricemia; CAG: Co-abundance group; ASV: Amplicon sequence variant.
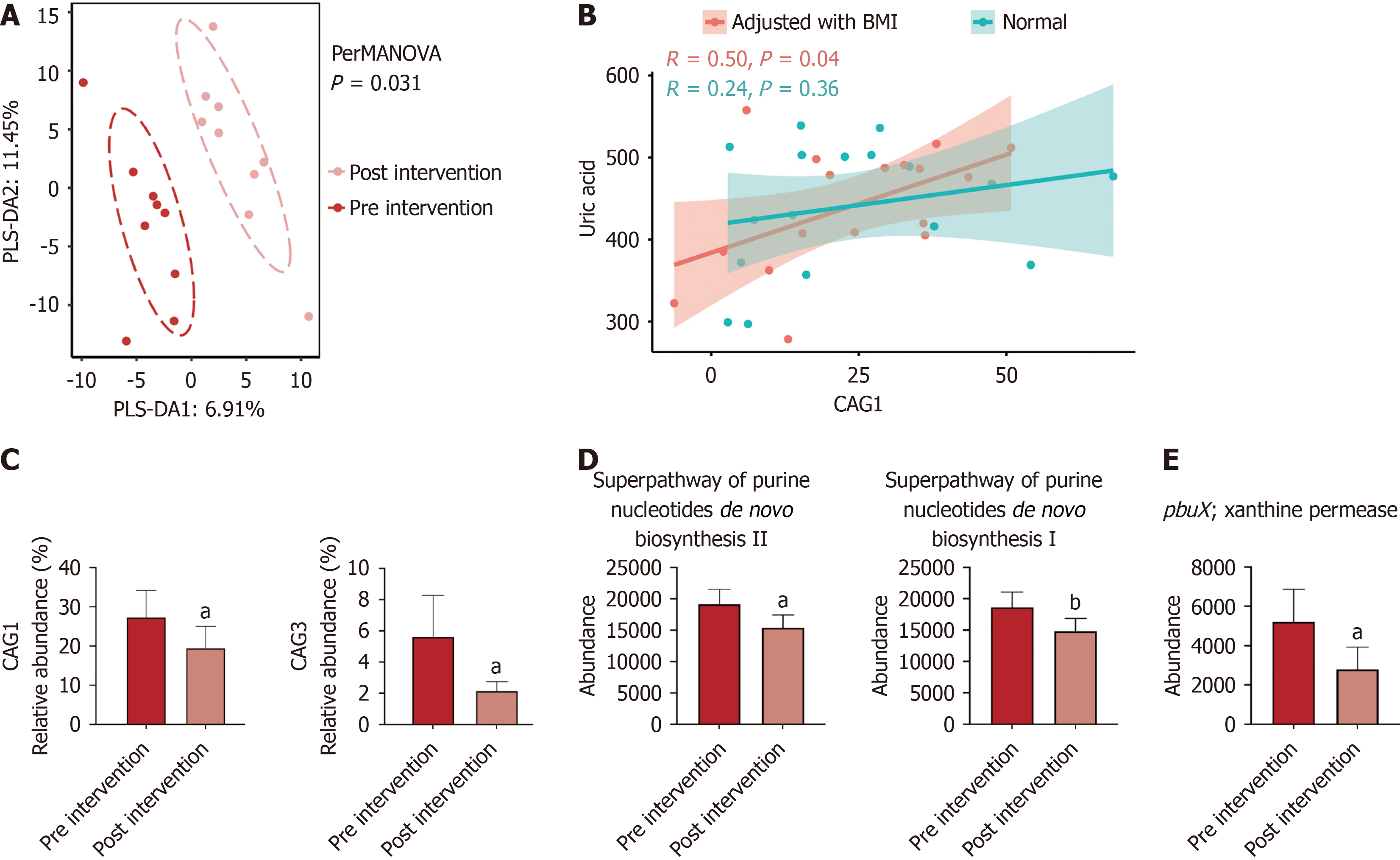
Figure 2 The characteristics of the microbiomes in the empagliflozin intervention study.
A: Partial least squares discrimination analysis was used to show the differences of microbiome structures, and the permutational multivariate analysis of variance test results based on Bray Curtis distance were labeled in the figure; B: Differences of microbiome co-abundance between groups were tested by the paired t test, and the threshold of significance was P < 0.05; C: The linear correlation between co-abundance group 1 and uric acid; D: Metabolic pathways related to the uric acid metabolism among the three groups; E: Kos related to the uric acid metabolism among the three groups. Differences were tested by the paired t test. aP < 0.05. bP < 0.01. PLS-DA: Partial least squares discrimination analysis; PERMANOVA: Permutational multivariate analysis of variance; BMI: Body mass index; CAG: Co-abundance group.
- Citation: Deng XR, Zhai YJ, Shi XY, Tang SS, Fang YY, Heng HY, Zhao LY, Yuan HJ. Characteristic dysbiosis in patients with type 2 diabetes and hyperuricemia, and the effect of empagliflozin on gut microbiota. World J Diabetes 2025; 16(4): 102970
- URL: https://www.wjgnet.com/1948-9358/full/v16/i4/102970.htm
- DOI: https://dx.doi.org/10.4239/wjd.v16.i4.102970