Published online Mar 28, 2017. doi: 10.4254/wjh.v9.i9.477
Peer-review started: June 13, 2016
First decision: July 20, 2016
Revised: September 19, 2016
Accepted: December 13, 2016
Article in press: December 14, 2016
Published online: March 28, 2017
Processing time: 288 Days and 11.6 Hours
To investigate the prevalence and virological characteristics of occult hepatitis B virus (HBV) infections in patients with hematological malignancies in South Egypt.
Serum samples were collected from 165 patients with hematological malignancies to monitor titers of HBV DNA, hepatitis B surface antigen (HBsAg), and antibodies to HBV core (anti-HBc) and surface antigens. Serum samples negative for HBsAg and positive for anti-HBc were subjected to nucleic acid extraction and HBV DNA detection by real-time polymerase chain reaction. DNA sequences spanning the S region were analyzed in cases with occult HBV infection. In vitro comparative study of constructed 1.24-fold wild type and S protein mutant HBV genotype D clones was further performed.
HBV DNA was detected in 23 (42.6%) of 54 patients with hematological malignancies who were HBsAg negative, but anti-HBc positive, suggesting the presence of occult HBV infection. The complete HBV genome was retrieved from 6 occult HBV patients, and P120T and S143L were detected in 3 and 2 cases, respectively. Site directed mutagenesis was done to produce 1.24-fold genotype D clones with amino acid mutations T120 and L143. The in vitro analyses revealed that a lower level of extracellular HBsAg was detected by chemiluminescence enzyme immunoassay (CLEIA) with the clone containing T120 mutation, compared with the wild type or the clone with S143L mutation despite the similar levels of extracellular and intracellular HBsAg detected by Western blot. Southern blot experiments showed that the levels of intracellular HBV DNA were not different between these clones.
Occult HBV infection is common in patients with hematological malignancies and associated with P120T and S143L mutations. 120T mutation impairs the detection of HBsAg by CLEIA.
Core tip: The present study was conducted to investigate the prevalence and virological characteristics of occult hepatitis B virus (HBV) infections in patients with hematological malignancies in Egypt. Serum samples were collected from 165 patients with different hematological malignancies and screened for occult HBV infection. In the present study, occult HBV infection was detected in 23 (42.6%) of 54 patients with hematological malignancies who were hepatitis B surface antigen (HBsAg) negative, but antibodies to HBV core (anti-HBc) positive. Based on in vitro study of clones inserted with 120T and 143L, it was found that the 120T mutation could impair HBsAg detection by changing its conformation. Patients with hematological malignancies should be screened and closely monitored for anti-HBc and HBV DNA.
- Citation: Elkady A, Iijima S, Aboulfotuh S, Mostafa Ali E, Sayed D, Abdel-Aziz NM, Ali AM, Murakami S, Isogawa M, Tanaka Y. Characteristics of escape mutations from occult hepatitis B virus infected patients with hematological malignancies in South Egypt. World J Hepatol 2017; 9(9): 477-486
- URL: https://www.wjgnet.com/1948-5182/full/v9/i9/477.htm
- DOI: https://dx.doi.org/10.4254/wjh.v9.i9.477
Occult hepatitis B virus (HBV) infection is defined by the presence of HBV DNA in the liver (with or without HBV DNA in the serum) in hepatitis B surface antigen (HBsAg) negative individuals[1]. Apart from posing diagnostic challenges, several studies indicated that occult HBV infection also associates with flares of liver disease in hepatitis C virus (HCV) infected patients who do not exhibit changes in HCV RNA levels and reduces the response rate to interferon therapy[2]. Furthermore, occult HBV infection is frequently detected in cryptogenic liver diseases and autoimmune hepatitis[3-5]. HBV reactivation is a well-known complication in patients with occult infection under immune suppression, such as anticancer therapy, and human immunodeficiency virus (HIV) infection[6,7]. In addition, occult HBV infection increases the risk of HBV transmission through blood transfusion[8].
Significant advances have been achieved in understanding the molecular basis for occult HBV infection, and several factors have been implicated in the pathogenesis of occult HBV infection[9,10]. A variety of mutations in HBsAg have been reported to affect in vitro antigen detection, in vivo immune recognition, HBV infectivity, cell tropism and virion morphology[11-14]. The aim of this study was to determine the prevalence of occult HBV infection in patients with hematological malignancies in South Egypt. An in vitro study was performed to assess the virological characteristics of prevalent HBsAg mutations detected in patients with occult HBV infection.
Serum samples were collected consecutively from 165 patients with hematological malignancies hospitalized in the Oncology Department of the Sohag Faculty of Medicine and South Egypt Cancer Institution from November 2010 to October 2011. All patients started their treatment regimen at the time of conduction of the study.
The serum samples were stored at -80 °C for future examination of HBsAg, antibodies to HBsAg (anti-HBs), antibodies to HBV core (anti-HBc), and HBV DNA.
HBsAg was measured by enzyme immunoassay (AxSYM; Abbott Japan, Tokyo, Japan) and chemiluminescence enzyme immunoassay (CLEIA) (Fujirebio, Tokyo, Japan). The IgG class of anti-HBc was determined by radioimmunoassay (Abbott Japan). Anti-HBs was tested by enzyme immunoassay (AxSYM; Abbott Japan, Tokyo Japan). Anti-HCV was tested by CLEIA (Fujirebio, Tokyo, Japan). All serologic assays were performed according to the manufacturer's instructions.
DNA was extracted from serum samples (200 μL) using a QIAamp DNA extraction kit (Qiagen, Hilden, Germany) and re-suspended in 100 μL of the storage buffer provided by the kit manufacturer.
HBV DNA sequences spanning the entire S gene were amplified by real-time polymerase chain reaction (PCR) according to a previously described protocol with a slight modification[15]. The detection limit of the assay was 100 copies/mL (equivalent to 20 IU/mL).
Extracted DNA was subjected to PCR for amplifying the complete genome of HBV as previously described[16]. Amplicons were sequenced directly using the ABI Prism Big Dye ver. 3.1 kit on the AMI 3100 DNA automated sequencer (Applied Biosystems, Foster City, CA, United States). All sequences were analyzed in both forward and reverse directions. HBV genotypes were determined by molecular evolution analysis. Target sequences were aligned by CLUSTALX with reference sequences retrieved from the DDBJ/EMBL/GenBank databases, and genetic distances were estimated by the 6-parameter method in the Hepatitis Virus Database (http://s2as02.genes.nig.ac.jp/)[17]. Based on the obtained distances, phylogenetic trees were constructed by neighbor-joining method with the mid-point rooting option. To confirm the reliability of the phylogenetic trees, bootstrap resampling tests were performed 1000 times for analysis by the ODEN program of the National Institute of Genetics.
Various plasmids were constructed based on a consensus clone named D-IND 60 in which 1.24-fold HBV DNA genome of wild type genotype D (nt 1413-3215/1-2185) was inserted into a pUC19 vector (Invitrogen Corp., Carlsbad, CA, United States)[18].
For site-directed mutagenesis, plasmid D-IND 60 was digested by HindIII and EcoO65I, and ligated with the fragments carrying P120T and S143L amino acid mutations to produce the 1.24-fold HBV genome. Cloned HBV DNA sequences were confirmed using ABI Prism Big-Dye (Applied Biosystems, Foster City, CA, United States) on an ABI 3100 automated sequencer.
The hepatoma derived cell line Huh-7 was maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum. For the standard replication assay, 1 × 106 Huh-7 cells were seeded onto a 10-cm-diamater dish, and 16 h later, transfected with 5 μg DNA constructs using a Fugene 6 transfection reagent (Roche Diagnostics, Indianapolis, IN, United States). Transfection efficiency was monitored by cotransfecting 0.5 μg of a reporter plasmid expressing secreted alkaline phosphatase (SEAP) and measuring SEAP activity in the culture supernatant. The supernatants and cell lysates of the transfected cells were collected 3 d after the transfection to analyze HBV markers. Three independent experiments were conducted for each clone[19].
HBsAg and HBcAg were determined by CLEIA using commercial assay kits (Fujirebio Inc., Tokyo, Japan). To detect the intracellular replicative intermediates of HBV, the core particle-associated HBV DNA in the cells was isolated and measured by Southern blot[20,21]. Briefly, cells were harvested and lysed in 1.5 mL of lysis buffer containing 50 mmol/L Tris-HCl (pH 7.4), 1 mmol/L EDTA and 1% IGEPAL CA-630 (Sigma-Aldrich, Japan G.K.). Total cell lysate was treated with 120 μg/mL of RNase A and 30 μg/mL of DNase I for 3 h at 37 °C, in the presence of 6 mmol/L magnesium acetate. HBV DNA was then released by proteinase K digestion, extracted with phenol, and ethanol precipitated. DNA was separated on a 1.2% w/v agarose gel and then transferred to a positive-charged nylon membrane (Roche Diagnostics). The membrane was hybridized with digoxigenin (DIG)-dUTP-labeled full-length HBV genotype C fragment, which was generated using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics GmbH), and then detected by alkaline phosphatase-labelled anti-DIG antibody according to the manufacturer’s instructions. Signals were analyzed using an ImageQuant LAS 4000 mini (GE Healthcare United Kingdom Ltd).
Huh7 cells (1 × 106) were transfected with genotype D wild type clone or its mutants using Fugene6 (Promega, Madison, WI, United States). Transfected Huh7 cells and culture supernatants were harvested 72 h post transfection. Cells were washed with phosphate-buffered saline twice and lysed with lysis buffer (CelLytic M Cell lysis reagent; Sigma). The culture supernatants and cell lysates were quantified for HBsAg and HBcrAg by CLEIA (Fujirebio, Japan). The protocol of Western blot was previously described[22]. To detect HBsAg and HBcAg in cell lysates and viral particles, we used monoclonal antibodies specific for PreS1 and Core (Institute of Immunology, Japan), respectively. Immunoreactive proteins were visualized using chemiluminescence reagents (Immobilon; Millipore).
Statistical analyses were performed by Fisher’s exact probability test and independent t-test for continuous variables using SPSS software (SPSS, Chicago, IL, United States). P-values (two-tailed) less than 0.05 were considered statistically significant.
This study was conducted in accordance with the guidelines of the Declaration of Helsinki and its subsequent amendments, and informed consent was obtained from all patients.
The baseline characteristics of 165 patients with hematological malignant diseases are shown in Table 1. The mean age of the studied cohort was 36.1 ± 23.1 years old. Among the 165 patients with hematological malignancies, 13 (7.9%) were found positive for HBsAg, 39 (23.6%) positive for anti-HCV, and 152 (92.1%) negative for HBsAg, of whom 54 (35.5%) were serologically positive for anti-HBc. Male predominance was observed in the studied cohort (55.6%). Overt HBV and HCV co-infections were not detected in the studied cohort. Eighty-eight (53.3%) patients were diagnosed with malignant lymphoma, 43 (26.1%) diagnosed with acute leukemia, 23 (13.9%) with chronic leukemia, 8 (4.8%) with Hodgkin’s disease, and 3 (1.8%) with multiple myeloma.
| Characteristic | Total (n = 165) |
| Age (mean ± SD) | 36.1 ± 23.1 |
| Gender (M) | 89 (55.6) |
| ALT (mean ± SD) | 29.7 ± 32.8 |
| AST (mean ± SD) | 38.2 ± 49.2 |
| Anti-HCV(+) | 39 (23.6) |
| HBsAg(+) | 13 (7.9) |
| Anti-HBc(+)/HBsAg(-) | 54 (32.7) |
| Clinical characteristics | |
| Malignant lymphoma | 88 (53.3) |
| Hodgkin’s disease | 8 (4.8) |
| Acute leukemia | 43 (26.1) |
| Chronic leukemia | 23 (13.9) |
| Multiple myeloma | 3 (1.8) |
| Steroid containing treatment | 110 (66.7) |
HBV DNA was detected in 42.6% (23/54) of the patients with hematological malignancies who were negative for HBsAg, but positive for anti-HBc, suggesting the presence of occult HBV infection. Anti-HCV was detected in 26.1% (6/23) of occult hepatitis B cases. The complete genome of HBV was successfully obtained from 6 cases with occult HBV infection. The clinical and HBV virological aspects of these 6 patients are summarized in Table 2. Four of these 6 patients were diagnosed with malignant lymphoma, and two patients were diagnosed with Hodgkin’s disease and ALL each (Table 2). Their age ranged between 5 and 80 years. Two patients were serologically anti-HBs positive (Samples ID; Egl6 and EGL4). Three patients were serologically negative for anti-HBs (samples ID; U79, D1 and D14). There was insufficient serum sample to measure anti-HBs levels in one case (sample ID; A79).
| Sample ID | Age | Gender | Diagnosis | Anti-HBs (mIU/L) | HBV DNA Log (copies/mL) | Mutation1 |
| EGL6 | 38 | M | Malignant lymphoma | (+)/15 | 3.5 | - |
| U31 | 55 | F | Malignant lymphoma | (-) | 3.8 | S143L |
| EGL4 | 80 | F | Malignant lymphoma | (+)/18.9 | 2.4 | P120S, S143L |
| A79 | 5 | M | ALL | NT | 4.4 | P120T |
| D1 | 70 | M | Malignant lymphoma | (-) | 3.9 | P120T |
| D14 | 5 | M | HD | (-) | 3.6 | P120T |
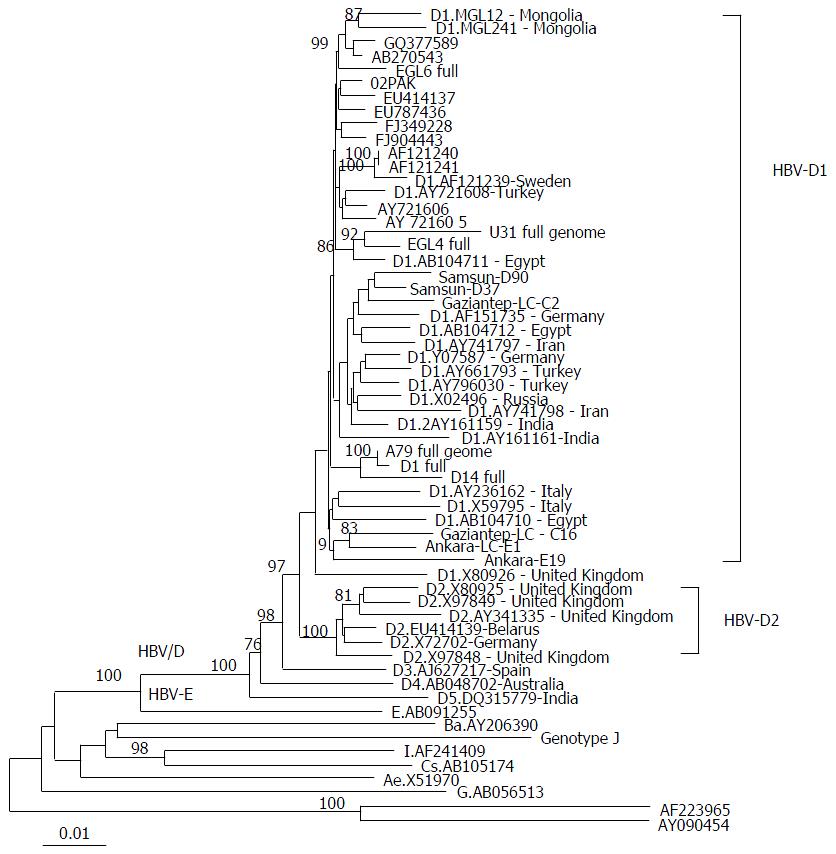
Phylogenetic analysis of the retrieved complete HBV genomic sequences indicated that all isolates were of genotype D subtype D1 (Figure 1). Further analysis was applied to the major hydrophilic region (MHR) of the HBV genome, revealing the presence of two prevalent amino acid substitutions; P120T in patients Egl4, A79, D1, and D79 and S143L in U31 and Egl4 (Table 2).
HBV genomic short sequences encompassing the “a” determinant region were available in 12 of 17 samples. Of the 12 samples, escape mutation Q129R was present in 7, while P120T was detected in 4 and S143L was present in 1, suggesting that the clones with 129R mutation were from a minor population with relatively low HBV-DNA levels.
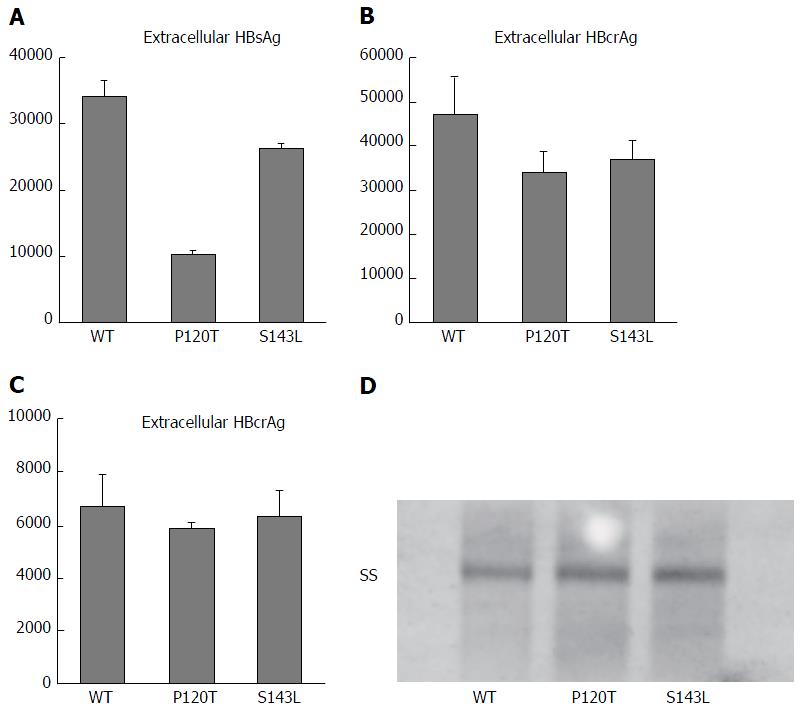
To elucidate the virological characteristics of P120T and S143L mutations obtained from the complete sequences in this study, these mutations were individually inserted by site mutagenesis into a 1.24-fold replication competent HBV clone based on genotype D. Wild type genotype D clone, and two variants containing P120T and S143L mutations were transfected to Huh7 cells, and supernatants (extracellular) and cell lysates (intracellular) were collected to compare HBsAg and HBcAg levels. As shown in Figure 2A, a lower concentration of HBsAg was detected in the supernatant of Huh7 cells transfected with the P120T variant compared with wild type or S143L variant transfected cells. No significant difference was observed in extracellular or intracellular HBcrAg levels between these three clones (Figure 2B and C). Furthermore, Southern blot experiments showed that the levels of intracellular HBV single stranded DNA were not different between these clones (Figure 2D). Taken together, these results indicate that P120T strongly reduces HBsAg levels as detected by CLEIA without affecting the HBV DNA replicative capacity.
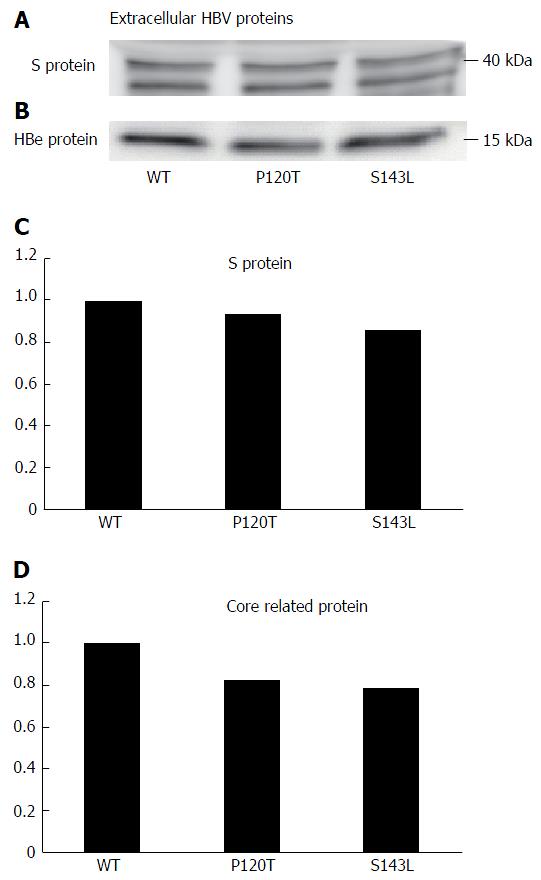
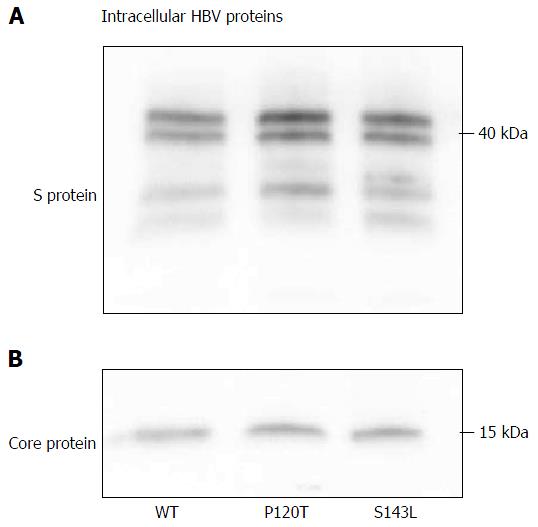
The above data suggest that the P120T mutation either prevents the secretion of HBsAg or reduces the antigenicity of HBsAg. To distinguish between these alternatives, we examined the extracellular and intracellular expression levels of denatured HBsAg by Western blot after transfecting Huh7 cell with the wild type genotype D clone, and both the P120T, and S143L mutants. As controls, we also monitored the extracellular and intracellular expression levels of the denatured HBcAg by Western blot. As shown in Figure 3, the extracellular expression levels of denatured HBsAg were not different between the wild type and mutants. The result suggests that the reduced HBsAg level detected by CLEIA in the supernatant of P120T mutant transfected cells was caused by reduced antigenicity in vitro rather than intracellular retention of the mutant HBsAg. In line with this notion, a similar amount of intracellular HBsAg was detected in cells transfected with either of the three clones (Figure 4). Collectively, these results suggest that the P120T mutation reduces the antigenicity of HBsAg in vitro.
Few studies have reported the characteristics of occult HBV infection with genotype D compared to reports of genotypes B and C. In the present study, we investigated the characteristics of occult HBV infections among patients with hematological malignancies in a population where genotype D infection is prevalent[23,24]. We detected two previously reported escape mutations P120T and S143L that were associated with occult HBV infection[25-28], and investigated the mechanism by which these mutations cause occult infection using an in vitro system.
No accurate estimate regarding the prevalence of occult HBV infections in different Egyptian cohorts has been reported, perhaps due to the high expense of PCR and low budget for scientific research in Egypt[29]. However, a high prevalence rate (17.2%; 52/303) of occult HBV was reported in Egyptian blood donors positive for anti-HBc[30]. Our results indicated that the frequency of occult HBV infection among patients with hematological malignancies was even higher. Therefore, patients with hematological malignancies might be more likely to develop occult HBV infections than those without. The highest prevalence of occult HBV was reported in patients with hepatocellular carcinoma (62.5%; 25/40) in tissue samples[8,31].
HCV infection was not uncommon in patients with occult HBV. In a similar Egyptian cohort (children with hematological malignancy), 38% (15/49) were HCV coinfected patients[32]. HCV and HBV viruses shared many risk factors and routes of transmission, and more likely parenteral antischistosomal therapy was responsible for transmission of HBV and HCV in many Egyptians[33].
The presence of anti-HBs in patients with occult HBV was in line with a recent study conducted on patients treated for TB, which showed that half of patients with occult HBV infection had both anti-HBc and anti-HBs[34]. Occult hepatitis B was also detected in blood units from healthy volunteer blood donors showing an adequate level of anti-HBs[30]. Possible explanation for the presence of such cases (anti-HBc+/anti-HBs+/HBV DNA+) in HBsAg- individuals is that anti-HBs antibody is poorly neutralizing due to loss of recognition, allowing these mutant viruses to escape neutralization even when antibody is present at protective levels[35-37].
Isolated anti-HBc positive individuals with undetectable anti-HBs or HBV DNA were observed in the studied cohort. In a cohort of blood donors with anti-HBc only, the observation of the anti-HBs kinetics after administration of Engerix HBV vaccine allowed the discrimination between the naïve HBV infection with likely false positive anti-HBc, subjects with resolved HBV infection, and subjects with persistent low level replication[38]. Coffin et al[39] believed that the long-term presence of anticore antibodies alone is a consequence of sustained restimulation of the immune system by virus nucleocapsid produced during low-level hepadnaviral assembly.
Several studies reported the higher prevalence of mutations in the MHR region of the S gene product in HBV isolates retrieved from occult HBV cases compared to overt HBV cases[40]. In addition, Martin et al[41] reported the potential virological differences between chronic HBV and occult HBV in HIV coinfected individuals and that positive selection immune pressures are acting against Pre-S and S regions in occult HBV, resulting in mutations that may adversely affect the production and/or detection of HBsAg. In concordance with previous findings, the high prevalence of mutations was observed in another immunocompromised group (patients with hematological malignancy).
The HBV complete genome was retrieved from 6 patients, and sequence and phylogenic analysis revealed that they were of genotype D1, the prevalent HBV genotype in Egypt regardless of the studied cohort. Two amino acid substitutions, P120T and S143L, were found in HBsAg.
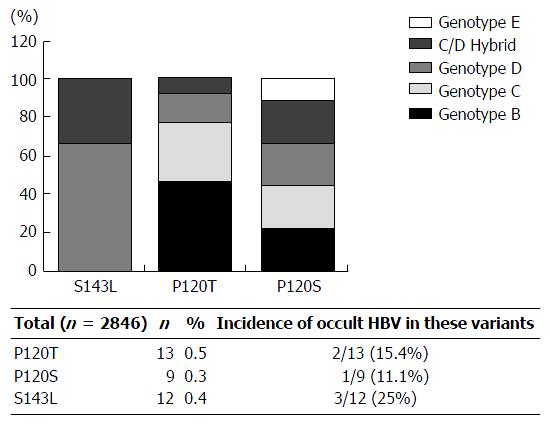
Inspection of 2846 HBV strains currently available in DDBJ, EMBL and GenBank genetic databases indicated that amino acid substitutions 120T and 143L are present in 0.5% (13/2846) and 0.4% (9/2846), respectively, of the total isolates examined. The 143L substitution is common in genotype D strains, while 120T is common in genotypes B, C, D, C/D hybrid, and E. Collectively, these results suggest that 143L specifically occurs in genotype D, while 120T is a genotype independent substitution (Figure 5). Interestingly, 15% (2/13) and 25% (3/12) of cases infected with T/S120 and 143L variants were occult HBV infection, respectively (Figure 5). In contrast, 129R was mainly found in the short sequences (low HBV DNA) in this study as well as genotypes A, B and C isolates as previously reported[26,42,43]. Taking previous data together, further in vitro analysis of 120T and 143L was applied to the current study.
Both mutations are located in the MHR that extends from amino acids 99 to 169 of the HBsAg. The MHR is exposed on the surface of the antigen and is the principal binding site of anti-HBs following natural infection, and after immunization[44]. In many cases, mutations in the MHR are frequently associated with occult HBV infection[45,46], because they can change immunogenicity and render the HBsAg unrecognizable by commercially available detection assays. Clones with such mutations are often referred to as “diagnostic-escape” mutants[47]. Consistent with this, the results obtained by in vitro experiments showed that extracellular HBsAg with P120T was less detectable than wild type HBsAg or the mutant with S143L by CLEIA. The lower extracellular HBsAg levels observed were not due to reduced HBV replication or the intracellular retention of HBsAg. Rather, the P120T mutation appeared to reduce the antigenicity of HBsAg. A similar result was also reported by Yen et al[48], but their study did not address the impact of 120T mutation on HBV replication or HBsAg secretion. In vitro studies described the impairment of virion and/or S protein secretion in both Huh7 cells and hydrodynamic injected mice by Q129R MHR mutation[49].
Our previous study demonstrated that HBV reactivation frequently occurs among patients with hematological malignancies under chemotherapy[50]. One important risk factor for the development of HBV reactivation in this critical group is the presence of occult HBV infection[50,51]. The present data suggest that patients with hematological malignancies should be screened and closely monitored for anti-HBc and HBV DNA.
In conclusion, the prevalence of occult hepatitis B was detected in patients with hematological malignancies in South Egypt in association with two mutations in the HBsAg, P120T and S143L. Neither of these mutations affected the replication activity, virion or S protein secretion but one of the mutations, P120T, interfered with detection by current commercial assays probably by inducing a conformational change. Our results highlight a challenge for detecting occult strains in developing countries.
Occult hepatitis B (OBI) is defined by the presence of hepatitis B virus (HBV) DNA in the serum or the liver in the absence of hepatitis B surface antigen (HBsAg) with or without anti-HBc or antibodies to HBsAg. Prevalence of OBI is different according to the endemicity of HBV. OBI is implicated in different clinical contexts including the progression of liver disease, the development of hepatocellular carcinoma, the risk for HBV reactivation, and the transmission of HBV infection. Both viral and host factors are implicated in the pathogenesis of OBI. Major hydrophilic region in genomic HBV extending from aa99 to aa169, clustered with a highly conformational epitope, is critical to the antigenicity of HBsAg and may affect the diagnosis of HBV in HBV screening tests.
In a cohort of 165 patients with hematological malignancies receiving cancer chemotherapy who were negative for HBsAg, 54 patients (35.5%) were serologically positive for antibodies to HBV core (anti-HBc). Occult HBV infection was determined in 42.6% (23/54) of patients with hematological malignancies who were negative for HBsAg, but positive for anti-HBc. The complete genome of HBV was successfully obtained from 6 cases with occult HBV infection and all were of genotype D subtype D1. Two prevalent amino acid substitutions P120T and S143L were associated with OBI in the present study. In vitro analysis of these two amino acid mutations revealed that P120T mutation reduces the antigenicity of HBsAg in vitro without affecting the HBV DNA replication capacity.
This study records the high prevalence of occult HBV infection in patients with hematological malignancies. Occult HBV infection is associated with 120T and 143L mutations, and 120T mutation might impair HBsAg detection by changing its conformation.
The study strongly recommends mandatory serological screening for anti-HBc and HBV DNA in patients with hematological malignancies.
In this manuscript, Abeer Elkady et al argue characteristics of escape mutations from occult hepatitis B virus infected patients with hematological malignancies in Egypt. The authors concluded that occult HBV infection is associated with P120T and S143L mutations and 120T mutation impairs the detection of HBsAg by chemiluminescence enzyme immunoassay. The aim of this study might be interesting and important.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Ahmed Said ZN, Matsui T, Rodriguez-Frias F S- Editor: Gong ZM L- Editor: Wang TQ E- Editor: Li D
| 1. | Raimondo G, Pollicino T, Cacciola I, Squadrito G. Occult hepatitis B virus infection. J Hepatol. 2007;46:160-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 386] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 2. | Fernandez-Rodriguez CM, Gutierrez ML, Lledó JL, Casas ML. Influence of occult hepatitis B virus infection in chronic hepatitis C outcomes. World J Gastroenterol. 2011;17:1558-1562. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (2)] |
| 3. | Larrubia JR. Occult hepatitis B virus infection: a complex entity with relevant clinical implications. World J Gastroenterol. 2011;17:1529-1530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 14] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Berasain C, Betés M, Panizo A, Ruiz J, Herrero JI, Civeira MP, Prieto J. Pathological and virological findings in patients with persistent hypertransaminasaemia of unknown aetiology. Gut. 2000;47:429-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 87] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 5. | Kitab B, Ezzikouri S, Alaoui R, Nadir S, Badre W, Trepo C, Chemin I, Benjelloun S. Occult HBV infection in Morocco: from chronic hepatitis to hepatocellular carcinoma. Liver Int. 2014;34:e144-e150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 6. | Lau GK. Hepatitis B reactivation after chemotherapy: two decades of clinical research. Hepatol Int. 2008;2:152-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 87] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 7. | Pei R, Grund S, Verheyen J, Esser S, Chen X, Lu M. Spontaneous reactivation of hepatitis B virus replication in an HIV coinfected patient with isolated anti-Hepatitis B core antibodies. Virol J. 2014;11:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 8. | Arababadi MK, Hassanshahi G, Pourfathollah AA, Zarandi ER, Kennedy D. Post-transfusion occult hepatitis B (OBI): a global challenge for blood recipients and health authorities. Hepat Mon. 2011;11:714-718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Ferraro D, Pizzillo P, Urone N, Iannitto E, Craxi A, Di Stefano R. Viral sequence analysis of occult HBV infection and its reactivation in immunosuppressed patients. J Biol Regul Homeost Agents. 2012;26:457-465. [PubMed] |
| 10. | Pondé RA. Molecular mechanisms underlying HBsAg negativity in occult HBV infection. Eur J Clin Microbiol Infect Dis. 2015;34:1709-1731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Kim H, Lee SA, Won YS, Lee H, Kim BJ. Occult infection related hepatitis B surface antigen variants showing lowered secretion capacity. World J Gastroenterol. 2015;21:1794-1803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 12. | Biswas R. Post-transfusion non-A, non-B hepatitis: significance of raised ALT and anti-HBc in blood donors. Vox Sang. 1989;56:63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 13. | Larralde O, Dow B, Jarvis L, Davidson F, Petrik J. Hepatitis B escape mutants in Scottish blood donors. Med Microbiol Immunol. 2013;202:207-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Martin CM, Welge JA, Rouster SD, Shata MT, Sherman KE, Blackard JT. Mutations associated with occult hepatitis B virus infection result in decreased surface antigen expression in vitro. J Viral Hepat. 2012;19:716-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 15. | Sugiyama M, Tanaka Y, Kurbanov F, Maruyama I, Shimada T, Takahashi S, Shirai T, Hino K, Sakaida I, Mizokami M. Direct cytopathic effects of particular hepatitis B virus genotypes in severe combined immunodeficiency transgenic with urokinase-type plasminogen activator mouse with human hepatocytes. Gastroenterology. 2009;136:652-62.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 16. | Sugauchi F, Orito E, Ichida T, Kato H, Sakugawa H, Kakumu S, Ishida T, Chutaputti A, Lai CL, Ueda R. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J Virol. 2002;76:5985-5992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 233] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 17. | Shin-I T, Tanaka Y, Tateno Y, Mizokami M. Development and public release of a comprehensive hepatitis virus database. Hepatol Res. 2008;38:234-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 18. | Sugiyama M, Tanaka Y, Kato T, Orito E, Ito K, Acharya SK, Gish RG, Kramvis A, Shimada T, Izumi N. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology. 2006;44:915-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 244] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 19. | Ozasa A, Tanaka Y, Orito E, Sugiyama M, Kang JH, Hige S, Kuramitsu T, Suzuki K, Tanaka E, Okada S. Influence of genotypes and precore mutations on fulminant or chronic outcome of acute hepatitis B virus infection. Hepatology. 2006;44:326-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 188] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 20. | Fujiwara K, Tanaka Y, Paulon E, Orito E, Sugiyama M, Ito K, Ueda R, Mizokami M, Naoumov NV. Novel type of hepatitis B virus mutation: replacement mutation involving a hepatocyte nuclear factor 1 binding site tandem repeat in chronic hepatitis B virus genotype E. J Virol. 2005;79:14404-14410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 21. | Hayashi S, Murakami S, Omagari K, Matsui T, Iio E, Isogawa M, Watanabe T, Karino Y, Tanaka Y. Characterization of novel entecavir resistance mutations. J Hepatol. 2015;63:546-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 22. | Ito K, Qin Y, Guarnieri M, Garcia T, Kwei K, Mizokami M, Zhang J, Li J, Wands JR, Tong S. Impairment of hepatitis B virus virion secretion by single-amino-acid substitutions in the small envelope protein and rescue by a novel glycosylation site. J Virol. 2010;84:12850-12861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 92] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 23. | Saudy N, Sugauchi F, Tanaka Y, Suzuki S, Aal AA, Zaid MA, Agha S, Mizokami M. Genotypes and phylogenetic characterization of hepatitis B and delta viruses in Egypt. J Med Virol. 2003;70:529-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 84] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 24. | Ragheb M, Elkady A, Tanaka Y, Murakami S, Attia FM, Hassan AA, Hassan MF, Shedid MM, Abdel Reheem HB, Khan A. Multiple intra-familial transmission patterns of hepatitis B virus genotype D in north-eastern Egypt. J Med Virol. 2012;84:587-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Bhattacharya S, Ijaz S, Ratnaraja N, Smith S, Osman H, Boxall E. Hepatitis B virus reactivation in a large haemodialysis unit: virological and infection control issues. J Clin Virol. 2009;46:101-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 26. | Darmawan E, Turyadi KE, Nursanty NK, Thedja MD, Muljono DH. Seroepidemiology and occult hepatitis B virus infection in young adults in Banjarmasin, Indonesia. J Med Virol. 2015;87:199-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 27. | Svicher V, Cento V, Bernassola M, Neumann-Fraune M, Van Hemert F, Chen M, Salpini R, Liu C, Longo R, Visca M. Novel HBsAg markers tightly correlate with occult HBV infection and strongly affect HBsAg detection. Antiviral Res. 2012;93:86-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 28. | Su H, Zhang Y, Xu D, Wang B, Zhang L, Li D, Xiao D, Li F, Zhang J, Yan Y. Occult hepatitis B virus infection in anti-HBs-positive infants born to HBsAg-positive mothers in China. PLoS One. 2013;8:e70768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 29. | Emara MH. Occult hepatitis B: the Egyptian situation. Trop Gastroenterol. 2012;33:242-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 30. | Said ZN, Sayed MH, Salama II, Aboel-Magd EK, Mahmoud MH, Setouhy ME, Mouftah F, Azzab MB, Goubran H, Bassili A. Occult hepatitis B virus infection among Egyptian blood donors. World J Hepatol. 2013;5:64-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 31. | Elbahrawy A, Alaboudy A, El Moghazy W, Elwassief A, Alashker A, Abdallah AM. Occult hepatitis B virus infection in Egypt. World J Hepatol. 2015;7:1671-1678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 32. | Said ZN, El-Sayed MH, El-Bishbishi IA, El-Fouhil DF, Abdel-Rheem SE, El-Abedin MZ, Salama , II . High prevalence of occult hepatitis B in hepatitis C-infected Egyptian children with haematological disorders and malignancies. Liver Int. 2009;29:518-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 33. | El-Sherif A, Abou-Shady M, Abou-Zeid H, Elwassief A, Elbahrawy A, Ueda Y, Chiba T, Hosney AM. Antibody to hepatitis B core antigen as a screening test for occult hepatitis B virus infection in Egyptian chronic hepatitis C patients. J Gastroenterol. 2009;44:359-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 34. | Trigo C, do Brasil PE, Costa MJ, de Castro L. Occult hepatitis B virus infection: clinical implications in tuberculosis treatment. J Viral Hepat. 2016;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 35. | Oluyinka OO, Tong HV, Bui Tien S, Fagbami AH, Adekanle O, Ojurongbe O, Bock CT, Kremsner PG, Velavan TP. Occult Hepatitis B Virus Infection in Nigerian Blood Donors and Hepatitis B Virus Transmission Risks. PLoS One. 2016;10:e0131912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 36. | Levicnik-Stezinar S, Rahne-Potokar U, Candotti D, Lelie N, Allain JP. Anti-HBs positive occult hepatitis B virus carrier blood infectious in two transfusion recipients. J Hepatol. 2008;48:1022-1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 101] [Article Influence: 5.9] [Reference Citation Analysis (1)] |
| 37. | Candotti D, Grabarczyk P, Ghiazza P, Roig R, Casamitjana N, Iudicone P, Schmidt M, Bird A, Crookes R, Brojer E. Characterization of occult hepatitis B virus from blood donors carrying genotype A2 or genotype D strains. J Hepatol. 2008;49:537-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 98] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 38. | Gessoni G, Beggio S, Barin P, Favarato M, Galli C, Valverde S, Nata MB, Salvadego MM, Marchiori G. Significance of anti-HBc only in blood donors: a serological and virological study after hepatitis B vaccination. Blood Transfus. 2014;12 Suppl 1:s63-s68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 39. | Coffin CS, Pham TN, Mulrooney PM, Churchill ND, Michalak TI. Persistence of isolated antibodies to woodchuck hepatitis virus core antigen is indicative of occult infection. Hepatology. 2004;40:1053-1061. |
| 40. | Yuan Q, Ou SH, Chen CR, Ge SX, Pei B, Chen QR, Yan Q, Lin YC, Ni HY, Huang CH. Molecular characteristics of occult hepatitis B virus from blood donors in southeast China. J Clin Microbiol. 2010;48:357-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 60] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 41. | Martin CM, Welge JA, Shire NJ, Rouster SD, Shata MT, Sherman KE, Blackard JT. Genomic variability associated with the presence of occult hepatitis B virus in HIV co-infected individuals. J Viral Hepat. 2010;17:588-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 42. | Jeantet D, Chemin I, Mandrand B, Tran A, Zoulim F, Merle P, Trepo C, Kay A. Cloning and expression of surface antigens from occult chronic hepatitis B virus infections and their recognition by commercial detection assays. J Med Virol. 2004;73:508-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 43. | Takahashi K, Akahane Y, Hino K, Ohta Y, Mishiro S. Hepatitis B virus genomic sequence in the circulation of hepatocellular carcinoma patients: comparative analysis of 40 full-length isolates. Arch Virol. 1998;143:2313-2326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 129] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 44. | Chen YC, Delbrook K, Dealwis C, Mimms L, Mushahwar IK, Mandecki W. Discontinuous epitopes of hepatitis B surface antigen derived from a filamentous phage peptide library. Proc Natl Acad Sci USA. 1996;93:1997-2001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 102] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 45. | Shizuma T, Hasegawa K, Ishikawa K, Naritomi T, Iizuka A, Kanai N, Ogawa M, Torii N, Joh R, Hayashi N. Molecular analysis of antigenicity and immunogenicity of a vaccine-induced escape mutant of hepatitis B virus. J Gastroenterol. 2003;38:244-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 46. | Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman AJ, Thomas HC. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 817] [Cited by in RCA: 772] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 47. | Ireland J, Hino K, Lau GK, Cheng CC, Carman WF. Failed adoptive immunity transfer: reactivation or reinfection? J Viral Hepat. 1999;6:73-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 48. | Yen TH, Huang CC, Lin HH, Huang JY, Tian YC, Yang CW, Wu MS, Fang JT, Yu CC, Chiang YJ. Does hepatitis C virus affect the reactivation of hepatitis B virus following renal transplantation? Nephrol Dial Transplant. 2006;21:1046-1052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 49. | Huang CH, Yuan Q, Chen PJ, Zhang YL, Chen CR, Zheng QB, Yeh SH, Yu H, Xue Y, Chen YX. Influence of mutations in hepatitis B virus surface protein on viral antigenicity and phenotype in occult HBV strains from blood donors. J Hepatol. 2012;57:720-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 162] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 50. | Elkady A, Aboulfotuh S, Ali EM, Sayed D, Abdel-Aziz NM, Ali AM, Murakami S, Iijima S, Tanaka Y. Incidence and characteristics of HBV reactivation in hematological malignant patients in south Egypt. World J Gastroenterol. 2013;19:6214-6220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 20] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 51. | Sugauchi F, Tanaka Y, Kusumoto S, Matsuura K, Sugiyama M, Kurbanov F, Ueda R, Mizokami M. Virological and clinical characteristics on reactivation of occult hepatitis B in patients with hematological malignancy. J Med Virol. 2011;83:412-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |