Published online Sep 28, 2016. doi: 10.4254/wjh.v8.i27.1128
Peer-review started: March 21, 2016
First decision: April 19, 2016
Revised: May 6, 2016
Accepted: July 29, 2016
Article in press: August 1, 2016
Published online: September 28, 2016
Processing time: 188 Days and 6.7 Hours
The intestinal microbiome (IM) is altered in patients with cirrhosis, and emerging literature suggests that this impacts on the development of complications. The PubMed database was searched from January 2000 to May 2015 for studies and review articles on the composition, pathophysiologic effects and therapeutic modulation of the IM in cirrhosis. The following combination of relevant text words and MeSH terms were used, namely intestinal microbiome, microbiota, or dysbiosis, and cirrhosis, encephalopathy, spontaneous bacterial peritonitis, hepatorenal syndrome, variceal bleeding, hepatopulmonary syndrome, portopulmonary hypertension and hepatocellular carcinoma. The search results were evaluated for pertinence to the subject of IM and cirrhosis, as well as for quality of study design. The IM in cirrhosis is characterized by a decreased proportion of Bacteroides and Lactobacilli, and an increased proportion of Enterobacteriaceae compared to healthy controls. Except for alcoholic cirrhosis, the composition of the IM in cirrhosis is not affected by the etiology of the liver disease. The percentage of Enterobacteriaceae increases with worsening liver disease severity and decompensation and is associated with bacteremia, spontaneous bacterial peritonitis and hepatic encephalopathy. Lactulose, rifaximin and Lactobacillus-containing probiotics have been shown to partially reverse the cirrhosis associated enteric dysbiosis, in conjunction with improvement in encephalopathy. The IM is altered in cirrhosis, and this may contribute to the development of complications associated with end-stage liver disease. Therapies such as lactulose, rifaximin and probiotics may, at least partially, reverse the cirrhosis-associated changes in the IM. This, in turn, may prevent or alleviate the severity of complications.
Core tip: There has recently been an increasing understanding of the importance of the intestinal microbiome (IM) in the physiology of cirrhosis and its complications. Novel sequencing techniques have enabled a better characterization of the bacteria in the IM of patients with cirrhosis, and how this differs from the microbiome in a healthy individual. Additionally, therapeutics for enteric dysbiosis in patients with cirrhosis have been studied, and have shown promise in reducing the morbidity of complications in cirrhosis. In this review, we will critically review the literature on characterization of the IM in cirrhosis, its role in complications, and the evidence for strategies to address enteric dysbiosis.
- Citation: Bhat M, Arendt BM, Bhat V, Renner EL, Humar A, Allard JP. Implication of the intestinal microbiome in complications of cirrhosis. World J Hepatol 2016; 8(27): 1128-1136
- URL: https://www.wjgnet.com/1948-5182/full/v8/i27/1128.htm
- DOI: https://dx.doi.org/10.4254/wjh.v8.i27.1128
Emerging literature has demonstrated that the intestinal microbiome (IM) plays an important role in health and disease. The intestine of a healthy adult harbors 100 trillion intestinal bacteria, and at least 500 different species have been identified with novel molecular biology techniques that allow for sequencing of whole genomes of the IM[1,2]. The healthy adult IM consists principally of Bacteroides and Firmicutes, which together comprise over 90% of the bacteria present in the colon[3]. The Bacteroides are Gram-negative, anaerobic, non-spore-forming bacteria, and especially produce carbohydrate-degrading enzymes, whereas the Firmicutes are Gram-positive, anaerobic, spore-forming bacteria, that ferment simple sugars leading to the production of short-chain fatty acids such as butyrate, acetate and propionate[4]. The concentrations of bacteria progressively increase from the proximal to the distal digestive tract, from a maximum of 103 bacteria/mL in the stomach to 1012 bacteria/mL in the colon[5]. The IM has various functions that affect biochemical, metabolic and physiologic processes both within the intestine and elsewhere in the body[6]. These physiologic functions include the digestion of nutrients, with bacterial disaccharidases transforming unabsorbed dietary sugars to short chain fatty acids (SCFA)[7]. These SCFA can be used as a source of energy by the human body, as they are absorbed through the colon. In addition, butyrate and acetate can be a source of fuel for the enterocytes[8], affect lipid metabolism[9], and have anti-inflammatory effects[10]. Intestinal bacteria can also produce vitamins such as folate and vitamin K[11,12], which are absorbed into the bloodstream[12]. Additionally, the presence of the physiologic IM within the intestinal milieu prevents colonization by pathogenic bacteria and decreases intestinal permeability[8]. Crosstalk between bacteria and enterocytes via binding sites and Toll-like receptors help distinguish commensal from pathogenic bacteria[13]. The microbiota then generate an immune response to pathogenic bacteria, and enable oral tolerance by preventing a reaction to dietary protein antigens. Many endogenous bacteria can produce bacteriocins that hinder replication of pathogenic bacterial species[14]. Additionally, commensal microbiota have been shown to promote regulatory T cell function[15] and maturation of natural killer T cells[16]. Finally, many medications including digoxin, opiates, hormones and various antibiotics are transformed into their active forms through intestinal bacterial metabolism. Bacterial deconjugation of glucuronide, sulfate and cysteine conjugates decreases the polarity of drugs, and enables enterohepatic circulation, reabsorption, and prolonged retention[17]. One prime example of a compound whose bacterial metabolism is essential to its activity is sulfasalazine, which is broken down into 5-aminosalicylic acid and sulfapyridine[18]. Additionally, the effects of anticancer immunotherapy can be modulated by the intestinal microbiota. The antitumor effects of CTLA4 blockade were shown to be dependent on specific Bacteroides species[19].
Bacterial growth and functions may be affected by several physiologic and anatomic conditions in the GI tract such as peristalsis (may inhibit mucosal attachment of ingested bacteria), the presence of gastric acid and bile (toxic effects), the presence of proteolytic enzymes (degradation of bacteria), the intestinal mucus layer (trapping of bacteria), the ileocecal valve preventing retrograde bacterial translocation[20,21], and secretory IgA inhibiting bacterial proliferation[22]. Changes in small intestinal and colonic motility, gastric acid secretion, bile flow/composition, and the intestinal innate immune response can impact bacterial composition and lead to overgrowth[23]. In addition, external factors such as diet[24], antibiotic use[25] and other environmental factors[26] affect IM composition.
IM composition can also be affected by disease states and vice versa, as shown in various types of autoimmune, metabolic and malignant conditions including colon and gynecologic cancers[3,27-30]. Intestinal microbial dysbiosis, with both qualitative and quantitative changes in bacterial species, has also been associated with the development of obesity[31], diabetes[32], metabolic syndrome[33] and inflammatory bowel disease[34]. In relation to liver disease, recent studies have reported differences in the IM associated with non-alcoholic fatty liver disease (NAFLD)[35,36], alcoholic liver disease[37] and liver cancer[38]. The IM composition in cirrhosis has been shown to be associated with the development of complications, particularly spontaneous bacterial peritonitis and encephalopathy. The goal of this review is to highlight the unique composition of the IM in cirrhosis, its underlying pathophysiology, and its association with clinical manifestations and complications of cirrhosis. Additionally, we will review therapeutic strategies in cirrhosis aimed at restoring a healthy microbiome and at reducing complications.
The PubMed database was searched from 2000 to May 2015 for studies on the causes, outcomes and modulation of the IM in cirrhosis. The following combination of relevant text words and MeSH terms were used: “IM”, microbiome, microbiota, or dysbiosis, and cirrhosis, encephalopathy, spontaneous bacterial peritonitis, hepatorenal syndrome, variceal bleeding, hepatopulmonary syndrome, portopulmonary hypertension and Hepatocellular carcinoma (HCC). Our search included both original and review articles as well as letters to the editor. We (Mamatha Bhat and Venkat Bhat) obtained 369 abstracts, manually searching the abstracts for pertinence to the subject of IM and cirrhosis. This resulted in 46 entries that provided information on the etiology, pathophysiology, characterization of the IM, and management of enteric dysbiosis in cirrhosis. Table 1 summarizes the results of our systematic review.
| Patient population | Changes in IM |
| Healthy patients | Comprised principally of Bacteroides and Firmicutes (over 90% of IM)[3] |
| Compensated cirrhosis | Higher Enterobacteriaceae, Staphylococcaeae, and Enterococcaceae[53,55,56] |
| Decreased Lachnospiraceae, Ruminococcaceae, Clostridiales XIV, Bacteroides, Faecalibacterium prausnitzii and Coprococcus comes[45,53,55,56] | |
| Alcoholic cirrhosis | Higher Enterobacteriaceae and endotoxemia compared to other cirrhosis[46] |
| Decompensated cirrhosis | Enterobacteriaceae species correlated with increasing MELD score, Ruminococcaceae species associated with lower MELD scores[56] |
| Overt hepatic encephalopathy | Higher Enterobacteriaceae[57] |
| Hepatorenal syndrome | No established data |
| Hepatocellular carcinoma | No established data |
| Therapeutic strategies and effects on IM | |
| Lactulose | No RCT or prospective studies of microbiome |
| Decreased urea-producing Klebsiella and Proteus species, increased non-urease-producing lactobacilli[70] | |
| Rifaximin | Improved cognitive function due to change in microbiome-metabolome correlation networks, particularly Enterobacteriaceae |
| Probiotics | Decreased risk of endotoxemia, TNF-α[74] |
| Enteric dysbiosis reduced, relatively decreased proportion of Enterobacteriaceae and Porphyromonadaceae[74,75] | |
| Fecal microbiota transplantation | Case report data[76] |
| Resolution of hepatic encephalopathy with healthy IM transfer, however IM not characterized |
Emerging literature suggests that the IM is not only altered in liver disease of various etiologies, but that this dysbiosis may play an etiopathogenetic role. For example, dysbiosis may contribute to NAFLD[35,39] by contributing to enhanced hepatic fat accumulation[39]. In a cross-sectional study, patients with non-alcoholic steatohepatitis (NASH) had a significantly lower percentage of Bacteroides in their stool[35] compared to patients with simple steatosis and healthy controls, although a cause-effect relationship could not be established. Mechanisms engendered by microbial genes, such as an increase in appetite signaling, energy extraction from the diet, and expression of lipogenic genes likely contribute to enhanced hepatic fat accumulation[31,40]. In addition, hepatic inflammation and fibrosis in patients with NASH are thought to occur due to bacterial translocation of intestine-derived microbial products (including endotoxin) and activation of Toll-like receptor (TLR) signaling[41,42]. This results in stimulation of hepatic stellate cell activity, with subsequent liver fibrogenesis[43]. The role of bacterial lipopolysaccharide (endotoxin) in fibrogenesis has been confirmed in mouse models, where TLR4 knockout significantly decreased expression of markers for liver fibrosis such as collagen, α-smooth muscle actin, procollagen-I, transforming growth factor-β1 and matrix metalloproteinase-2[44]. It is thought that enteric dysbiosis in the context of liver disease of any etiology contributes through the above mechanism to liver disease progression.
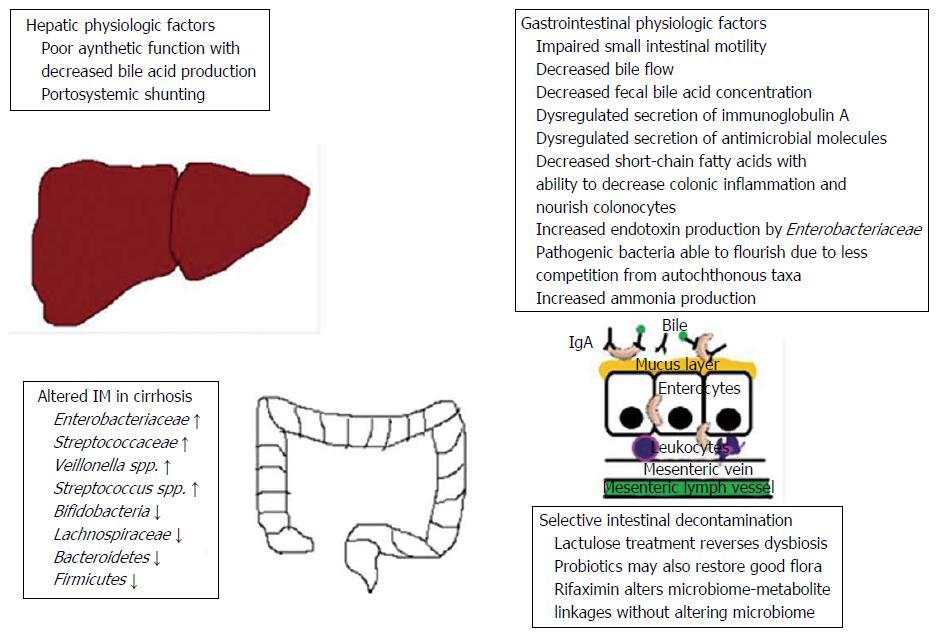
Patients with cirrhosis have both qualitative and quantitative changes in their gut microbiota[45-47]. Small intestinal bacterial overgrowth, defined as > 105 CFU/mL and/or the presence of colonic bacteria in upper jejunal aspirate, is present in 48% to 73% of cirrhotic patients[48,49]. Impaired small intestinal motility[50], decreased bile flow[51], and dysregulated secretion of immunoglobulin A[51] and antimicrobial molecules[52] all contribute to small intestinal bacterial overgrowth in patients with cirrhosis (Figure 1).
In addition to this increased bacterial burden, taxonomic differences in the fecal microbial communities have been demonstrated[51,53-56]. Patients with cirrhosis commonly have decreased proportions of beneficial, autochthonous taxa, such as Lachnospiraceae, Ruminococcaceae and Clostridiales XIV. There is a relative overgrowth of potentially pathogenic bacteria such as Enterobacteriaceae, Staphylococcaeae, and Enterococcaceae, whose abundance correlates with disease progression and endotoxemia[53,55,56]. A recent study comparing the microbiome in cirrhosis vs healthy controls revealed that the beneficial Bacteroides genus was significantly decreased in patients with cirrhosis[24]. Additional bacterial species that enrich the health and diversity of the microbiome, such as Faecalibacterium prausnitzii (anti-inflammatory properties) and Coprococcus comes (butyrate production) were found to occupy a relatively lower proportion of the microbiome in cirrhosis[41]. Most studies have shown that the etiology of cirrhosis does not affect taxonomic composition, with similar fecal microbial communities across the spectrum of liver disease etiologies[51,57]. Recently however, the pattern of dysbiosis has been reported to be slightly different in alcoholic cirrhosis, with higher Enterobacteriaceae and a higher proportion of patients with endotoxemia compared to cirrhotic patients of non-alcoholic etiologies. This held true after adjusting for severity of disease (MELD score) and abstinence[42].
The beneficial, autochthonous taxa of the IM generate SCFA that sustain colonocytes and decrease inflammation, in addition to anti-bacterial peptides that help prevent colonization by pathogenic taxa and reinforce the intestinal barrier[58]. The decreased presence of certain benign bacteria is thought to be due to decreased bile acid production in cirrhosis. This environment allows pathogenic bacteria to thrive and outgrow the “beneficial” species[6]. The combination of a decreased, “leaky” intestinal barrier with increased endotoxin production by pathogenic taxa such as the Enterobacteriaceae can lead to endotoxemia[41].
The IM composition appears to vary with the severity of cirrhosis and the presence of complications. The increased presence of the Streptococaceae taxon has been correlated with worsening Child-Pugh score, whereas the Lachnospiraceae taxon was associated with less severe disease[53]. However, the Child-Pugh score includes hepatic encephalopathy as a component, and encephalopathy itself is associated with a distinct IM as described further below. Later studies of the IM in cirrhotics have therefore employed the MELD score, in order to allow for simple correlation of the IM with severity of liver dysfunction. Enterobacteriaceae species have been reported to be associated with higher MELD scores, whereas the Ruminococcaceae species have been associated with lower MELD scores[56]. A study of the IM in patients with advanced liver disease revealed that decreased abundance of Bifidobacterium and increased abundance of Enterococcus were associated with increasing liver dysfunction[59]. The term “cirrhosis dysbiosis ratio” was coined to describe the ratio of autochthonous taxa (taxa that are benign and usually present in the gut such as Ruminococcaceae, Lachnospiraceae, and Clostridiales) to non-autochthonous ones (Enterobacteriaceae and Bacteroidaceae). This ratio was highest among healthy individuals as expected, and decreased with worsening MELD score and degree of hepatic decompensation[57]. Those with compensated cirrhosis had a cirrhosis-dysbiosis ratio of 0.89, whereas those with decompensated cirrhosis had a ratio of 0.66, and patients hospitalized for cirrhosis related complications had a ratio of 0.32 (P < 0.0001). An increase in the relative abundance of pathogenic bacteria was associated with the development of complications such as hepatic encephalopathy. Liver disease stability over months was associated with a stable cirrhosis-dysbiosis ratio[57].
Interestingly, salivary dysbiosis is concomitantly present with enteric dysbiosis, with a relative increased abundance of Enterobacteriaceae and decrease in autochthonous species[60]. Dysbiosis of the salivary microbiome was particularly pronounced in patients requiring 90-d liver-related hospitalizations. Thus, the salivary microbiome may serve as a substitute for the IM, and would be an easier sample to obtain.
Patients with cirrhosis show changes in both serum and fecal bile acids, which results from decreased liver synthetic function, altered enterohepatic circulation and altered IM composition. Overgrowth of Enterobacteriaceae leads to impaired conversion of primary to secondary bile acids[61]. This results in a decreased ratio of secondary to primary bile acids, along with a reduced overall fecal bile acid concentration, which correlate with increasing severity of liver disease. These findings are accompanied by a concomitant increase in serum bile acids[61].
Complications of end-stage liver disease, such as spontaneous bacterial peritonitis and hepatic encephalopathy, have been linked to pathological bacterial translocation. The translocation of bacteria or their products (such as muramyl-dipeptides, lipopolysaccharides (endotoxin), peptidoglycans and bacterial DNA) from the intestine to the mesenteric lymph nodes is a normal physiological process that bolsters host immunity[62]. Pathological bacterial translocation occurs due to an increase in the rate or degree of translocation. This is the case in cirrhosis, given the leaky intestinal barrier and relatively immunodeficient state[49]. The bacteria causing SBP are mostly gram-negative bacilli such as Escherichia coli and other members of the Enteriobacteriaceae family (Proteus, Klebsiella and Enterobacter), which are present in higher abundance in the gut microbiota of cirrhotic patients[51,53,55,56]. Migration of these bacteria to the peritoneal cavity or systemic circulation results in peritonitis and bacteremia, respectively. Pathological bacterial translocation triggers inflammation and the hyperdynamic circulation of cirrhosis that contributes to portal hypertension. These in turn result in serious systemic infections with up to 38% mortality[6,63]. Therefore, translocation of bacteria from an altered IM represents an important determinant of mortality in cirrhotic patients.
The IM contributes to development of hepatic encephalopathy through ammoniagenesis and an endotoxin-driven inflammatory response. Additional compounds produced by the microbiota, such as mercaptans, phenols, short- and medium-chain fatty acids and benzodiazepine-like compounds, potentially contribute as well[2]. In a metagenomic study of the microbiome in cirrhosis, Qin et al[64] performed in-depth assessment of functions of the microbiome enriched in liver cirrhosis. In cirrhosis, bacterial genes involved in the assimilation or dissimilation of nitrate to or from ammonia, denitrification, gamma-aminobutyric acid biosynthesis, and amino acid transport were highly represented[64]. Additionally, manganese-related transport system modules were enriched in the IM of patients with cirrhosis[64]. This may be associated with manganese accumulation within the basal ganglia of cirrhotic patients, which is thought to contribute to hepatic encephalopathy[65]. A patient discrimination index was developed based on a group of 15 bacterial species, and it was highly accurate as a biomarker for cirrhosis. In addition to the enteric dysbiosis described above, altered intestinal permeability results in translocation of bacteria and their products, which has an important effect on the progression of cirrhosis[66].
HCC is another complication of cirrhosis whose development may be influenced by the altered IM in the cirrhotic patient[67], although there is no concrete evidence as yet. It is well known that chronic inflammation can foster the initiation and progression of malignancies. Translocation of intestinal bacteria can lead to hepatic inflammation, with release of key inflammatory mediators such as NF-κB and TLR4. Downregulation of the NF-κB signaling pathway in vivo (by ablating the protein that activates this transcription factor) was shown to sequentially induce NAFLD, fibrosis and finally HCC[68]. TLR4 and the IM contributed to tumor progression in an HCC mouse model, and have therefore been proposed as chemopreventive targets[69]. This study suggests that the IM may have adverse effects on hepatic stellate cell function, activating the release of inflammatory mediators that promote HCC development.
The longstanding practice of treating hepatic encephalopathy with lactulose not only decreases ammonia absorption, but also results in modulation of the IM with decreased ammonia production[70]. Lactulose acidifies the colonic pH, which renders the environment hostile to the urease-producing Klebsiella and Proteus species. Conversely, the intestinal lumen becomes friendlier to non-urease-producing lactobacilli and bifidobacteria. The end-result of these changes in the microbiome is decreased ammonia production[71].
Antibiotics such as neomycin, metronidazole and ciprofloxacin have been used in the past to treat hepatic encephalopathy, although the IM was never characterized in this context. More recently, rifaximin has been offered to patients with lactulose-resistant hepatic encephalopathy. In a prospective, open-label study, 20 cirrhotic patients with minimal hepatic encephalopathy were treated with rifaximin 550 mg twice daily for 8 wk[72]. The IM, serum metabolome, and cognitive function were assessed before and after rifaximin treatment in all 20 patients. Although a significant improvement in cognitive function and endotoxemia was seen, the composition of the IM was not distinctly different. Rather, there was a shift from pathogenic to beneficial metabolite linkages around pathogenic bacterial species (Enterobacteriaceae and Porphyromonadaceae). Therefore, although the IM composition itself was not altered, the metabolic profile produced by the pathogenic species was more beneficial. On the other hand, the correlation networks around the autochthonous bacteria (looking at the interactions between the microbiome and metabolome) remained the same. This study therefore illustrated how rifaximin could alter intestinal microbial linkages with metabolites, without any significant effect on microbial composition or abundance per se[72].
The effect of probiotic therapy on the IM in cirrhotic patients has also been studied[73]. One appealing benefit of probiotics is their excellent safety profile. A phase I, 8-wk, randomized controlled trial of the probiotic Lactobacillus GG in 30 patients with minimal hepatic encephalopathy revealed that it was safe and well-tolerated, while decreasing the risk of endotoxemia and lowering TNF-α in the serum, plasma and liver[74]. Enteric dysbiosis was reduced, with a relatively decreased proportion of Enterobacteriaceae and Porphyromonadaceae (both associated with worse disease in cirrhosis). Conversely, the abundance of autochthonous species like Lachnospiraceae, Ruminococcaceae, and Clostridiales XIV increased. There was no change in the Lactobacillaceae abundance, and it was hypothesized that this species either promoted colonization by beneficial microbiota or enhanced intestinal epithelial function and the immune system, thereby displacing pathogens[74]. Additionally, changes in metabolites related to amino acid, vitamin and secondary bile acid were found. Cognition however was not improved, although this trial was a phase I study without the statistical power to determine this outcome[72,74].
A second randomized, double-blind, placebo-controlled trial of VSL#3 daily for 6 mo assessed the probiotic’s efficacy in preventing recurrent encephalopathy, reducing severity of liver disease and reducing hospitalizations[75]. There was a tendency towards decreased episodes of recurrent encephalopathy (primary outcome), with 34.8% in the probiotic group vs 51.6% in the placebo group (P = 0.12). In addition, there was a significantly reduced risk of hospitalization, as well as improved Child-Pugh and MELD scores with daily use of VSL#3.
This is a potentially interesting approach to addressing enteric dysbiosis, although the only evidence to date is a single case report where healthy gut microbiota transfer was used to treat hepatic encephalopathy[76]. This was recently described in a case report of a patient with Grade 1-2 encephalopathy not responsive to lactulose, and unable to afford rifaximin. Fecal microbiota from a healthy stool donor was transplanted into the patient by colonoscopy and by retention enemas weekly over a 5-wk period. The patient’s alertness, as well as his performance on measures of encephalopathy (inhibitory control test and Stroop test) significantly improved and normalized. This case demonstrates that fecal microbiota transplantation is a plausible strategy in treating mild encephalopathy by correcting enteric dysbiosis, although further larger-scale studies are required.
In summary, the IM is significantly altered in cirrhosis, with a decrease in beneficial, autochthonous bacterial species such as Bacteroides, and an increase in pathogenic bacteria such as the Enterobacteriaceae. Except for alcoholic liver disease, IM composition appears to be similar across etiologies of hepatic cirrhosis. The dysregulated IM likely is associated with and may contribute to the development of complications of end-stage liver disease, including hyperdynamic circulation, portal hypertension, bacteremia, spontaneous bacterial peritonitis, and encephalopathy. The role of the IM in the development of hepatorenal syndrome and HCC is suspected, but not yet elucidated. Treatment with lactulose, antibiotics, and probiotics may be effective in preventing or improving these complications by targeting the enteric dysbiosis.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Canada
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Dirchwolf M, Elalfy H, Moller S, Sunami Y S- Editor: Qiu S L- Editor: A E- Editor: Li D
| 1. | Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. Metagenomic pyrosequencing and microbial identification. Clin Chem. 2009;55:856-866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 389] [Cited by in RCA: 318] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 2. | Minemura M, Shimizu Y. Gut microbiota and liver diseases. World J Gastroenterol. 2015;21:1691-1702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 135] [Cited by in RCA: 129] [Article Influence: 12.9] [Reference Citation Analysis (1)] |
| 3. | Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9101] [Cited by in RCA: 7837] [Article Influence: 522.5] [Reference Citation Analysis (4)] |
| 4. | Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 364] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 5. | Vanderhoof JA. Etiology and pathogenesis of small intestinal bacterial overgrowth. [accessed 2015 Mar]. Available from: http://www.uptodate.com.proxy3.library.mcgill.ca/contents/etiology-and-pathogenesis-of-small-intestinal-bacterial-overgrowth?source=machineLearning&search=bacterialovergrowth&selectedTitle=3~122§ionRank=5&anchor=H2. |
| 6. | Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513-1524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 615] [Cited by in RCA: 751] [Article Influence: 68.3] [Reference Citation Analysis (0)] |
| 7. | Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring). 2010;18:190-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1670] [Cited by in RCA: 1787] [Article Influence: 119.1] [Reference Citation Analysis (0)] |
| 8. | Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, Bultman SJ. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517-526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1507] [Cited by in RCA: 1352] [Article Influence: 96.6] [Reference Citation Analysis (0)] |
| 9. | Awad AB, Horvath PJ, Andersen MS. Influence of butyrate on lipid metabolism, survival, and differentiation of colon cancer cells. Nutr Cancer. 1991;16:125-133. [PubMed] |
| 10. | Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2007] [Cited by in RCA: 2397] [Article Influence: 149.8] [Reference Citation Analysis (0)] |
| 11. | LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 790] [Cited by in RCA: 960] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 12. | Conly JM, Stein K, Worobetz L, Rutledge-Harding S. The contribution of vitamin K2 (menaquinones) produced by the intestinal microflora to human nutritional requirements for vitamin K. Am J Gastroenterol. 1994;89:915-923. [PubMed] |
| 13. | Wells JM, Rossi O, Meijerink M, van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4607-4614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 408] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 14. | Gorbach SL. Probiotics and gastrointestinal health. Am J Gastroenterol. 2000;95:S2-S4. [PubMed] |
| 15. | Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204-12209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1484] [Cited by in RCA: 1674] [Article Influence: 111.6] [Reference Citation Analysis (0)] |
| 16. | Zeissig S, Blumberg RS. Commensal microbial regulation of natural killer T cells at the frontiers of the mucosal immune system. FEBS Lett. 2014;588:4188-4194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Mikov M. The metabolism of drugs by the gut flora. Eur J Drug Metab Pharmacokinet. 1994;19:201-207. [PubMed] |
| 18. | Peppercorn MA. Sulfasalazine. Pharmacology, clinical use, toxicity, and related new drug development. Ann Intern Med. 1984;101:377-386. [PubMed] |
| 19. | Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079-1084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1834] [Cited by in RCA: 2542] [Article Influence: 254.2] [Reference Citation Analysis (0)] |
| 20. | Phillips SF, Quigley EM, Kumar D, Kamath PS. Motility of the ileocolonic junction. Gut. 1988;29:390-406. [PubMed] |
| 21. | Roland BC, Ciarleglio MM, Clarke JO, Semler JR, Tomakin E, Mullin GE, Pasricha PJ. Low ileocecal valve pressure is significantly associated with small intestinal bacterial overgrowth (SIBO). Dig Dis Sci. 2014;59:1269-1277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 22. | Riordan SM, McIver CJ, Wakefield D, Duncombe VM, Thomas MC, Bolin TD. Small intestinal mucosal immunity and morphometry in luminal overgrowth of indigenous gut flora. Am J Gastroenterol. 2001;96:494-500. [PubMed] |
| 23. | Bures J, Cyrany J, Kohoutova D, Förstl M, Rejchrt S, Kvetina J, Vorisek V, Kopacova M. Small intestinal bacterial overgrowth syndrome. World J Gastroenterol. 2010;16:2978-2990. [PubMed] |
| 24. | Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4098] [Cited by in RCA: 4541] [Article Influence: 324.4] [Reference Citation Analysis (1)] |
| 25. | Morgun A, Dzutsev A, Dong X, Greer RL, Sexton DJ, Ravel J, Schuster M, Hsiao W, Matzinger P, Shulzhenko N. Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut. 2015;64:1732-1743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 211] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 26. | Hollister EB, Gao C, Versalovic J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology. 2014;146:1449-1458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 326] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 27. | Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258-1270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2236] [Cited by in RCA: 2558] [Article Influence: 196.8] [Reference Citation Analysis (0)] |
| 28. | Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1:653-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 501] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 29. | Chase D, Goulder A, Zenhausern F, Monk B, Herbst-Kralovetz M. The vaginal and gastrointestinal microbiomes in gynecologic cancers: a review of applications in etiology, symptoms and treatment. Gynecol Oncol. 2015;138:190-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 98] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 30. | Sheflin AM, Whitney AK, Weir TL. Cancer-promoting effects of microbial dysbiosis. Curr Oncol Rep. 2014;16:406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 199] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 31. | Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027-1031. [PubMed] |
| 32. | Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3971] [Cited by in RCA: 4828] [Article Influence: 371.4] [Reference Citation Analysis (1)] |
| 33. | Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1496] [Cited by in RCA: 1588] [Article Influence: 105.9] [Reference Citation Analysis (0)] |
| 34. | Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 687] [Cited by in RCA: 643] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 35. | Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, McGilvray ID, Allard JP. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 555] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 36. | Wieland A, Frank DN, Harnke B, Bambha K. Systematic review: microbial dysbiosis and nonalcoholic fatty liver disease. Aliment Pharmacol Ther. 2015;42:1051-1063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 164] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 37. | Hartmann P, Seebauer CT, Schnabl B. Alcoholic liver disease: the gut microbiome and liver cross talk. Alcohol Clin Exp Res. 2015;39:763-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 222] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 38. | Usami M, Miyoshi M, Kanbara Y, Aoyama M, Sakaki H, Shuno K, Hirata K, Takahashi M, Ueno K, Hamada Y. Analysis of fecal microbiota, organic acids and plasma lipids in hepatic cancer patients with or without liver cirrhosis. Clin Nutr. 2013;32:444-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 39. | Abu-Shanab A, Quigley EM. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2010;7:691-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 354] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 40. | Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718-15723. [PubMed] |
| 41. | Lin RS, Lee FY, Lee SD, Tsai YT, Lin HC, Lu RH, Hsu WC, Huang CC, Wang SS, Lo KJ. Endotoxemia in patients with chronic liver diseases: relationship to severity of liver diseases, presence of esophageal varices, and hyperdynamic circulation. J Hepatol. 1995;22:165-172. [PubMed] |
| 42. | Frasinariu OE, Ceccarelli S, Alisi A, Moraru E, Nobili V. Gut-liver axis and fibrosis in nonalcoholic fatty liver disease: an input for novel therapies. Dig Liver Dis. 2013;45:543-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 43. | Alisi A, Carsetti R, Nobili V. Pathogen- or damage-associated molecular patterns during nonalcoholic fatty liver disease development. Hepatology. 2011;54:1500-1502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 44. | Csak T, Velayudham A, Hritz I, Petrasek J, Levin I, Lippai D, Catalano D, Mandrekar P, Dolganiuc A, Kurt-Jones E. Deficiency in myeloid differentiation factor-2 and toll-like receptor 4 expression attenuates nonalcoholic steatohepatitis and fibrosis in mice. Am J Physiol Gastrointest Liver Physiol. 2011;300:G433-G441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 198] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 45. | Gómez-Hurtado I, Such J, Sanz Y, Francés R. Gut microbiota-related complications in cirrhosis. World J Gastroenterol. 2014;20:15624-15631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 37] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 46. | Giannelli V, Di Gregorio V, Iebba V, Giusto M, Schippa S, Merli M, Thalheimer U. Microbiota and the gut-liver axis: bacterial translocation, inflammation and infection in cirrhosis. World J Gastroenterol. 2014;20:16795-16810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 156] [Cited by in RCA: 169] [Article Influence: 15.4] [Reference Citation Analysis (1)] |
| 47. | Macnaughtan J, Jalan R. Clinical and pathophysiological consequences of alterations in the microbiome in cirrhosis. Am J Gastroenterol. 2015;110:1399-1410; quiz 1411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 48. | Bauer TM, Steinbrückner B, Brinkmann FE, Ditzen AK, Schwacha H, Aponte JJ, Pelz K, Kist M, Blum HE. Small intestinal bacterial overgrowth in patients with cirrhosis: prevalence and relation with spontaneous bacterial peritonitis. Am J Gastroenterol. 2001;96:2962-2967. [PubMed] |
| 49. | Wiest R, Lawson M, Geuking M. Pathological bacterial translocation in liver cirrhosis. J Hepatol. 2014;60:197-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 562] [Article Influence: 51.1] [Reference Citation Analysis (0)] |
| 50. | Chang CS, Chen GH, Lien HC, Yeh HZ. Small intestine dysmotility and bacterial overgrowth in cirrhotic patients with spontaneous bacterial peritonitis. Hepatology. 1998;28:1187-1190. [PubMed] |
| 51. | Lu H, Wu Z, Xu W, Yang J, Chen Y, Li L. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Intestinal microbiota of HBV cirrhotic patients. Microb Ecol. 2011;61:693-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 179] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 52. | Teltschik Z, Wiest R, Beisner J, Nuding S, Hofmann C, Schoelmerich J, Bevins CL, Stange EF, Wehkamp J. Intestinal bacterial translocation in rats with cirrhosis is related to compromised Paneth cell antimicrobial host defense. Hepatology. 2012;55:1154-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 148] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 53. | Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 674] [Cited by in RCA: 797] [Article Influence: 56.9] [Reference Citation Analysis (3)] |
| 54. | Xu M, Wang B, Fu Y, Chen Y, Yang F, Lu H, Chen Y, Xu J, Li L. Changes of fecal Bifidobacterium species in adult patients with hepatitis B virus-induced chronic liver disease. Microb Ecol. 2012;63:304-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 55. | Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, Monteith P, Noble NA, Sikaroodi M, Gillevet PM. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675-G685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 429] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 56. | Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, Sikaroodi M, Gillevet PM. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302:G168-G175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 423] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 57. | Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 659] [Cited by in RCA: 836] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 58. | Nava GM, Stappenbeck TS. Diversity of the autochthonous colonic microbiota. Gut Microbes. 2011;2:99-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 59. | Grąt M, Hołówko W, Wronka KM, Grąt K, Lewandowski Z, Kosińska I, Krasnodębski M, Wasilewicz M, Gałęcka M, Szachta P. The relevance of intestinal dysbiosis in liver transplant candidates. Transpl Infect Dis. 2015;17:174-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 60. | Bajaj JS, Betrapally NS, Hylemon PB, Heuman DM, Daita K, White MB, Unser A, Thacker LR, Sanyal AJ, Kang DJ. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology. 2015;62:1260-1271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 248] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 61. | Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 619] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 62. | Berg RD, Garlington AW. Translocation of certain indigenous bacteria from the gastrointestinal tract to the mesenteric lymph nodes and other organs in a gnotobiotic mouse model. Infect Immun. 1979;23:403-411. [PubMed] |
| 63. | Arvaniti V, D’Amico G, Fede G, Manousou P, Tsochatzis E, Pleguezuelo M, Burroughs AK. Infections in patients with cirrhosis increase mortality four-fold and should be used in determining prognosis. Gastroenterology. 2010;139:1246-1256, 1256.e1-e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 720] [Cited by in RCA: 837] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 64. | Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1230] [Cited by in RCA: 1536] [Article Influence: 139.6] [Reference Citation Analysis (40)] |
| 65. | Krieger D, Krieger S, Jansen O, Gass P, Theilmann L, Lichtnecker H. Manganese and chronic hepatic encephalopathy. Lancet. 1995;346:270-274. [PubMed] |
| 66. | Wiest R, Garcia-Tsao G. Bacterial translocation (BT) in cirrhosis. Hepatology. 2005;41:422-433. [PubMed] |
| 67. | Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. 2013;10:656-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 772] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 68. | Luedde T, Beraza N, Kotsikoris V, van Loo G, Nenci A, De Vos R, Roskams T, Trautwein C, Pasparakis M. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2007;11:119-132. [PubMed] |
| 69. | Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 854] [Cited by in RCA: 1028] [Article Influence: 79.1] [Reference Citation Analysis (0)] |
| 70. | Prasad S, Dhiman RK, Duseja A, Chawla YK, Sharma A, Agarwal R. Lactulose improves cognitive functions and health-related quality of life in patients with cirrhosis who have minimal hepatic encephalopathy. Hepatology. 2007;45:549-559. [PubMed] |
| 71. | Riordan SM, Williams R. Treatment of hepatic encephalopathy. N Engl J Med. 1997;337:473-479. [PubMed] |
| 72. | Bajaj JS, Heuman DM, Sanyal AJ, Hylemon PB, Sterling RK, Stravitz RT, Fuchs M, Ridlon JM, Daita K, Monteith P. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One. 2013;8:e60042. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 254] [Cited by in RCA: 339] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 73. | Qamar AA. Probiotics in Nonalcoholic Fatty Liver Disease, Nonalcoholic Steatohepatitis, and Cirrhosis. J Clin Gastroenterol. 2015;49 Suppl 1:S28-S32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 74. | Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, Puri P, Sterling RK, Luketic V, Stravitz RT, Siddiqui MS, Fuchs M. Randomised clinical trial: Lactobacillus GG modulates gut microbiome, metabolome and endotoxemia in patients with cirrhosis. Aliment Pharmacol Ther. 2014;39:1113-1125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 222] [Article Influence: 20.2] [Reference Citation Analysis (1)] |
| 75. | Dhiman RK, Rana B, Agrawal S, Garg A, Chopra M, Thumburu KK, Khattri A, Malhotra S, Duseja A, Chawla YK. Probiotic VSL#3 reduces liver disease severity and hospitalization in patients with cirrhosis: a randomized, controlled trial. Gastroenterology. 2014;147:1327-1337.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 255] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 76. | Kao D, Roach B, Park H, Hotte N, Madsen K, Bain V, Tandon P. Fecal microbiota transplantation in the management of hepatic encephalopathy. Hepatology. 2016;63:339-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 102] [Article Influence: 11.3] [Reference Citation Analysis (0)] |