Copyright
©The Author(s) 2020.
World J Diabetes. May 15, 2020; 11(5): 182-192
Published online May 15, 2020. doi: 10.4239/wjd.v11.i5.182
Published online May 15, 2020. doi: 10.4239/wjd.v11.i5.182
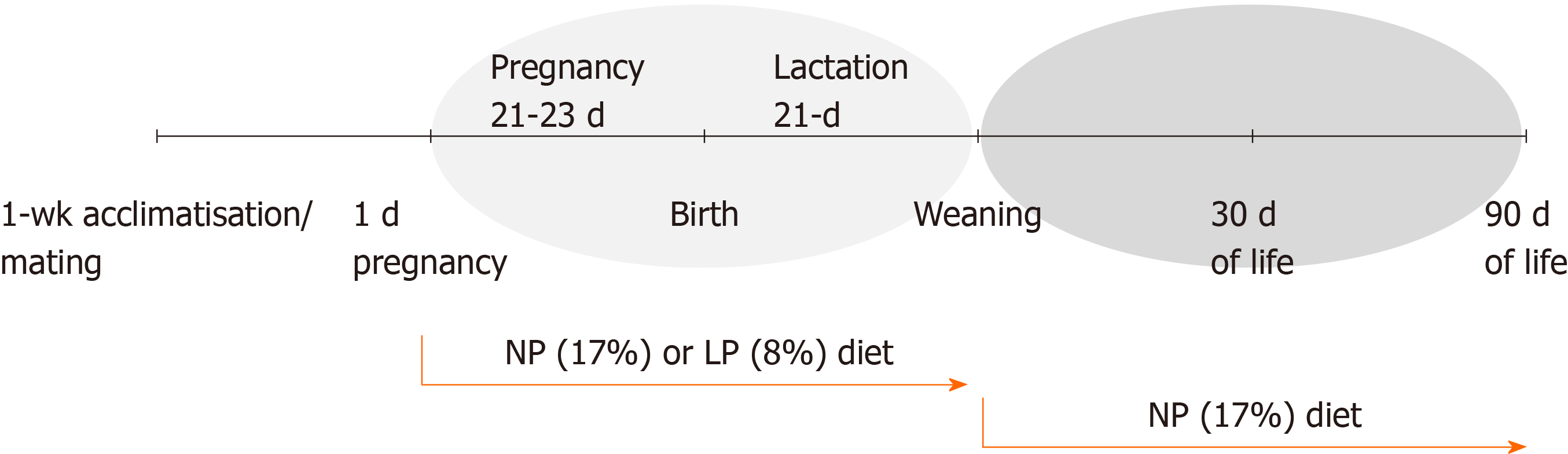
Figure 1 Schematic diagram of the experimental protocol in Wistar rats.
Rats were exposed to maternal low protein diet during gestation and lactation and then switched to a normal protein diet. NP: Normal protein; LP: Low protein.
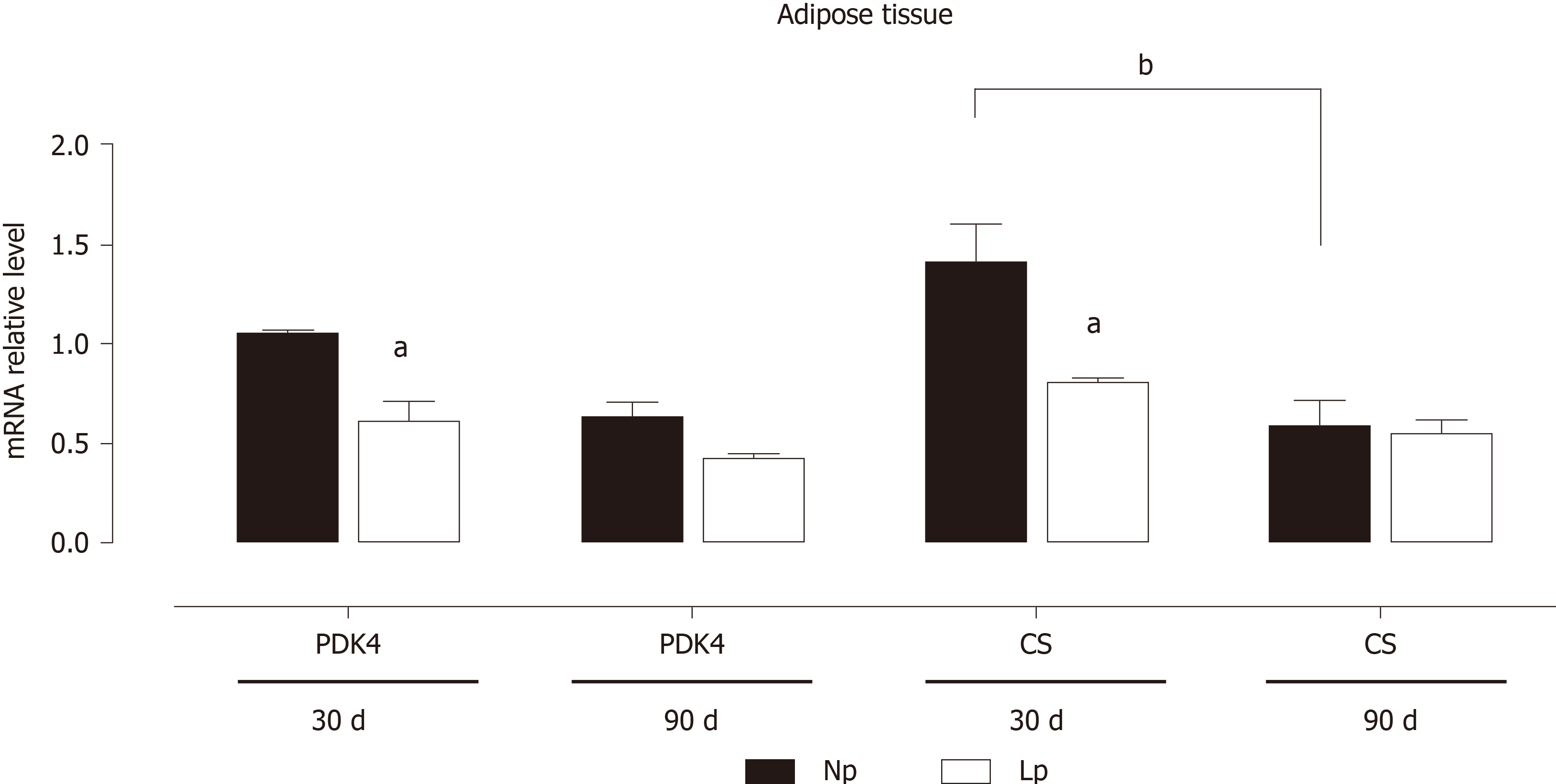
Figure 2 Expression of pyruvate dehydrogenase kinase 4 and citrate synthase mRNA in adipose tissue of rats.
Rats were exposed to maternal low protein diet during gestation and lactation, and their RNA extracted from adipose tissue was analyzed by quantitative reverse transcription PCR. Data are shown as mean ± standard error means analyzed by two-way ANOVA with the mother’s diet (normal protein; low protein) and age (30 d, 90 d) as variable factors. Bonferroni’s post-hoc test was used. Differences between diet groups are indicated by an asterisks; differences between ages are indicated by bars. aP < 0.05, bP < 0.01. NP: Normal protein; LP: Low protein; PDK4: Pyruvate dehydrogenase kinase 4; CS: Citrate synthase.
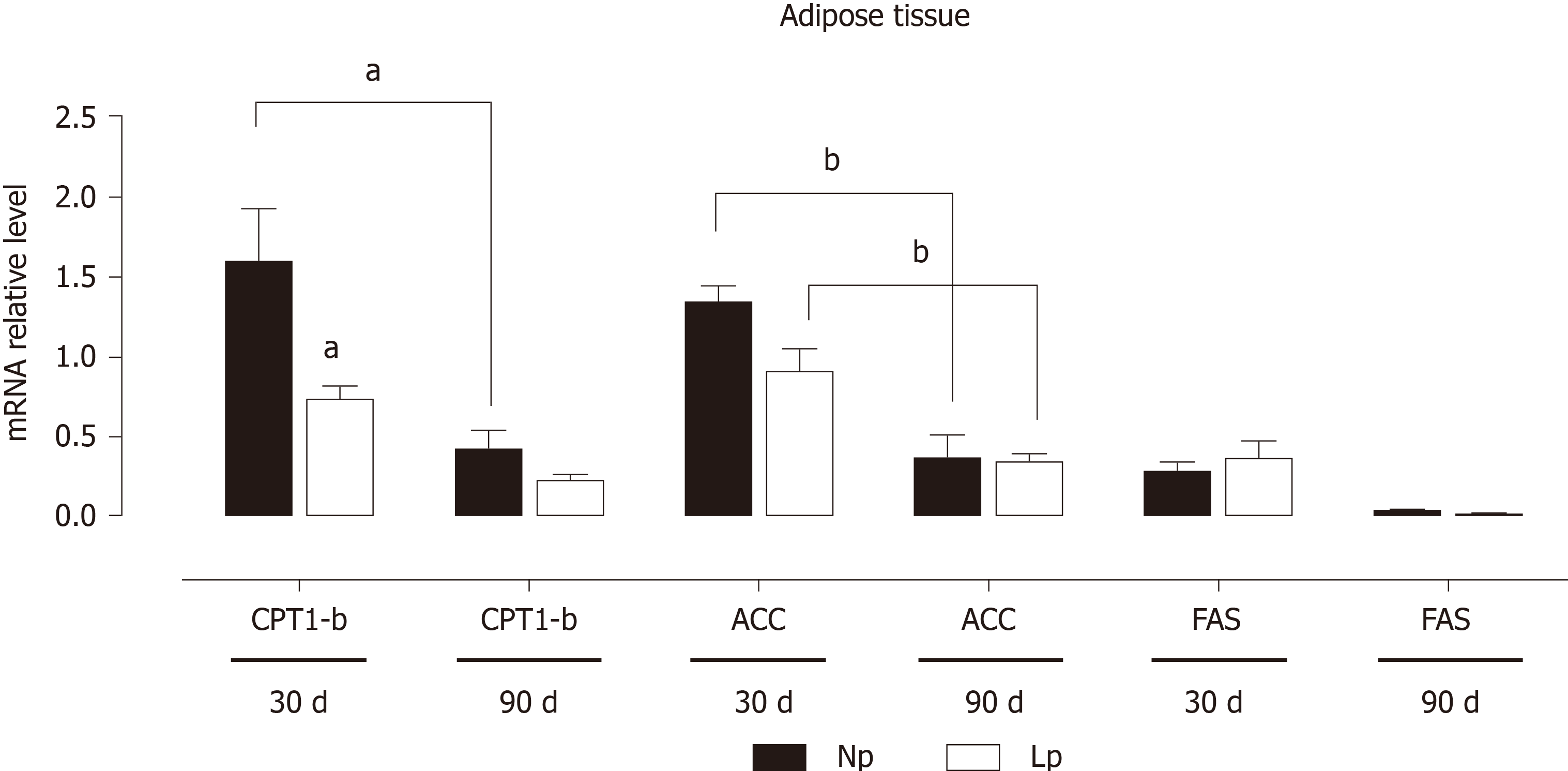
Figure 3 Expression of adipogenic and lipolytic genes in the adipose tissue of rats.
Rats were exposed to maternal low protein diet during gestation and lactation, and gene expression of carnitine palmitoylacyltransferase 1, acetyl-CoA carboxylase and fatty acid synthase in adipose tissue was analyzed by quantitative reverse transcription PCR. Data are shown as mean ± standard error means and analyzed by two-way ANOVA with the mother’s diet (normal protein; low protein) and age (30 d, 90 d) as variable factors. Bonferroni’s post hoc test was used. Differences between diet groups are indicated by an asterisks; differences between ages are indicated by bars. aP < 0.05, bP < 0.01. NP: Normal protein; LP: Low protein; FAS: Fatty acid synthase; CPT1-b: Carnitine palmitoylacyltransferase 1b; ACC: Acetyl-CoA carboxylase.
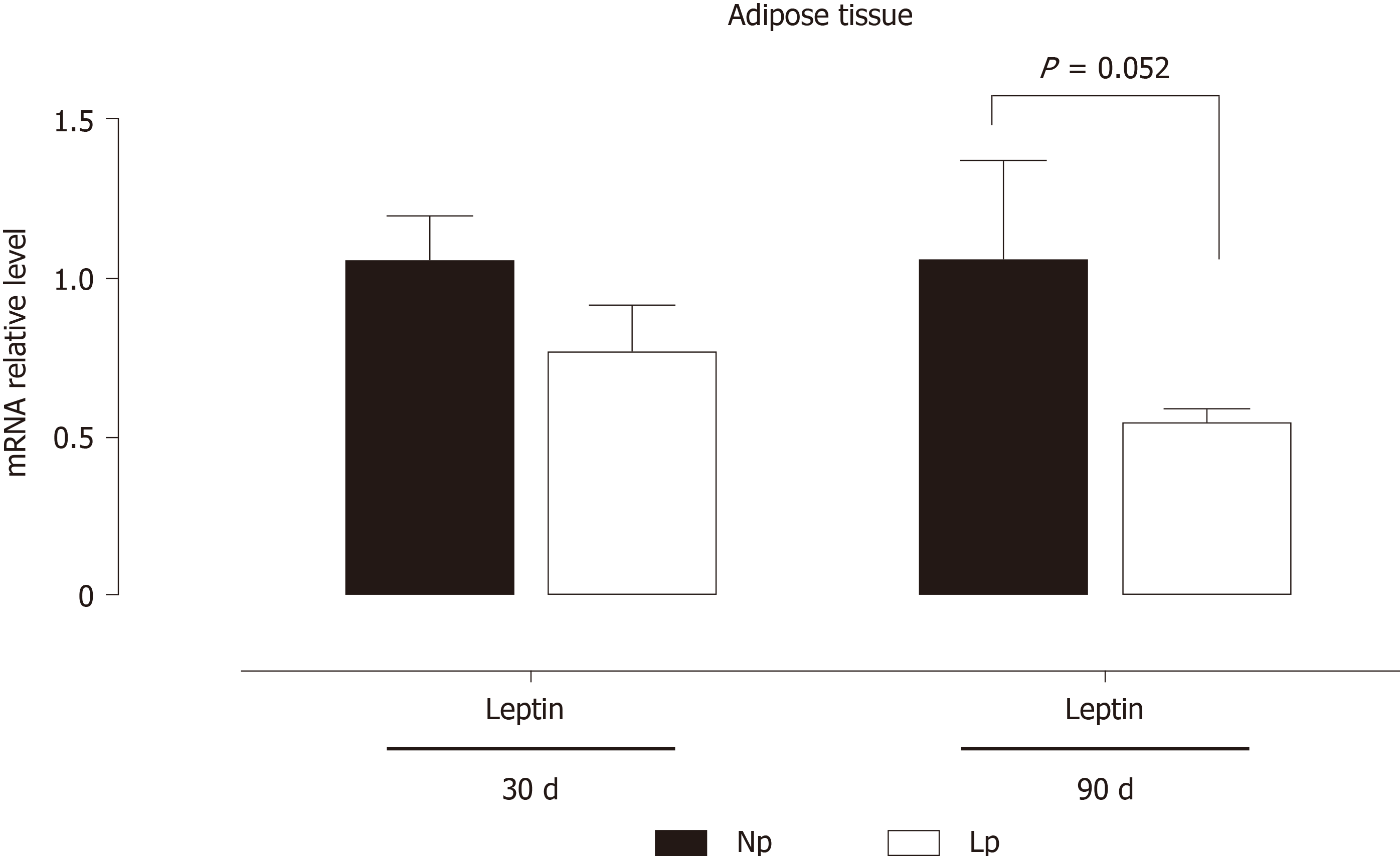
Figure 4 Gene expression of leptin in adipose tissue of rats.
Rats were exposed to maternal low protein diet during gestation and lactation, and gene expression of leptin from adipose tissue was analyzed by quantitative reverse transcription PCR. Data are shown as mean ± standard error means and analyzed by two-way ANOVA with the mother’s diet (normal protein; low protein) and age (30 d, 90 d) as variable factors. Bonferroni’s post hoc test was used. Differences between diet groups are indicated by asterisks. NP: Normal protein; LP: Low protein.
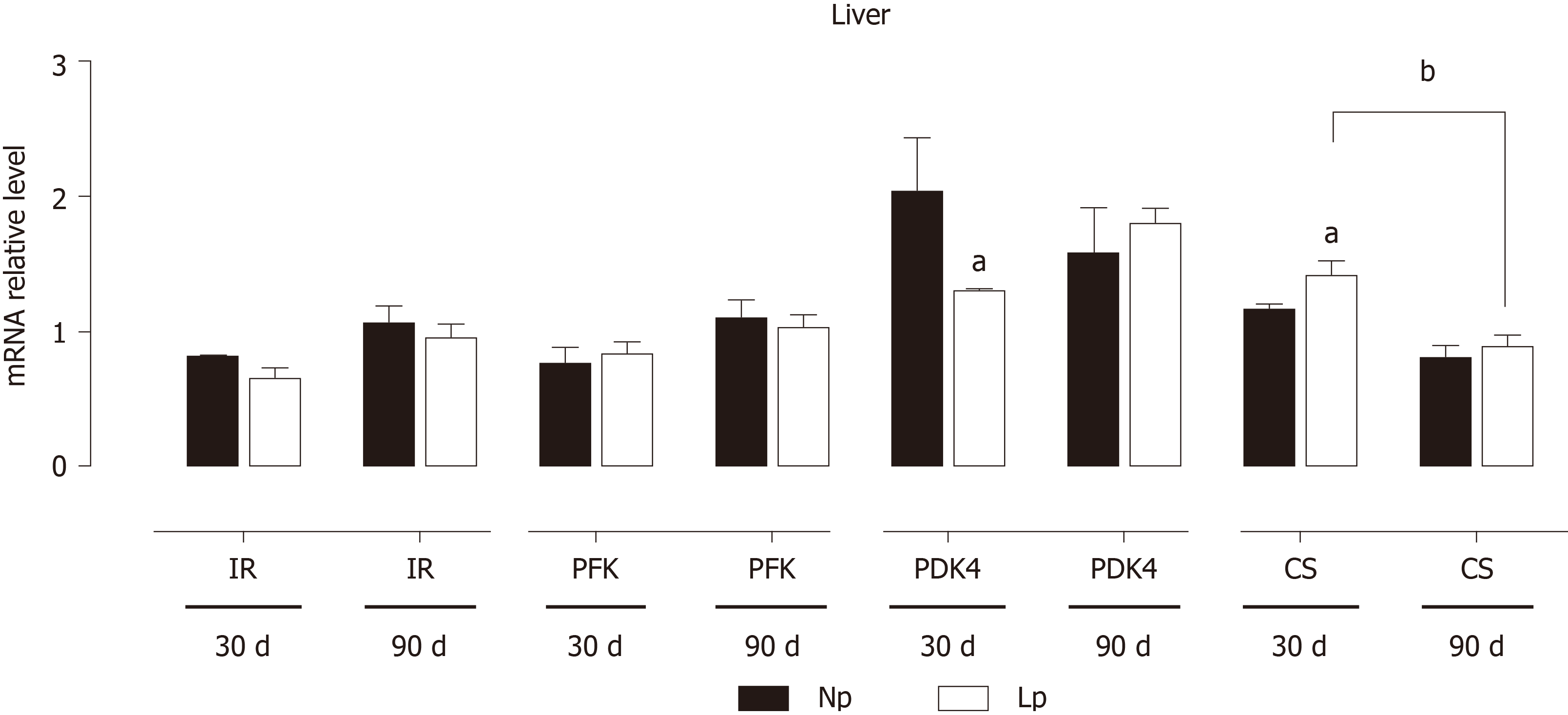
Figure 5 Expression of insulin receptor, glycolytic genes and Krebs cycle genes in the liver of rats.
Rats were exposed to maternal low protein diet during gestation and lactation, and gene expression of insulin receptor, phosphofructokinase, pyruvate dehydrogenase kinase 4 and citrate synthase was analyzed by quantitative reverse transcription PCR. Data are shown as mean ± standard error means and analyzed by two-way ANOVA with the mother’s diet (normal protein; low protein) and age (30 d, 90 d) as variable factors. Bonferroni’s post hoc test was used. Differences between diet groups are indicated by asterisks; differences between ages are indicated by bars. aP < 0.05, bP < 0.01. NP: Normal protein; LP: Low protein; CS: Citrate synthase; PDK4: Pyruvate dehydrogenase kinase 4; PFK: Phosphofructokinase; IR: Insulin receptor.
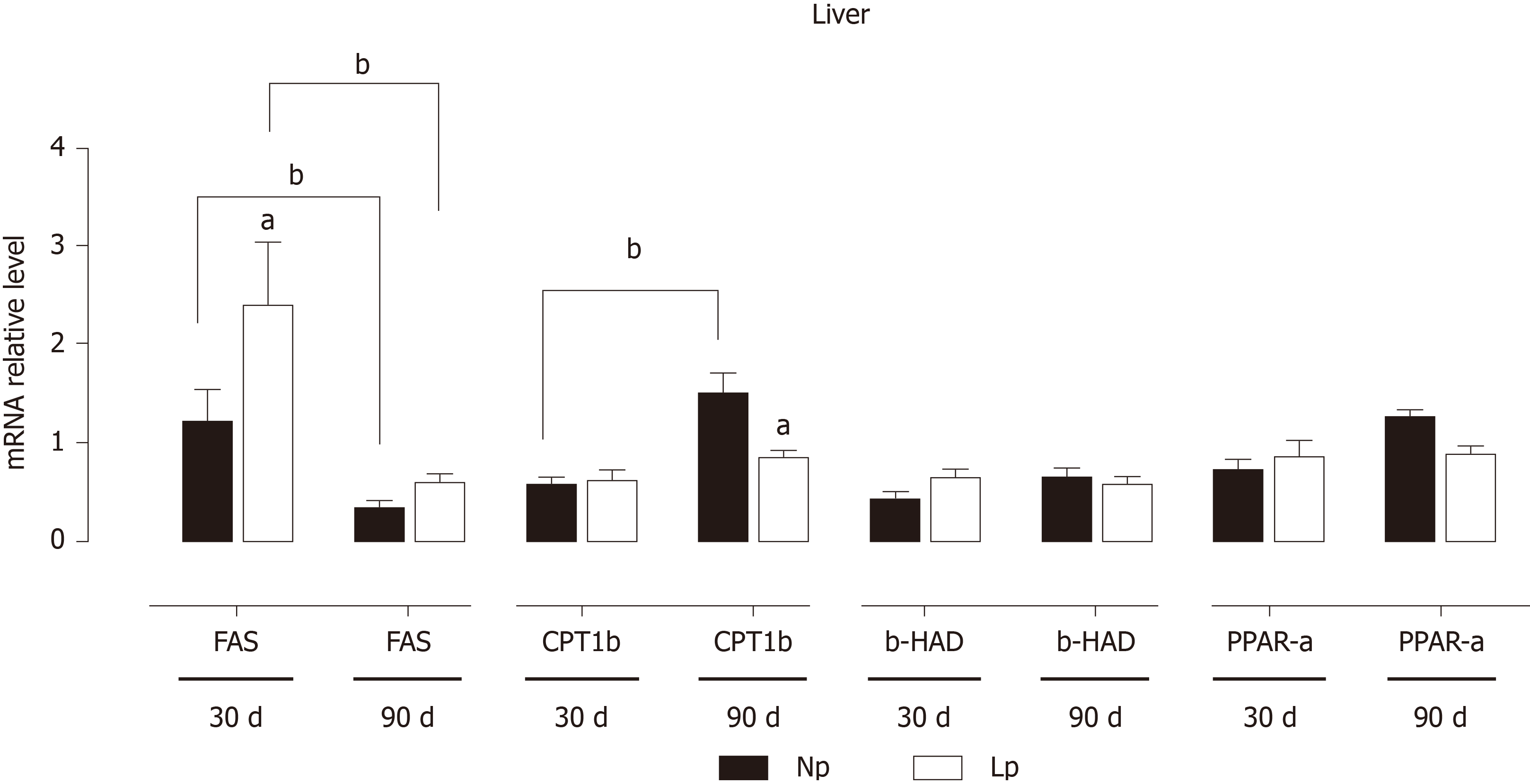
Figure 6 Expression of genes related to lipid metabolism in the liver of rats.
Rats were exposed to maternal low protein diet during gestation and lactation, and gene expression was analyzed by quantitative reverse transcription PCR. Data are shown as mean ± standard error means and analyzed by two-way ANOVA with the mother’s diet (normal protein; low protein) and age (30 d, 90 d) as variable factors. Bonferroni’s post hoc test was used. Differences between diet groups are indicated by asterisks; differences between ages are indicated by bars. aP < 0.05, bP < 0.01. NP: Normal protein; LP: Low protein; FAS: Fatty acid synthase; CPT1-b: Carnitine palmitoylacyltransferase 1b; b-HAD: Beta hydroxyacyl-coenzyme-A dehydrogenase; PPAR-a: Peroxisome proliferator-activated receptor-alpha.
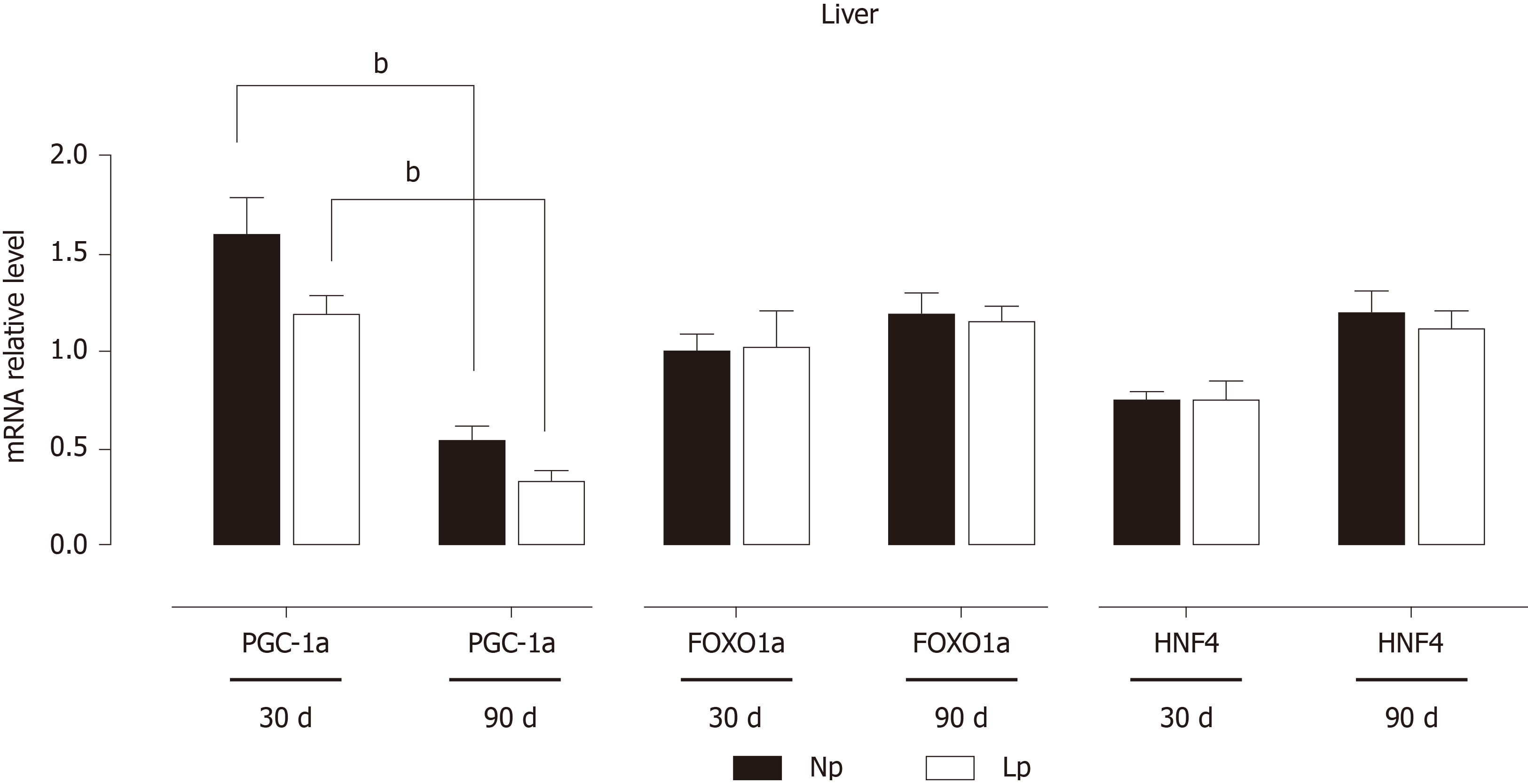
Figure 7 Expression of key transcription factors in the liver of rats.
Rats were exposed to maternal low protein diet during gestation and lactation, and gene expression of peroxisome proliferator-activated receptor-alpha coactivator 1 alpha, forkhead box protein O1 and hepatocyte nuclear factor 4 was analyzed by quantitative reverse transcription PCR. Data are shown as mean ± standard error means and analyzed by two-way ANOVA with the mother’s diet (normal protein; low protein) and age (30 d, 90 d) as variable factors. Bonferroni’s post hoc test was used. Differences between diet groups are indicated by asterisks; differences between ages are indicated by bars. aP < 0.05, bP < 0.01. NP: Normal protein; LP: Low protein; PGC-1α: Peroxisome proliferator-activated receptor-alpha coactivator 1 alpha; FOXO1a: Forkhead box protein O1; HNF4: Hepatocyte nuclear factor 4.
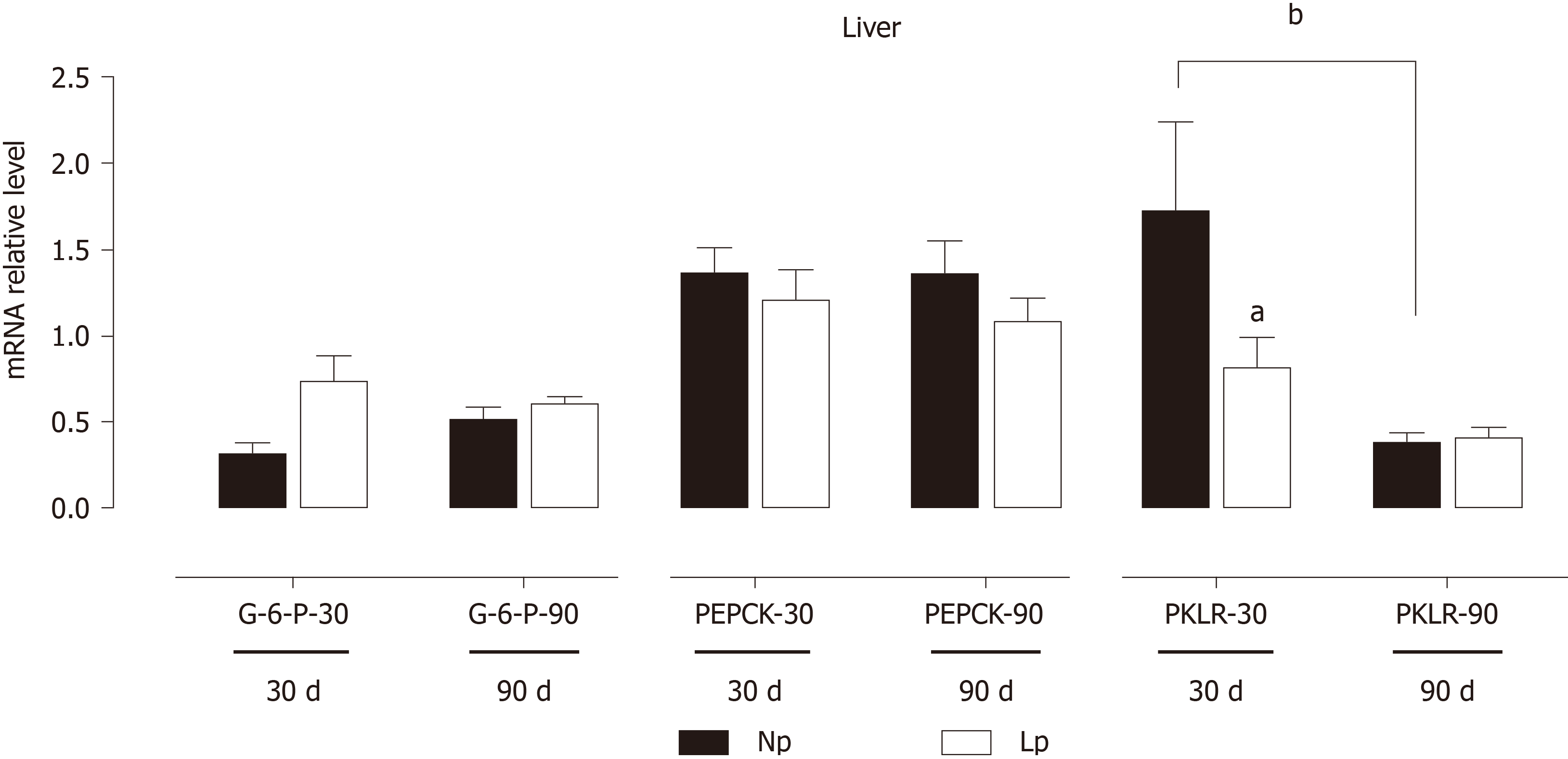
Figure 8 Expression of gluconeogenic genes in the liver of rats.
Rats were exposed to maternal low protein diet during gestation and lactation, and gene expression of glucose 6-phosphatase, phosphoenolpyruvate carboxykinase and pyruvate kinase L/R was analyzed by quantitative reverse transcription PCR. Data are shown as mean ± standard error means and analyzed by two-way ANOVA with the mother’s diet (normal protein; low protein) and age (30 d, 90 d) as variable factors. Bonferroni’s post hoc test was used. Differences between diet groups are indicated by asterisks; differences between ages are indicated by bars. aP < 0.05, bP < 0.01. NP: Normal protein; LP: Low protein; G6Pase: Glucose 6-phosphatase; PEPCK: Phosphoenolpyruvate carboxykinase; PKLR: Pyruvate kinase L/R.
- Citation: de Oliveira Lira A, de Brito Alves JL, Pinheiro Fernandes M, Vasconcelos D, Santana DF, da Costa-Silva JH, Morio B, Góis Leandro C, Pirola L. Maternal low protein diet induces persistent expression changes in metabolic genes in male rats. World J Diabetes 2020; 11(5): 182-192
- URL: https://www.wjgnet.com/1948-9358/full/v11/i5/182.htm
- DOI: https://dx.doi.org/10.4239/wjd.v11.i5.182