Copyright
©The Author(s) 2024.
World J Diabetes. Feb 15, 2024; 15(2): 275-286
Published online Feb 15, 2024. doi: 10.4239/wjd.v15.i2.275
Published online Feb 15, 2024. doi: 10.4239/wjd.v15.i2.275
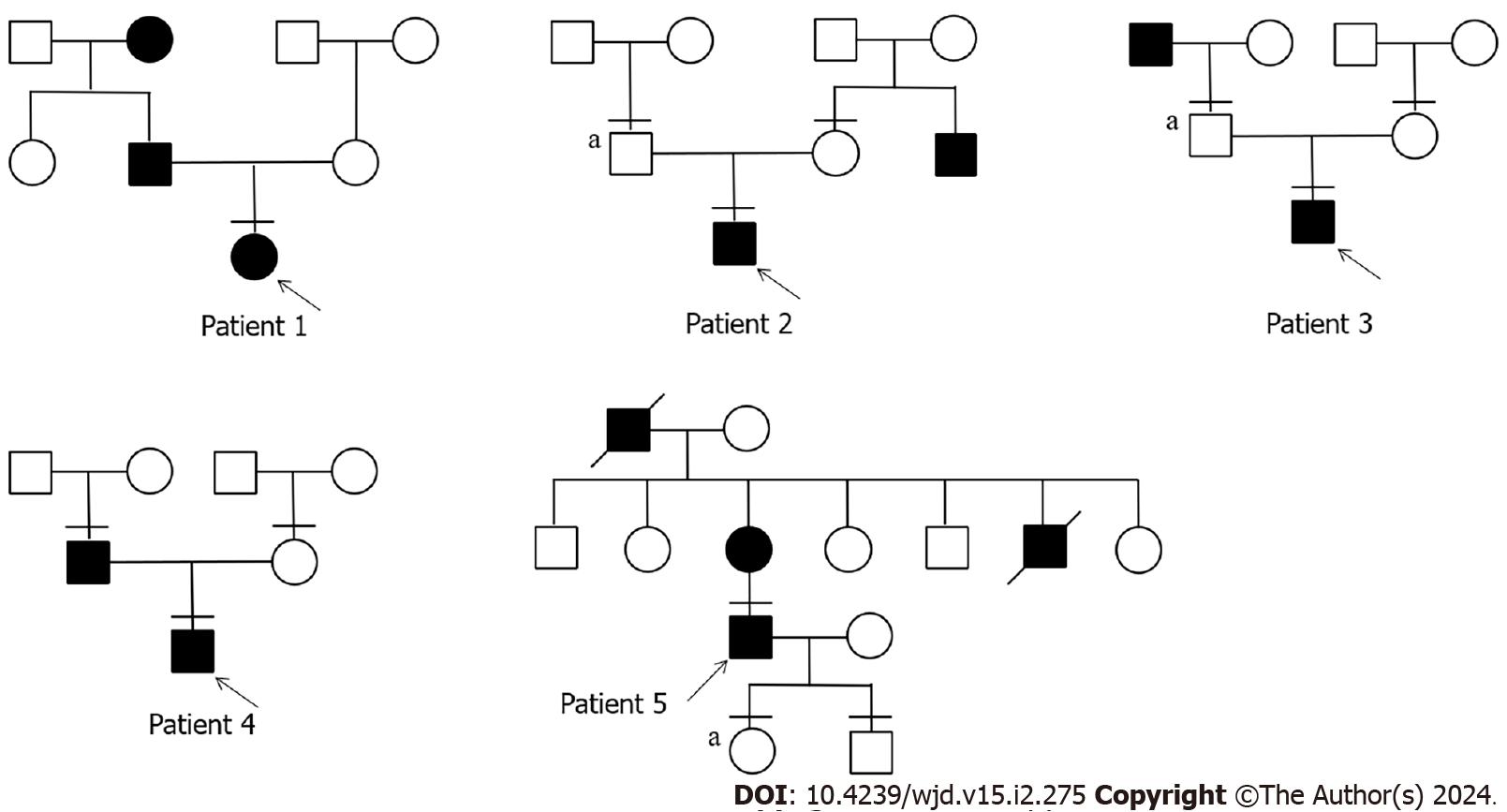
Figure 1 Pedigree of 5 diabetes patients with novel adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 variants.
Males and females are represented by squares and circles, respectively. The black padding suggests that the patient has diabetes, the arrow represents the progenitor, and the horizontal line indicates a patient who has undergone full exon sequencing. “a” indicates that the patient has the same mutation as the proband.
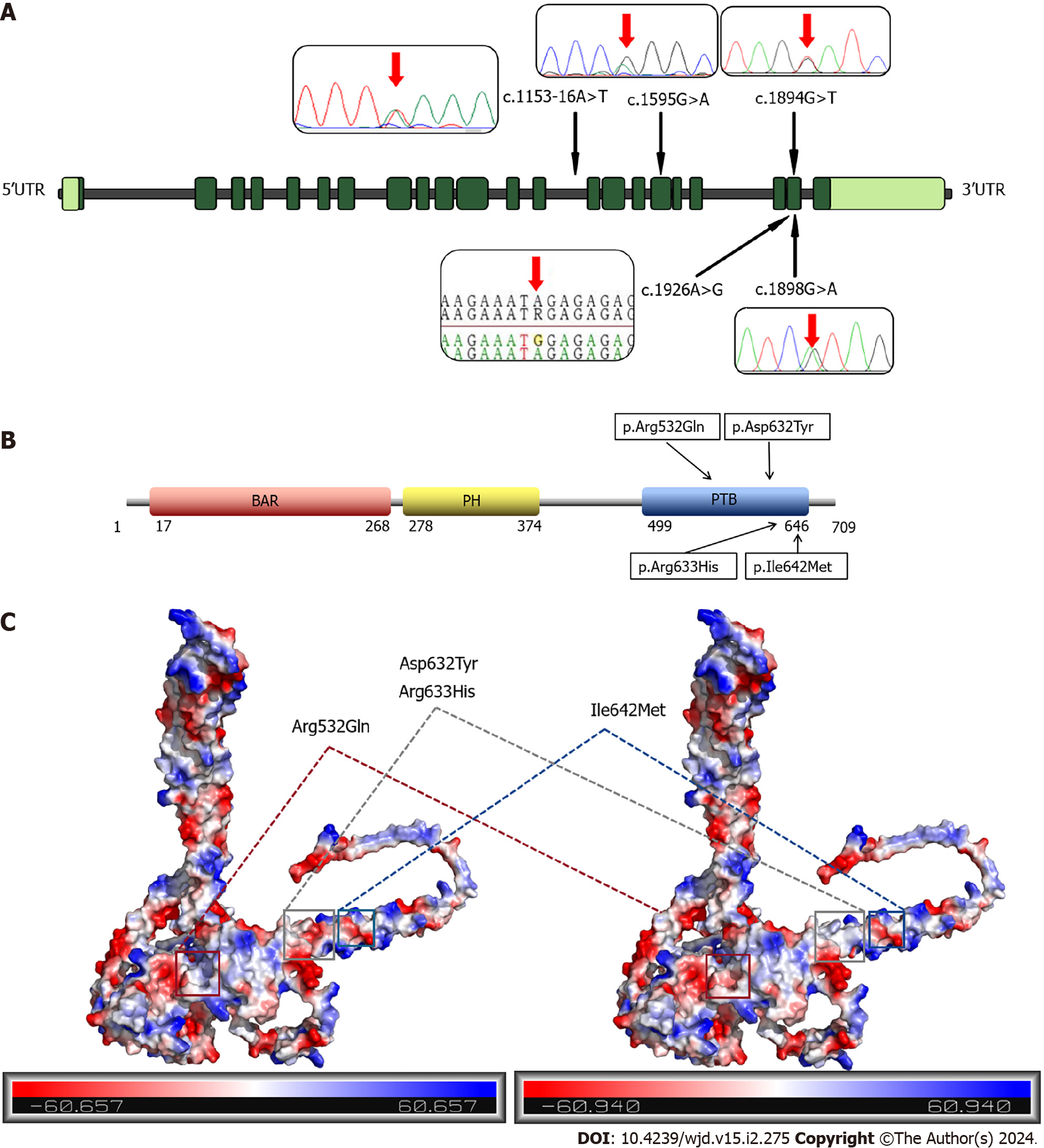
Figure 2 Distribution of mutation sites in adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 and adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 protein and potential changes in mutation sites.
A: Exon and mutation site distribution of adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 (APPL1) gene; B: Domain and mutation site distribution of APPL1 protein; C: Potential change of mutated APPL1 protein. BAR: Bin-Amphiphysin-Rvs; PH: Pleckstrin homology; PTB: Phosphotyrosine-binding; UTR: Untranslated region.
Figure 3 Conservation of mutation sites in multiple species.
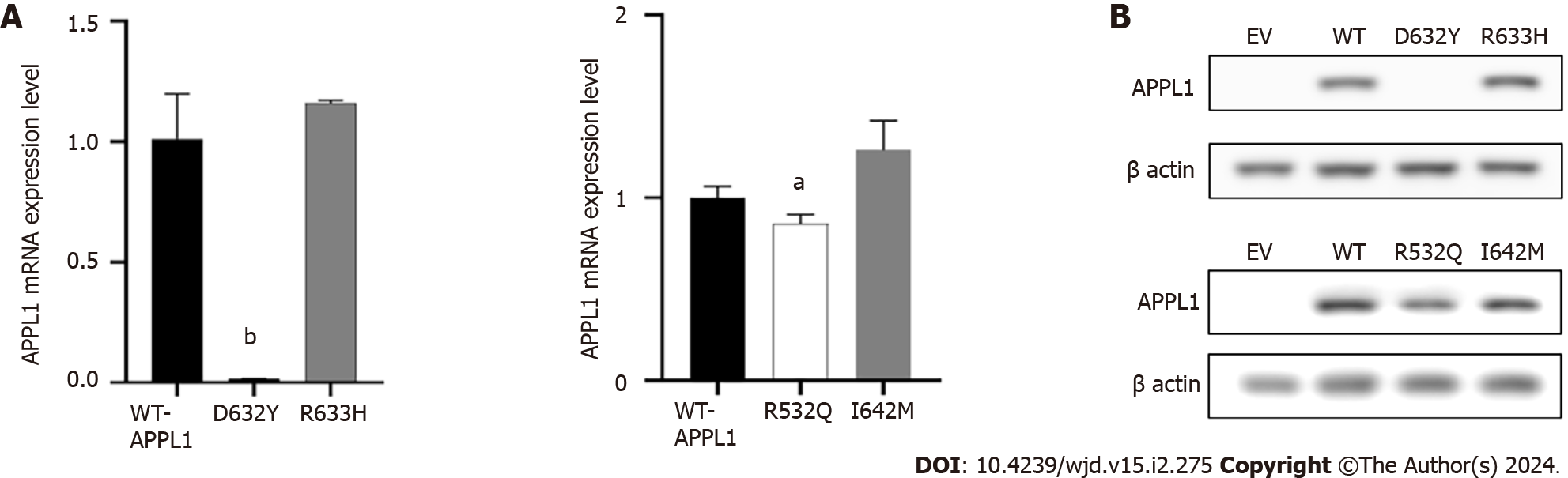
Figure 4 mRNA and protein expression at mutation sites.
A: mRNA expression at the mutation site; B: Protein expression at the mutation site. aP < 0.05; bP < 0.001. APPL1: Adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1; EV: Empty vector; WT: Wild-type.
Figure 5 Adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1-related protein networks and the docking between adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 and insulin receptor.
A The adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 (APPL1)-related protein network. The degree of binding between proteins is indicated by the colors from yellow to orange on the nodes. The larger the node, the darker the color, and the higher the degree of binding to the APPL1 protein. The edge shows the association of protein-protein; B: The binding of the phosphotyrosine binding domain of APPL1 to insulin receptor proteins. Red is the phosphotyrosine binding domain, and blue is the insulin receptor protein.
Figure 6 Role of adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 in the insulin pathway.
AKT: Serine/threonine kinase Akt; APPL1: Adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1; AS160: Akt substrate of 160 kDa; GLUT4: Glucose transporter type 4; IRS1/2: Insulin receptor substrate 1/2; PDK1: Pyruvate dehydrogenase (acetyl-transferring) kinase isozyme 1; PI3K: Phosphatidylinositol 3 - kinase; PIP3: Phosphatidylinositol-3,4,5-trisphosphate; TRB3: Tribble homolog 3.
- Citation: Shi P, Tian Y, Xu F, Liu LN, Wu WH, Shi YZ, Dai AQ, Fang HY, Li KX, Xu C. Assessment of pathogenicity and functional characterization of APPL1 gene mutations in diabetic patients. World J Diabetes 2024; 15(2): 275-286
- URL: https://www.wjgnet.com/1948-9358/full/v15/i2/275.htm
- DOI: https://dx.doi.org/10.4239/wjd.v15.i2.275