Published online Oct 8, 2016. doi: 10.4254/wjh.v8.i28.1194
Peer-review started: May 21, 2016
First decision: July 4, 2016
Revised: July 8, 2016
Accepted: August 27, 2016
Article in press: August 29, 2016
Published online: October 8, 2016
Processing time: 131 Days and 1.5 Hours
To clarify whether Agtr1a methylation is involved in the development of nonalcoholic steatohepatitis (NASH)-related liver fibrosis in adult rats.
A choline-deficient amino acid (CDAA) diet model was employed for methylation analysis of NASH-related liver fibrosis. Agtr1a methylation levels were measured in the livers of CDAA- and control choline-sufficient amino acid (CSAA)-fed rats for 8 and 12 wk using quantitative methylation-specific PCR. Hepatic stellate cells (HSCs) were isolated by collagenase digestion of the liver, followed by centrifugation of the crude cell suspension through a density gradient. Agtr1a methylation and its gene expression were also analyzed during the activation of HSCs.
The mean levels of Agtr1a methylation in the livers of CDAA-fed rats (11.5% and 18.6% at 8 and 12 wk, respectively) tended to be higher (P = 0.06 and 0.09, respectively) than those in the livers of CSAA-fed rats (2.1% and 5.3% at 8 and 12 wk, respectively). Agtr1a was not methylated at all in quiescent HSCs, but was clearly methylated in activated HSCs (13.8%, P < 0.01). Interestingly, although Agtr1a was hypermethylated, the Agtr1a mRNA level increased up to 2.2-fold (P < 0.05) in activated HSCs compared with that in quiescent HSCs, suggesting that Agtr1a methylation did not silence its expression but instead had the potential to upregulate its expression. These findings indicate that Agtr1a methylation and its upregulation of gene expression are associated with the development of NASH-related liver fibrosis.
This is the first study to show that DNA methylation is potentially involved in the regulation of a renin-angiotensin system-related gene expression during liver fibrosis.
Core tip: We report the first study to show that Agtr1a methylation occurred during the development of nonalcoholic steatohepatitis-related liver fibrosis. Interestingly, Agtr1a gene expression was upregulated during liver fibrosis, although Agtr1a was methylated. This study demonstrates for the first time that renin-angiotensin system-related gene expression is regulated by DNA methylation during liver fibrosis. This finding raises expectations about the therapeutic application of demethylating agents for the treatment of liver fibrosis.
- Citation: Asada K, Aihara Y, Takaya H, Noguchi R, Namisaki T, Moriya K, Uejima M, Kitade M, Mashitani T, Takeda K, Kawaratani H, Okura Y, Kaji K, Douhara A, Sawada Y, Nishimura N, Seki K, Mitoro A, Yamao J, Yoshiji H. DNA methylation of angiotensin II receptor gene in nonalcoholic steatohepatitis-related liver fibrosis. World J Hepatol 2016; 8(28): 1194-1199
- URL: https://www.wjgnet.com/1948-5182/full/v8/i28/1194.htm
- DOI: https://dx.doi.org/10.4254/wjh.v8.i28.1194
Liver fibrosis is a characteristic feature of chronic liver disease regardless of the etiology. Cirrhosis is the terminal condition of chronic liver diseases, and hepatic failure due to liver cirrhosis is caused by progressive fibrosis that ultimately results in nodular regeneration with loss of function[1-3]. Considering that hepatocellular carcinoma (HCC) also develops from liver fibrosis, it is necessary to investigate the molecular mechanisms underlying liver fibrosis development to reduce the morbidity and mortality of chronic liver disease.
The renin-angiotensin system (RAS) is continually activated in patients with chronic liver diseases, such as cirrhosis[4]. Angiotensin II (AT-II), an octapeptide produced mainly via the enzymatic cleavage of angiotensin I by angiotensin I-converting enzyme, reportedly plays an important role in chronic liver disease progression. AT-II activates a series of signal transduction pathways in activated hepatic stellate cells (HSCs) by binding to the AT-II type 1 receptor (AT1-R)[5]. We previously reported that AT1-R blockers significantly attenuate experimental liver fibrosis development with the suppression of activated HSC proliferation[6-8]. However, the molecular mechanisms regulating RAS-related gene expression remain unelucidated.
Epigenetic alterations, including DNA methylation, are involved in the progression of liver fibrosis and HCC in human and animal studies[9-11]. Recently, Chen et al[12] reported that RAS-related genes, especially Agtr1a encoding rat AT1-R, are methylated in rats born to mothers fed a methyl donor-deficient diet during gestation and lactation. They showed that Agtr1a methylation can be a surrogate marker to predict susceptibility in developing nonalcoholic fatty liver disease (NAFLD) later in life. However, it is unclear whether Agtr1a methylation is associated with the development of nonalcoholic steatohepatitis (NASH)-related liver fibrosis.
Here we employed choline-deficient amino acid (CDAA)-fed rats to evaluate the importance of Agtr1a methylation in the development of NASH-related liver fibrosis. Our results demonstrate that Agtr1a methylation is potentially associated with liver fibrosis development and HSC activation.
Six-week-old male Fisher 344 rats (CLEA Japan, Inc., Osaka, Japan) were housed in a room under a controlled temperature and a 12/12-h light-dark cycle. The animals were divided into the following four experimental groups: (1) choline-sufficient amino acid diet (CSAA) for 8 wk (n = 4); (2) CSAA for 12 wk (n = 11); (3) CDAA for 8 wk (n = 10); and (4) CDAA for 12 wk (n = 12). Initially, sample sizes for group (1)-(5) were 5, 12, 10, and 12, respectively, but two animals (one for CSAA-diet for 8 wk and the other for CSAA-diet for 12 wk) were dropped out because of entry in another experiment. All animal procedures were performed in accordance with standard protocols and following the standard recommendations for the appropriate care and use of laboratory animals. This study was approved by the animal experiment ethical committee at the Nara Medical University (protocol number: 9354).
HSCs were isolated by the collagenase digestion of the liver of a 6-week-old male Fisher 344 rat using a perfusion system, followed by the centrifugation of the crude cell suspension through a density gradient, as described previously[13]. Genomic DNA and total RNA were isolated from freshly isolated HSCs in a quiescent state. Thereafter, HSCs were activated in a culture on a plastic dish for 5 d.
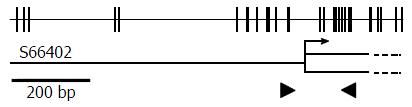
Genomic DNA was isolated using a DNeasy® Blood and Tissue Kit (Qiagen, Hilden, Germany). Fully methylated control DNA was prepared by methylating genomic DNA with SssI methylase (New England Biolabs, Beverly, MA), and completely unmethylated control DNA was purchased from EpigenDx (Hopkinton, MA). Bisulfite modification was performed using an EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA). Agtr1a genomic structure is illustrated in Figure 1. An aliquot of 1 μL was used for quantitative real-time methylation-specific PCR (qMSP) with primers specific to a methylated sequence of Agtr1a (forward 5′-GGT TGG AAT TTG TAG AGT AGC GAC-3′, reverse 5′-CAA CGC TAA TAC CGA CCT CG-3′) and to a B2 repeat sequence, regardless of the methylation status, as demonstrated in a previous report[14].
qMSP was performed by real-time PCR using a Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA) and a StepOnePlusTM Real-Time PCR® (Thermo Fisher Scientific, Waltham, MA). The methylation level was calculated as the methylation percentage obtained as follows: {[number of DNA molecules methylated at a target CpG island (CGI) in a sample]/(number of B2 repeats in the sample)}/[(number of DNA molecules methylated at the target CGI in completely methylated control DNA)/(number of B2 repeats in the completely methylated control DNA)] × 100, as described previously[15].
Total RNA was extracted using an RNeasy® Mini Kit (Qiagen, Hilden, Germany). cDNA was synthesized from 1 μg of total RNA using a High Capacity RNA to cDNA Master Mix (Thermo Fisher Scientific, Waltham, MA). Agtr1a mRNA level was measured by quantitative PCR using the StepOnePlusTM Real-Time PCR® (Thermo Fisher Scientific, Waltham, MA). Primer sequences for Agtr1a and for Ppia were reported previously[14,16]. The number of Agtr1a cDNA molecules was normalized to that of Ppia cDNA molecules.
The difference in mean methylation levels was analyzed using Welch’s t-test. The results were considered significant with a P value of < 0.05.
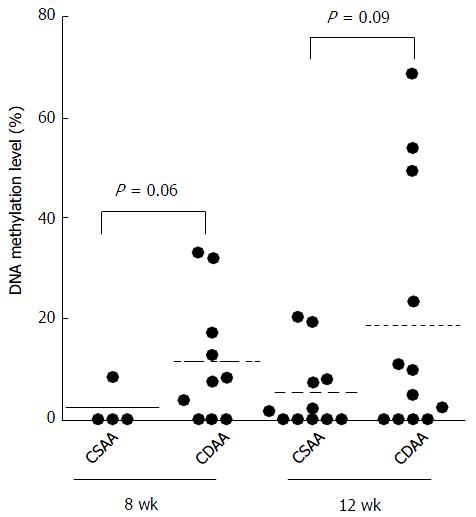
To evaluate the status of Agtr1a methylation in the whole liver, we performed qMSP using the liver samples of CSAA- and CDAA-fed rats after the two feeding periods, 8 and 12 wk. The mean levels of Agtr1a methylation in the livers of CDAA-fed rats were 11.5% and 18.6% at 8 and 12 wk, respectively, whereas those in the livers of CSAA-fed rats were 2.1% and 5.3% at 8 and 12 wk, respectively. These findings suggested that the levels of Agtr1a methylation in the livers of CDAA-fed rats tended to be higher than those in the livers of CSAA-fed rats at 8 and 12 wk (P = 0.06 and 0.09, respectively; Figure 2).
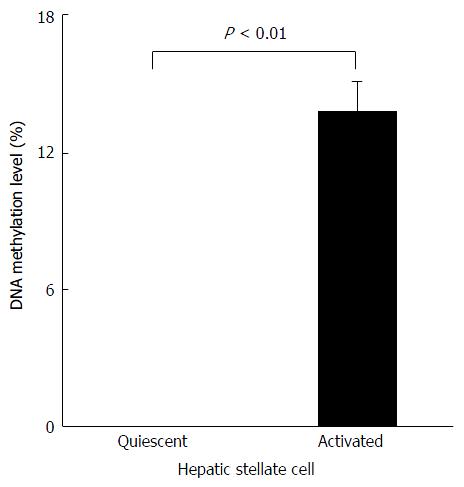
Next, we evaluated the level of Agtr1a methylation during HSC activation in vitro. We found that Agtr1a methylation was not detected at all in quiescent HSCs, but was clearly observed in activated HSCs (13.8%, P < 0.01; Figure 3). Taken together with the in vivo results, our findings indicate that Agtr1a is hypermethylated in accordance with the development of NASH-related liver fibrosis.
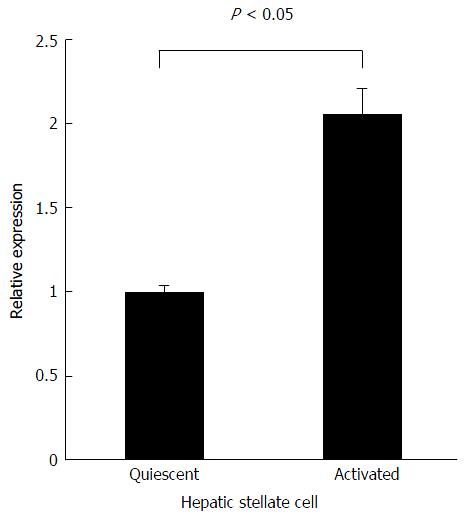
To address the contribution of Agtr1a methylation to its gene expression, we performed quantitative real-time reverse transcription PCR using quiescent and activated HSCs. Agtr1a expression was observed in quiescent HSCs in which Agtr1a was unmethylated. Unexpectedly, Agtr1a expression increased up to 2.2-fold (P < 0.05) in the activated HSCs compared with that in quiescent HSCs, although Agtr1a was methylated (Figure 4). Interestingly, in contrast to the general relationship between promoter CGIs and gene expression, Agtr1a methylation did not silence its expression but instead had the potential to upregulate its expression.
In this study, we found that Agtr1a methylation occurred during the development of NASH-related liver fibrosis. Agtr1a, which encodes rat AT1-R, the receptor for AT-II, is an important factor in liver fibrosis development[17,18]. Our previous reports demonstrated that both AT-II and AT1-R gene expressions were upregulated during fibrosis development in rat liver, and the blockage of AT-II/AT1-R signaling could attenuate liver fibrosis[6-8]. Considering that Agtr1a methylation upregulates its gene expression, Agtr1a demethylation can suppress liver fibrosis.
Agtr1a methylation was first demonstrated in the liver of rats born to mothers fed a methyl donor-deficient diet during gestation and lactation, and it was reported that rat pups with Agtr1a methylation have a high risk of developing NAFLD[12]. Epigenetics derived from mother-pup interaction is a prominent research field, and epigenetic susceptibility to phenotypes and diseases, such as yellow coat color, stress response, and breast cancer in offspring, has been identified[19-21]. However, few studies have focused on whether these epigenetic changes responsible for susceptibility to particular diseases occur when the diseases actually develop in adults. Here we found that Agtr1a methylation, associated with susceptibility to NAFLD in pups, occurs in liver fibrosis development in adult NASH model rats.
As an experimental NASH model, we employed the CDAA model in this study. In the CDAA model, liver fibrosis develops at 8 wk and severely progresses at 12 wk[22,23]. This model has an advantage of histological progression of liver fibrosis, which is very similar to human NASH. However, there are critical disadvantages of this model. For examples, obesity, glucose intolerance, and insulin resistance, which are common features in human NASH, are not observed in this model. It remains to be elucidated whether Agtr1a methylation is induced in other experimental NASH models.
In CDAA model, Agtr1a methylation in the livers of CDAA-fed rats tended to be higher than that in the livers of CSAA-fed rats, but it was not statistically significant. We consider that methylation levels are highly variable in each diet group and the difference between CDAA- and CSAA-fed rats appears to be small. This variability depends on individual differences in rats and tissue heterogeneity in each sample, but both of them are hardly avoided. On the other hand, in HSC analysis, Agtr1a methylation and upregulation was clearly observed. Even in the CDAA model, it would be better to isolate HSC from the livers of CDAA-fed rats to obtain clear methylation changes.
Agtr1a hypermethylation was associated with Agtr1a upregulation. As for the promoter CGI, hypermethylation is generally considered to be strongly associated with gene silencing[24]. On the other hand, in the case of a CGI at the gene body, hypermethylation occasionally contributes to overexpression[25]. In the Agtr1a gene, 5′-CGI was not located at the promoter region but was just downstream of the transcription initiation site (Figure 1), which might contribute to gene overexpression. It is hoped that the mechanism by which gene body methylation induces overexpression can be demonstrated.
In conclusion, this study demonstrates for the first time that RAS-related gene expression is regulated by DNA methylation during liver fibrosis. This finding raises expectations about the therapeutic application of demethylating agents for the treatment of liver fibrosis.
The renin-angiotensin system (RAS) plays a crucial role in the development of liver fibrosis. Among the RAS-related genes, the methylation of Agtr1a, the rat Angiotensin II type 1 receptor gene, is a potential risk marker for the development of nonalcoholic fatty liver disease in rat pups. However, it remains to be elucidated whether Agtr1a methylation occurs in liver fibrosis development in adult rats with nonalcoholic steatohepatitis (NASH).
Epigenetics derived from mother-pup interaction is a prominent research field. However, few studies have focused on whether these epigenetic changes responsible for susceptibility to particular diseases occur when the diseases actually develop in adults.
This study demonstrates for the first time that the expression of Agtr1a, a RAS-related gene, is regulated by DNA methylation during liver fibrosis.
The authors finding raises expectations about the therapeutic application of demethylating agents for the treatment of liver fibrosis.
Epigenetics refers to heritable marks regulating tissue-specific gene expression without changes in the DNA sequence. Prominent epigenetic marks consist of DNA methylation and histone modifications. Aberrant epigenetic changes are involved in various diseases, including cancer.
This manuscript addresses the role of DNA methylation of angiotensin II receptor in fibrosis development in a rat model of NASH. The study is original and well designed.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): E
P- Reviewer: Bridle KR, Hamidi C, Safer AM S- Editor: Qi Y L- Editor: A E- Editor: Li D
| 1. | Albanis E, Friedman SL. Hepatic fibrosis. Pathogenesis and principles of therapy. Clin Liver Dis. 2001;5:315-334, v-vi. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 159] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 2. | Olaso E, Friedman SL. Molecular regulation of hepatic fibrogenesis. J Hepatol. 1998;29:836-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 186] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 3. | Friedman SL. Liver fibrosis -- from bench to bedside. J Hepatol. 2003;38 Suppl 1:S38-S53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1199] [Cited by in RCA: 1294] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 4. | Helmy A, Jalan R, Newby DE, Hayes PC, Webb DJ. Role of angiotensin II in regulation of basal and sympathetically stimulated vascular tone in early and advanced cirrhosis. Gastroenterology. 2000;118:565-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 62] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Bataller R, Ginès P, Nicolás JM, Görbig MN, Garcia-Ramallo E, Gasull X, Bosch J, Arroyo V, Rodés J. Angiotensin II induces contraction and proliferation of human hepatic stellate cells. Gastroenterology. 2000;118:1149-1156. [PubMed] |
| 6. | Yoshiji H, Kuriyama S, Yoshii J, Ikenaka Y, Noguchi R, Nakatani T, Tsujinoue H, Fukui H. Angiotensin-II type 1 receptor interaction is a major regulator for liver fibrosis development in rats. Hepatology. 2001;34:745-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 309] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 7. | Yoshiji H, Kuriyama S, Fukui H. Blockade of renin-angiotensin system in antifibrotic therapy. J Gastroenterol Hepatol. 2007;22 Suppl 1:S93-S95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 8. | Yoshiji H, Noguchi R, Ikenaka Y, Namisaki T, Kitade M, Kaji K, Shirai Y, Yoshii J, Yanase K, Yamazaki M. Losartan, an angiotensin-II type 1 receptor blocker, attenuates the liver fibrosis development of non-alcoholic steatohepatitis in the rat. BMC Res Notes. 2009;2:70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 9. | Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis--A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 196] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 10. | Arai E, Ushijima S, Gotoh M, Ojima H, Kosuge T, Hosoda F, Shibata T, Kondo T, Yokoi S, Imoto I. Genome-wide DNA methylation profiles in liver tissue at the precancerous stage and in hepatocellular carcinoma. Int J Cancer. 2009;125:2854-2862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 11. | Asada K, Kotake Y, Asada R, Saunders D, Broyles RH, Towner RA, Fukui H, Floyd RA. LINE-1 hypomethylation in a choline-deficiency-induced liver cancer in rats: dependence on feeding period. J Biomed Biotechnol. 2006;2006:17142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Chen G, Broséus J, Hergalant S, Donnart A, Chevalier C, Bolaños-Jiménez F, Guéant JL, Houlgatte R. Identification of master genes involved in liver key functions through transcriptomics and epigenomics of methyl donor deficiency in rat: relevance to nonalcoholic liver disease. Mol Nutr Food Res. 2015;59:293-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Yoshiji H, Kuriyama S, Yoshii J, Ikenaka Y, Noguchi R, Nakatani T, Tsujinoue H, Yanase K, Namisaki T, Imazu H. Tissue inhibitor of metalloproteinases-1 attenuates spontaneous liver fibrosis resolution in the transgenic mouse. Hepatology. 2002;36:850-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 172] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 14. | Hattori N, Okochi-Takada E, Kikuyama M, Wakabayashi M, Yamashita S, Ushijima T. Methylation silencing of angiopoietin-like 4 in rat and human mammary carcinomas. Cancer Sci. 2011;102:1337-1343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, Ichinose M, Tatematsu M, Ushijima T. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430-1440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 311] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 16. | Vaswani K, Chan HW, Verma P, Dekker Nitert M, Peiris HN, Wood-Bradley RJ, Armitage JA, Rice GE, Mitchell MD. The rat placental renin-angiotensin system - a gestational gene expression study. Reprod Biol Endocrinol. 2015;13:89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Granzow M, Schierwagen R, Klein S, Kowallick B, Huss S, Linhart M, Mazar IG, Görtzen J, Vogt A, Schildberg FA. Angiotensin-II type 1 receptor-mediated Janus kinase 2 activation induces liver fibrosis. Hepatology. 2014;60:334-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 108] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 18. | Rong X, Li Y, Ebihara K, Zhao M, Naowaboot J, Kusakabe T, Kuwahara K, Murray M, Nakao K. Angiotensin II type 1 receptor-independent beneficial effects of telmisartan on dietary-induced obesity, insulin resistance and fatty liver in mice. Diabetologia. 2010;53:1727-1731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293-5300. [PubMed] |
| 20. | Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4264] [Cited by in RCA: 3785] [Article Influence: 180.2] [Reference Citation Analysis (0)] |
| 21. | Govindarajah V, Leung YK, Ying J, Gear R, Bornschein RL, Medvedovic M, Ho SM. In utero exposure of rats to high-fat diets perturbs gene expression profiles and cancer susceptibility of prepubertal mammary glands. J Nutr Biochem. 2016;29:73-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 22. | Douhara A, Moriya K, Yoshiji H, Noguchi R, Namisaki T, Kitade M, Kaji K, Aihara Y, Nishimura N, Takeda K. Reduction of endotoxin attenuates liver fibrosis through suppression of hepatic stellate cell activation and remission of intestinal permeability in a rat non-alcoholic steatohepatitis model. Mol Med Rep. 2015;11:1693-1700. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 23. | Aihara Y, Yoshiji H, Noguchi R, Kaji K, Namisaki T, Shirai Y, Douhara A, Moriya K, Kawaratani H, Fukui H. Direct renin inhibitor, aliskiren, attenuates the progression of non-alcoholic steatohepatitis in the rat model. Hepatol Res. 2013;43:1241-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 24. | Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2107] [Cited by in RCA: 2226] [Article Influence: 159.0] [Reference Citation Analysis (0)] |
| 25. | Kulis M, Queirós AC, Beekman R, Martín-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta. 2013;1829:1161-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 165] [Article Influence: 13.8] [Reference Citation Analysis (0)] |