Copyright
©The Author(s) 2022.
World J Gastrointest Endosc. May 16, 2022; 14(5): 311-319
Published online May 16, 2022. doi: 10.4253/wjge.v14.i5.311
Published online May 16, 2022. doi: 10.4253/wjge.v14.i5.311
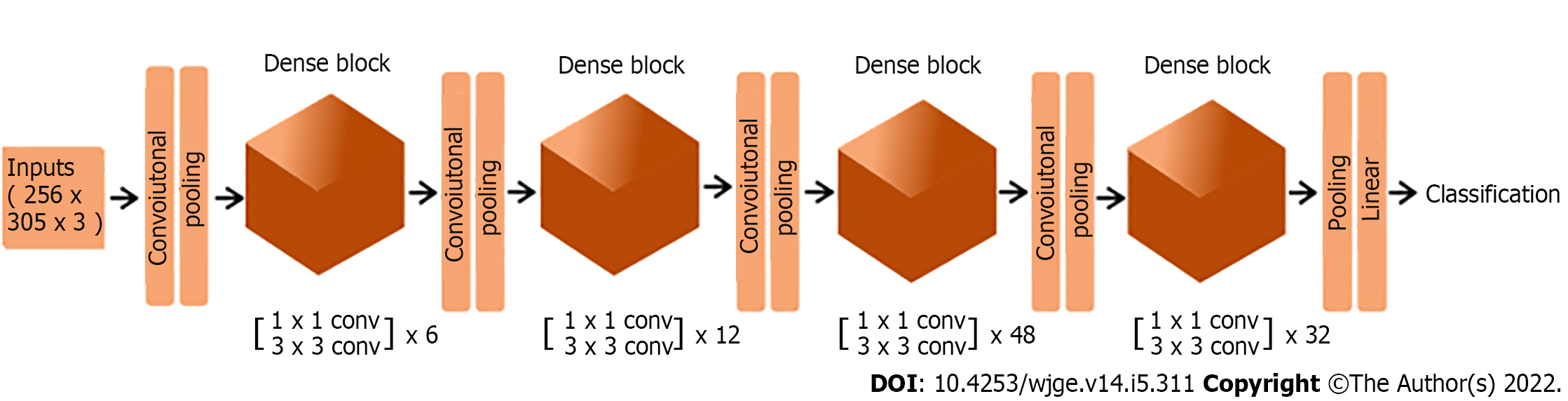
Figure 1 Representation of model’s final architecture.
In the proposed model, each image is used as an input for a deep neural network composed of four blocks of densely connected convolutional layers, together with convolutional and pooling transition layers. The network output is a binary classification.
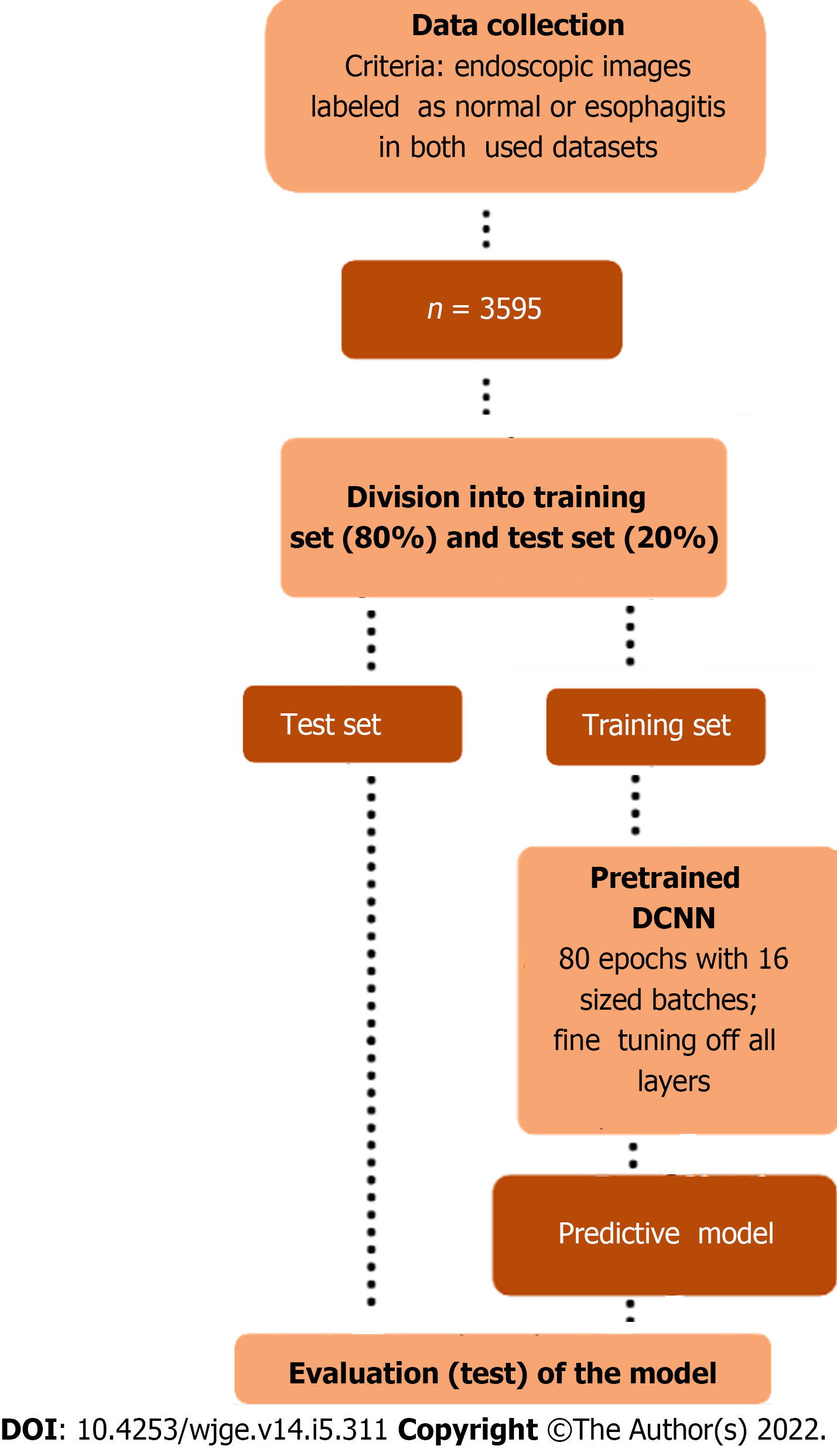
Figure 2 Methodological design of the study.
The proposed workflow encompasses selective collection of endoscopic images from the datasets, splitting and pre-processing of the data, iterative training of the classificatory model, and finally evaluation of its performance. DCNN: Dense convolutional neural network.
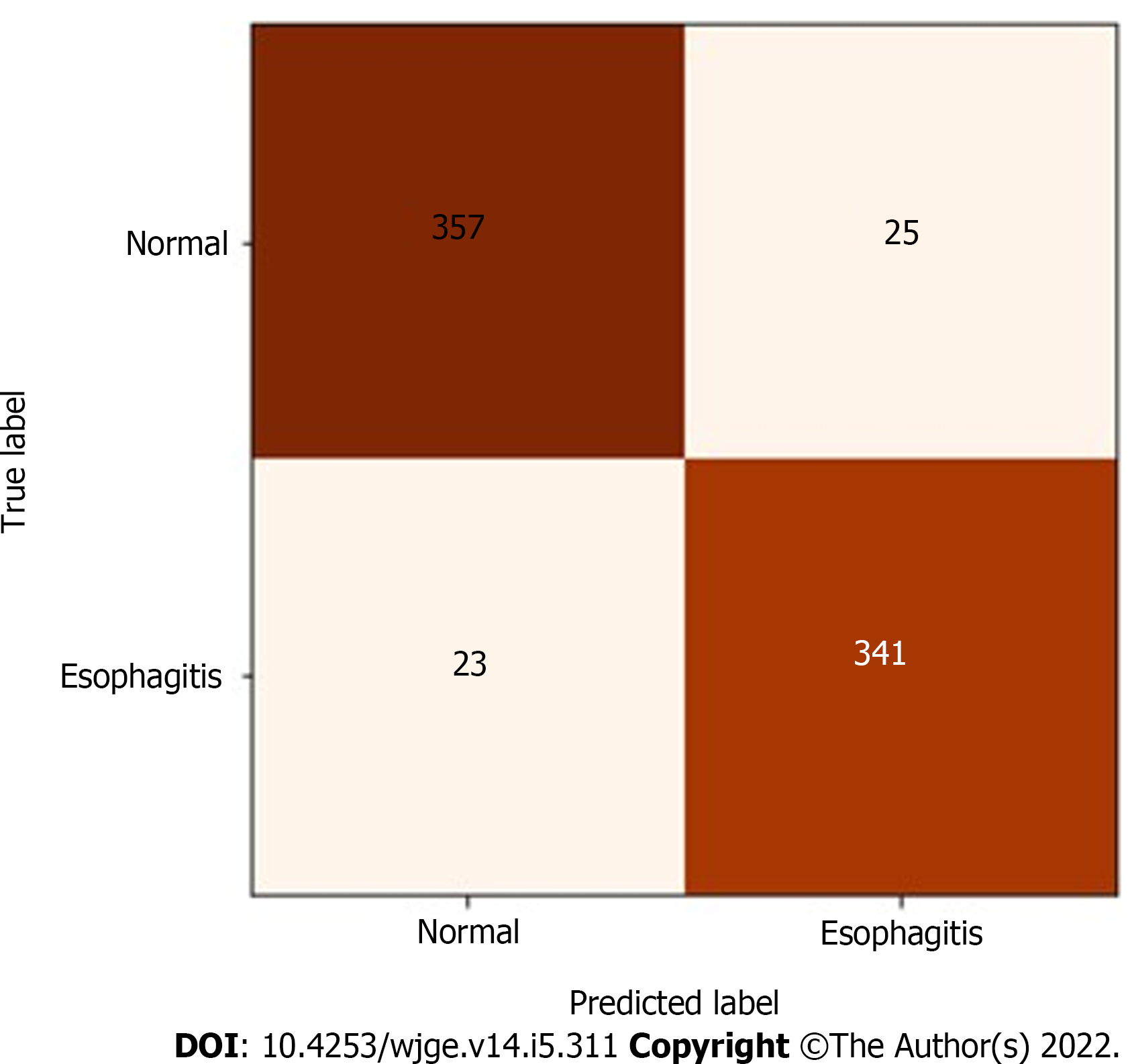
Figure 3 Confusion matrix for the predictive model.
As illustrated, the model was able to accurately classify 314 of 337 esophagitis images and 357 of 382 normal images, with true positive and false positive rates of 93.2% and 93.5%, respectively.
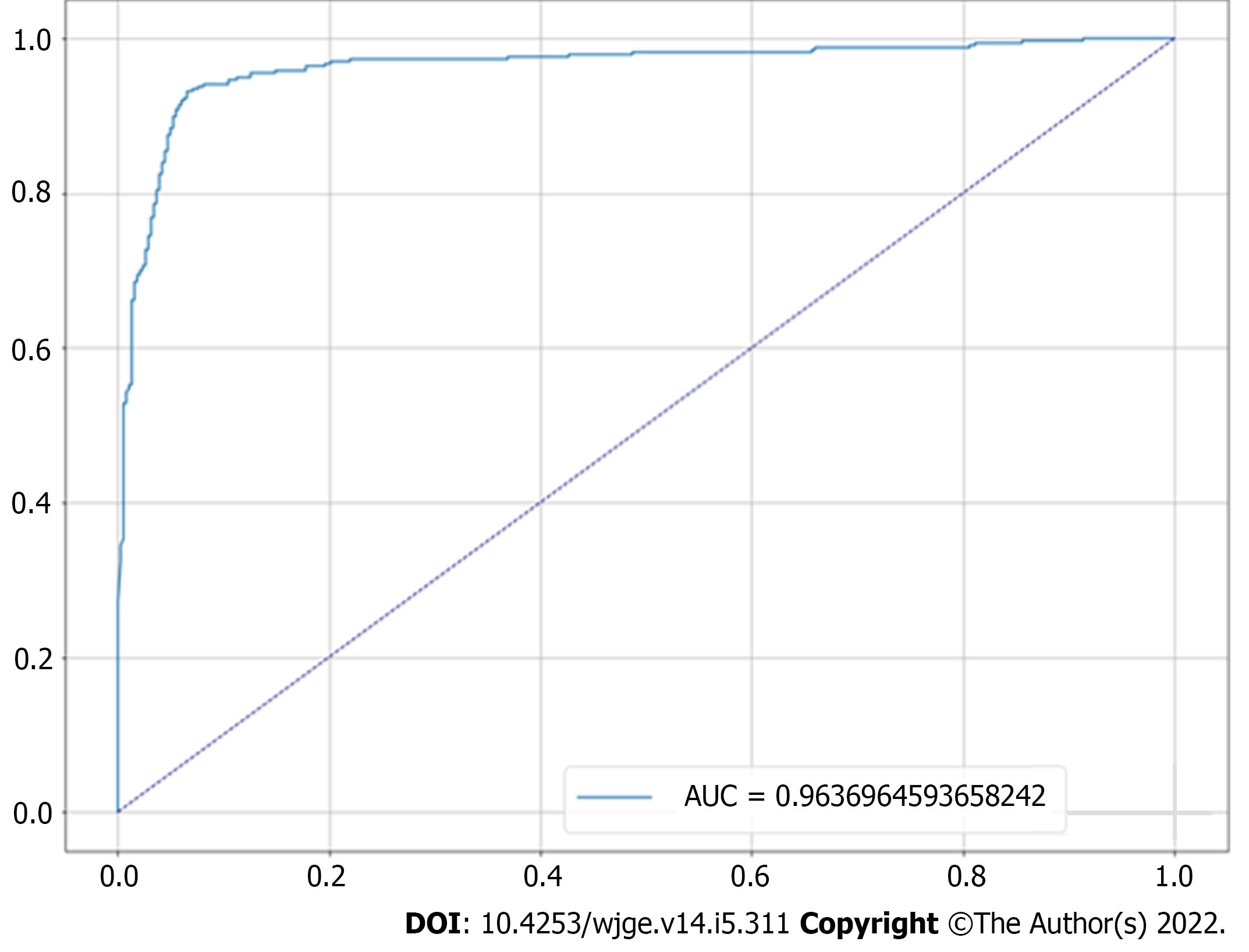
Figure 4 Receiver operating characteristic curve for the proposed predictive model.
The graph shows the resulting curve relating the true and false positive rates, giving an area on the curve of 96.4%. AUC: Area under the receiver operating characteristic curve.
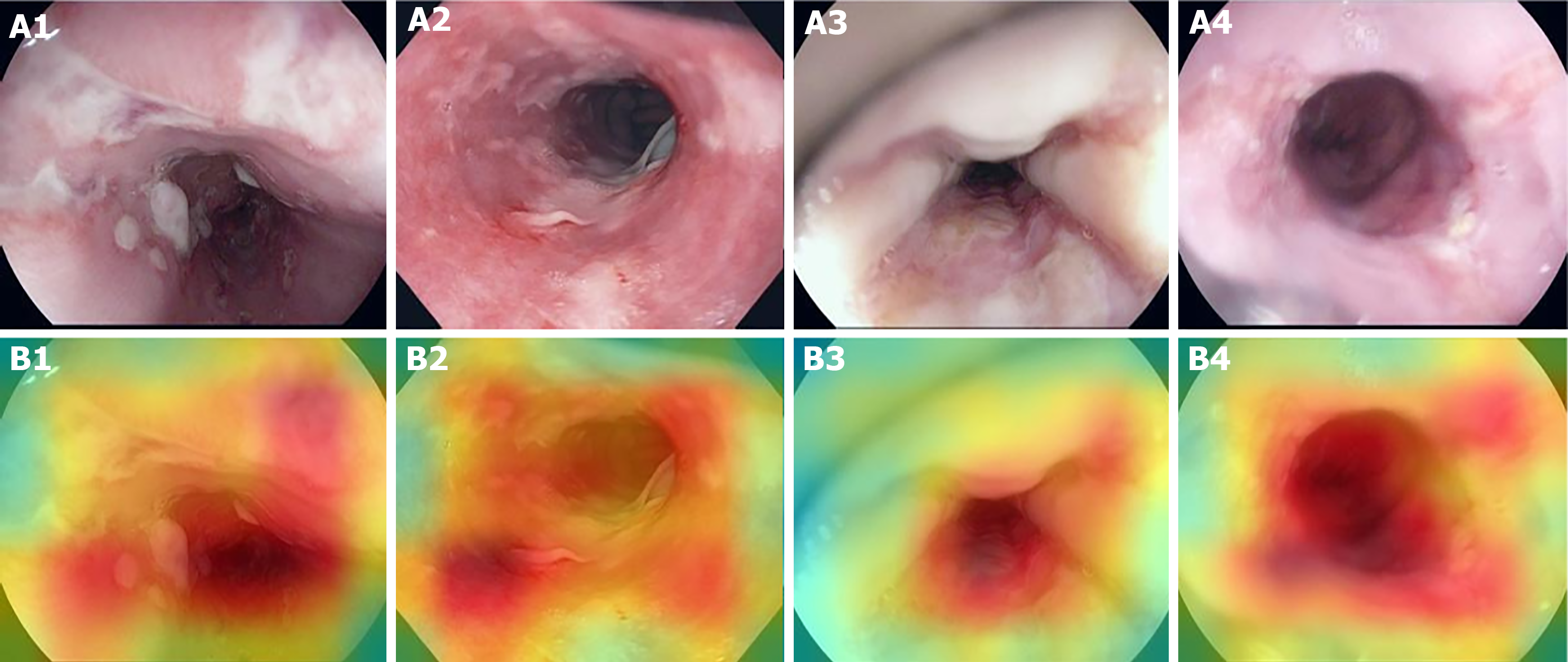
Figure 5 Heatmaps for spatial predictive relevance in esophagitis endoscopic images from test set.
A and B: Images A1-A4 represent examples esophagitis endoscopic images used to test our predictive model, while images B1-B4 represent the corresponding heatmaps indicating, for each image, the areas with the greatest influence on the prediction. A1-A4: Citation: Pogorelov K, Randel K, Griwodz C, Eskeland S, Lange T, Johansen D, Spampinato C, Dang-Nguyen D, Lux M., Schmidt P, Riegler M, Halvorsen P. Kvasir: A Multi-Class Image Dataset for Computer Aided Gastrointestinal Disease Detection. MMSys'17 Proceedings of the 8th ACM on Multimedia Systems Conference (MMSYS); 2017 June 20-23; Taipei, Taiwan. New York: Association for Computing Machinery, 2017: 164-169. Copyright © Simula Research Laboratory 2017. Published by Association for Computing Machinery[12]. The authors have obtained the permission for figure using from the Simula Research Laboratory (Supplementary material). Citation: Borgli H, Thambawita V, Smedsrud PH, Hicks S, Jha D, Eskeland SL, Randel KR, Pogorelov K, Lux M, Nguyen DTD, Johansen D, Griwodz C, Stensland HK, Garcia-Ceja E, Schmidt PT, Hammer HL, Riegler MA, Halvorsen P, de Lange T. HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci Data 2020; 7: 283. Copyright © Simula Research Laboratory 2020. Published by Nature Publishing Group[13]. The authors have obtained the permission for figure using from the Simula Research Laboratory (Supplementary material).
- Citation: Caires Silveira E, Santos Corrêa CF, Madureira Silva L, Almeida Santos B, Mattos Pretti S, Freire de Melo F. Recognition of esophagitis in endoscopic images using transfer learning. World J Gastrointest Endosc 2022; 14(5): 311-319
- URL: https://www.wjgnet.com/1948-5190/full/v14/i5/311.htm
- DOI: https://dx.doi.org/10.4253/wjge.v14.i5.311