Published online Apr 28, 2015. doi: 10.4254/wjh.v7.i6.831
Peer-review started: August 14, 2014
First decision: September 16, 2014
Revised: January 29, 2015
Accepted: February 9, 2015
Article in press: February 11, 2015
Published online: April 28, 2015
Processing time: 260 Days and 21.1 Hours
Hepatitis C virus (HCV) has infected over 170 million people worldwide and creates a huge disease burden due to chronic, progressive liver disease. HCV is a single-stranded, positive sense, RNA virus, member of the Flaviviridae family. The high error rate of RNA-dependent RNA polymerase and the pressure exerted by the host immune system, has driven the evolution of HCV into 7 different genotypes and more than 67 subtypes. HCV evolves by means of different mechanisms of genetic variation. On the one hand, its high mutation rates generate the production of a large number of different but closely related viral variants during infection, usually referred to as a quasispecies. The great quasispecies variability of HCV has also therapeutic implications since the continuous generation and selection of resistant or fitter variants within the quasispecies spectrum might allow viruses to escape control by antiviral drugs. On the other hand HCV exploits recombination to ensure its survival. This enormous viral diversity together with some host factors has made it difficult to control viral dispersal. Current treatment options involve pegylated interferon-α and ribavirin as dual therapy or in combination with a direct-acting antiviral drug, depending on the country. Despite all the efforts put into antiviral therapy studies, eradication of the virus or the development of a preventive vaccine has been unsuccessful so far. This review focuses on current available data reported to date on the genetic mechanisms driving the molecular evolution of HCV populations and its relation with the antiviral therapies designed to control HCV infection.
Core tip: Hepatitis C virus (HCV) is the major causative agent of parenterally-acquired hepatitis. To date there is no preventive vaccine, and though antiviral therapy has been improved in the past few years, not all patients eradicate the virus as a result of it. The main reason lies in the intrinsic genetic variability that characterises RNA viruses, such as HCV, whose RNA polymerase lacks proof-reading activity, leading to a high mutation rate and the generation of a wide range of genome variants better known as a quasispecies. Therefore this review summarises current data on HCV quasispecies dynamics, antiviral therapy and recombination events.
- Citation: Echeverría N, Moratorio G, Cristina J, Moreno P. Hepatitis C virus genetic variability and evolution. World J Hepatol 2015; 7(6): 831-845
- URL: https://www.wjgnet.com/1948-5182/full/v7/i6/831.htm
- DOI: https://dx.doi.org/10.4254/wjh.v7.i6.831
Hepatitis C virus (HCV) has infected over 170 million people worldwide and therefore creates a huge disease burden due to chronic, progressive liver disease[1]. Infections with HCV have become a major cause of liver cancer and one of the most common indications for liver transplantation[2-4]. The fact that chronic infection with HCV can lead to cirrhosis and hepatocellular carcinoma creates the need to develop drugs that effectively eradicate the infection[5] and a prophylactic vaccine that prevents its dissemination. Unfortunately, to date there is no effective vaccine available[6]. Currently, the standard of care (SOC) therapy involves pegylated interferon α (INF-α-peg) and ribavirin (RBV)[7]. In addition, the new SOC (NSOC) therapy of protease inhibitors boceprevir or telaprevir in combination with INF-α-peg and RBV have been approved for the eradication of HCV genotype 1 in the United States, Europe and Japan[8-11]. Unfortunately, interferon is not widely available globally, not always well tolerated and some genotypes of HCV respond better than others causing that not all patients achieve a sustained virological response (SVR)[12]. Other adverse events such as rash have also been associated with the NSOC[13].
The main route of transmission is direct or indirect exposure to contaminated blood, either through blood transfusions or blood products, through the consumption of intravenous drugs, use of surgical material poorly sterilized, organ transplants, accidents in health centres, vertical transmission from mother to child, etc.[12].
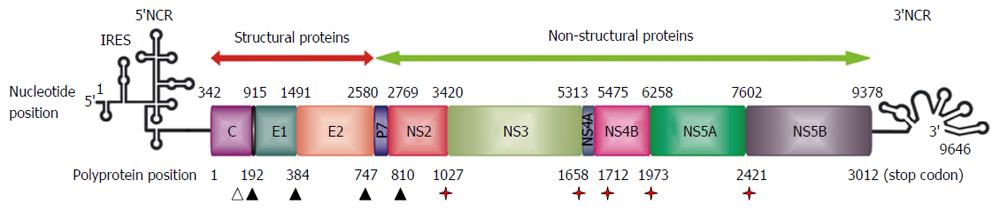
HCV is a member of the family Flaviviridae, although it differs from other members of this family in many details of its genome organization[1]. HCV is a single-stranded, positive sense, RNA virus with a genome of approximately 9600 nucleotides in length. Most of the genome carries a single open reading frame that encodes three structural (core, E1, E2) and seven non-structural (p7, NS2, NS3, NS4A, NS4B, NS5A, NS5B) proteins (Figure 1)[14,15]. In addition, alternative translation products (F protein) have been detected from a reading frame overlapping the core gene (core + 1/ARFP)[16,17]. Possible roles in regulation of gene expression, cell signalling and apoptosis have been suggested[18-20]. Short untranslated regions at each end of the genome (5’-NCR and 3’-NCR) are required for its translation and replication[21,22]. The mechanism of translation initiation is dependent on an internal ribosomal entry site in the 5’-NCR, which interacts directly with the 40S ribosomal subunit[23].
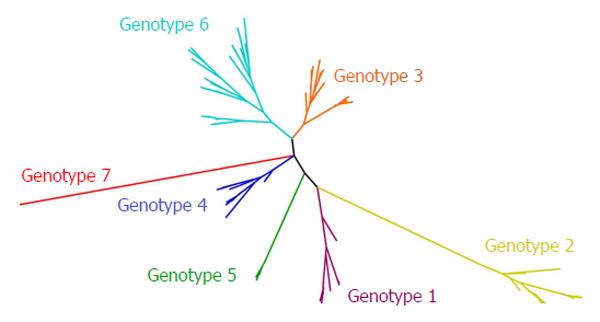
The high error rate of RNA-dependent RNA polymerase and the pressure exerted by the host immune system, has driven the evolution of HCV towards the development of a global diversity that revealed the existence of seven genetic lineages (genotypes 1 to 7) (Figure 2)[24]. On average, the complete genome differs in 31%-33% of nucleotide sites[4]. Genotypes 1 to 6 of HCV contain a series of more closely related sub-types (67 accepted subtypes and many more to be confirmed) that typically differ from each other by at least 15% in nucleotide positions within the coding region[24]. Subtypes 1a, 1b and 3a are widely distributed and account for the vast majority of infections in Western countries[1].
HCV genetic variability is not evenly distributed across the viral genome. The regions of the genome that correspond to essential viral functions (such as those involved in translation and replication) or those with major structural domains (5’-NCR and 3’-NCR) are the most conserved. The 5’-NCR region is the most conserved region of the genome with 90% sequence identity between distant strains[25,26]. The region encoding the viral capsid is also highly conserved with 81%-88% sequence identity between different isolates. The most variable region of the genome is the one that codes for the membrane glycoproteins E1 and E2[27]. The sequences belonging to the hypervariable regions 1 and 2 (HVR1 and HVR2) of E2 gene are the ones that show less sequence homology with only 50% identity between different isolates[28]. Factors that may contribute to high genetic variability of these viruses include large population sizes, short generation times and high replication rates[29].
An important breakthrough in the treatment of chronic HCV infection was undoubtedly the introduction of alpha interferon (IFN-α) plus RBV as combination therapy[27]. However, the rate of sustained virological response is still unsatisfactory[30,31], particularly in patients infected with genotype 1[32], the most prevalent in many geographic regions of the world[4,33]. Although IFN-α is effective in reducing the viral load, complete eradication of the virus is achieved in less than 20% of patients treated with IFN-α alone[32]. In those patients who initially respond to IFN-α, ribavirin helps increasing the frequency of virus eradication, yet its effect on non-responder patients is still limited[7]. Although viral genotype and viral load, as well as serum HCV RNA clearance during therapy are definitely related to response, further insight into viral factors involved in therapeutic responsiveness is still necessary[27]. The different genotypes and subtypes vary in their responses to treatment with INF-α or INF-α/RBV. As mentioned above, only 10%-20% of individuals chronically infected with HCV genotype 1 treated with IFN-α monotherapy and 40%-50% of those treated with combination therapy (INF-α/RBV) exhibit a complete and permanent disappearance of the virus. These percentages are lower than rates of 50 and 70%-80%, respectively, observed in the treatment of patients infected with HCV genotypes 2 or 3[3,34]. Despite these facts, the use of INF-α-peg has been associated with a significant increase in these rates[35].
Two inhibitors of the NS3/4A serine protease, boceprevir (BOC) and telaprevir (TVR) have demonstrated potent inhibition of HCV genotype 1 replication and markedly improved SVR rates in treatment-naïve and treatment-experienced patients[36]. Nowadays the NSOC therapy for genotype 1, chronic HCV infection is the use of BOC or TVR in combination with INF-α-peg and RBV[37-40]. Additionally, two other direct antiviral drugs (DAAs), simeprevir (protease-inhibitor)[41] and sofosbuvir (nucleotide analogue of the NS5B RNA-polymerase)[42] have been recently approved for triple therapy in the United States and Europe. Despite these new advances in what respects to triple therapy, it is worth to note that a wide range of different DAAs are currently under clinical trials aiming at all-oral IFN-free regimens[43-47].
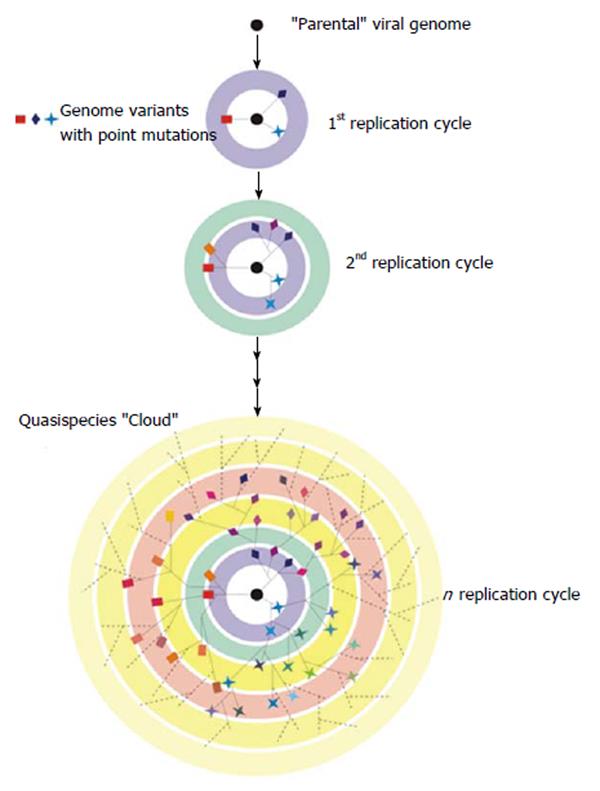
HCV evolution is a highly dynamic process[48]. Like most RNA viruses, HCV exploits all possible mechanisms of genetic variation to ensure its survival. Mutation at the nucleotide level seems to be the main cause of genetic variation in RNA viruses, such as HCV. These mutations are primarily generated by an error-prone, non-proofreading RNA-dependent RNA-polymerase which directs the replication of the virus genetic material[1,49]. The mutation rate of HCV, estimated at 10-4 substitutions per site and round of replication[50], is among the highest for RNA viruses including retroviruses[51], and would seem to be high enough to generate all the genetic variation found in this virus. Due to this feature and to the high replication rate of HCV, a large number of different but closely related viral variants are continuously produced during infection. These circulate in vivo as a complex population commonly referred to as a quasispecies[52-58].
The idea of quasispecies was first used by Eigen et al[59,60] to refer to the first self-replicating structures. Originally conceived as a mathematical framework formulated to explain the evolution of life in the ‘‘pre-cellular RNA world”[61], quasispecies theory is based on classical population genetics, but seeks to explore the consequences of error-prone replication and near-infinite population sizes for genome evolution. More recently, quasispecies theory has been used to describe the evolutionary dynamics of RNA viruses[62]. These structured populations also possess a high mutation rate which would lead to the existence of a complex mixture of different but related genomes that would behave as a selection unit (Figure 3). At a particular point of infection, the HCV quasispecies viral distribution reflects the balance between the continued generation of new variants, the need to preserve essential viral functions and the positive selective pressure exerted by the environment[63]. It is important to highlight that multiple viral quasispecies co-exist in infected individuals at different replicative sites and in consecutive times, which offers a rich environment for intra and inter quasispecies interactions.
The theory of quasispecies predicts that a particular mutant surrounded by a more favourable mutant spectrum (more related) can dominate another one with better fitness. This phenomenon has been called suppressive effect. This suppressive or interfering effect of the mutant spectrum rises along with the mutation rates in viral quasispecies, as has been suggested by the strong suppressive effect in infectivity that viral populations have near to extinction[64]. It has been proposed that interference generated by defective genomes contributes to viral extinction due to an increased mutagenesis rate. We can also see the opposite effect: genome complementation between different components of the mutant spectrum, which demonstrates that the behaviour of quasispecies cannot be understood solely as the sum of individual behaviours[65]. Because of this, quasispecies is defined as a dynamic distribution of genomes subject to variation, competition and selection, and that act as a selection unit. This means that the quasispecies as a whole, instead of a particular viral variant, is the target of the selection process[62,64,66].
As mentioned before, low fitness variants can be preserved at higher than expected frequencies just because they are coupled to a well represented higher fitness genotype in sequence space. One of the defining characteristics of a quasispecies is the phenomenon of mutational coupling, as it places individual mutants within a functional network of variants[62]. The elevated mutation rates in RNA viruses mean that a fast replicator will generate genetically diverse progeny, many of which will be significantly less fit than the parent. As a result, quasispecies theory predicts that slower replicators will be favoured if they give rise to a fitter progeny[67].
One of the consequences of quasispecies dynamics is the existence of a threshold error to the preservation of genetic information[64]. When the error rate exceeds a tolerable limit (related to genome size, fitness and population size of the quasispecies) distribution collapses and the nucleotide sequence loses its information. This transition is known as the entrance to catastrophic error and its application to viral extinction through mutagenesis is called lethal mutagenesis[64]. There are experimental evidences that show that RNA viruses replicate very closely to this threshold error and that an increase in mutation rates can have a negative impact on the viability of viral populations.
The theory of evolution predicts that, in a dynamic environment, high mutation rates are favoured, and therefore viral error rates may have been optimized by natural selection[68,69]. For RNA viruses, low replicative fidelity generates a diverse population of variants. Even though many of these variants are generally less fit, they may take over if an unexpected change in environment occurs, such as immune pressure, shifting the corresponding fitness landscape. On the contrary, a homogeneous population, generated by high replicative fidelity, would lack this flexibility and might be less successful in the dynamic host environment. Experimental support for this hypothesis has been provided by two different groups[70,71]. They isolated a poliovirus variant, resistant to ribavirin, which had a single amino acid substitution in the viral polymerase. This mutant exhibited a moderate resistance to lethal mutagenesis and assays for selectable markers indicated that this population had a lower mutation rate and it consequently displayed less genetic diversity. More recently, Vignuzzi et al[72] (2008) proposed that this attenuated high fidelity variant could be employed for vaccine development.
High mutation rates and quasispecies dynamics confer great adaptability to RNA viruses and represent one of the major obstacles for the control and prevention of RNA viral diseases[15,73]. The great quasispecies variability of HCV has also therapeutic implications since, by means of generating and selecting fitter variants within the quasispecies cloud, viruses might escape control by antiviral drugs[74]. The way HCV quasispecies evolves is highly dynamic, and for this reason, the complexity of the genetic information gathered from quasispecies populations cannot be accurately analysed by a unique analytical tool[75]. The three parameters most commonly used to determine the complexity of the quasispecies mutant spectrum are: mutation frequency (defined as the proportion of mutated nucleotides within a genome distribution relative to the consensus sequence), Shannon entropy (defined as the proportion of different genomes within a mutant distribution) and Hamming distance (defined as the number of mutations that differentiate two sequences within the mutant spectrum)[27,64]. The average of all the comparisons between possible pairs reflects the genetic complexity of quasispecies[62]. As stated previously, a fundamental feature of viral quasispecies, predicted from quasispecies theory, is that the target of selection is the mutant distribution as a whole rather than an individual genome. Selective transmission of predominant and minor HCV quasispecies has been shown in humans[76-80] and in experimentally infected chimpanzees[81,82]. Further understanding of quasispecies dynamics in infected individuals is necessary to gain knowledge on how to apply virus-specific drugs and to identify key parameters that are critical for the development of effective antiviral strategies[55]. The genetic variability within quasispecies level has been frequently used as a predictor of the response to antiviral therapy[83]. Several studies have reported that genetic diversity within different regions of HCV genome before combined therapy with INF/RBV was higher in non-responders than in responders[53,83-86]. Nevertheless, Cristina et al[15] (2007) showed that the response to antiviral therapy is independent from genetic variability within quasispecies populations at the beginning of therapy. Ueda et al[86] (2004) reported that during combined therapy with INF/RBV, HCV quasispecies significantly decreased in non-responders. However, another study showed that populations fluctuate along therapy, in treated patients that achieved a sustained virological response as well as in those non-responders. This would indicate that these fluctuations are intrinsic to each patient and that HCV follows different evolution paths in different patients[15]. The variations in the results obtained by the different research groups might be caused by different methodological approaches used to study the real variability within circulating quasispecies populations[87]. Another explanation to this might be that in viral populations with a high degree of genetic variability, the probability of finding variants that would replicate effectively in the presence of antiviral drugs is high. Despite this, those populations with lower variability degree might also present these types of resistant variants, though in lower probability. Likewise, given the stochastic nature of this phenomenon, the opposite situation might as well occur in these two same cases[88]. These correlations can probably mask the expected one between genetic variability and response to treatment, thus providing no conclusive results on these matters.
In addition to the multiplicity of viral genetic factors reported so far, host factors have also shown to be involved in the development of HCV infection. Since 2009, several single nucleotide polymorphisms (SNP) have been reported near the interleukin 28B gene (IL-28B) that influence response to dual antiviral therapy[89-94]. However, it was not until 2011 that the relationship between HCV quasispecies diversity and the host IL-28B genotype was investigated[95]. Their results evidenced a clear association between IL-28B risk allele (SNP rs8099917 - G allele) and a lower NS3 protease amino acid quasispecies diversity in infected patients, hence suggesting that IL-28B risk allele carriers exert less positive selection pressure on the NS3/4 protease[95]. However, it would be interesting to address whether the lower amino acid quasispecies diversification in patients with an IL-28B risk allele is restricted to the NS3 coding region or if it affects other viral genomic regions. Interestingly, also unfavourable rs8099917 genotypes were found linked to time-dependent changes in the core coding region, specifically the shift to residue 70Q associated with hepatocellular carcinoma[96]. These results might indicate that the IL-28B genotype influences viral evolution and disease outcome in addition to the behaviour of the innate immune system. More recently, Yuan et al[94] (2012) found that, despite exhibiting similar baseline viral loads, more chronically-infected patients with the rs12979860-CC polymorphism had amino acid substitutions in NS5A compared to non-CC patients. This result suggests that patients with the CC genotype undergo early viral evolution probably as a consequence of the selective pressure exerted by the use of interferon at the beginning of treatment. These studies raise the question of whether host genetics shapes viral evolution in response to immunity, and if the differences observed in the evolution of HCV quasispecies are implicated in the mechanism by which the IL-28B genotype influences the outcome of acute HCV infection and treatment response[97].
As previously mentioned, RNA virus populations exist as a cloud of sequence variants (continuously being generated by mutation) strongly related, therefore a better understanding of these populations within infected individuals is needed in order to apply antiviral strategies and to define critical parameters to the development of new and more effective antiviral therapies[62]. Nevertheless, to date, our ability to know in depth the quasispecies cloud from the study of isolated clones is limited. The recent techniques of deep sequencing allow us, for the first time to overcome these obstacles and observe quasispecies as a whole. These techniques are already being used on clinical samples to study different viral models[98-100].
Since 2005, the development of high throughput, or so-called next generation sequencing technologies (NGS), has allowed a huge increase in capacity to sequence genomes at a reasonably low cost and in a short time frame[101]. NGS comprises a set of high-throughput sequencing technologies, which make it possible to sequence several genomes from individual templates in a parallel fashion[102]. The current NGS technologies are known as second generation technologies (Table 1), to distinguish them from the first generation (Sanger sequencing), and the third generation (based on single molecule sequencing)[103].
| Platform | Run time (h) | Read length (bp) | Throughput per run (Mb) | Typical errors | Main biological applications | Company URL |
| Roche 454 FLX + | 23 | 700, up to 1000 | 700 | Insertions/deletions (indels) at homopolymer regions | Microbial genome sequencing, human genome sequencing, transcriptomics, metagenomics | http://www.my454.com/ |
| Illumina HySeq 1000 MySeq 2000 V3 | 8 10 | 2 × 100 2 × 150 | 400000 < 600000 | Indels, especially end of reads | Microbial genome sequencing, human genome sequencing, transcriptomics, metagenomics | http://www.illumina.com/systems.ilmn |
| SOLiD 4 | 12 | 50 × 35 | 71000 | End of read substitution errors | Microbial genome sequencing, human genome sequencing, transcriptomics, metagenomics | http://www.appliedbiosystems.com/ absite/us/en/home.html |
| Ion torrent PGM 318 Chip | 3 | 200 | 1000 | Indels at homopolymer regions | Microbial genome sequencing, human genome sequencing, transcriptomics, metagenomics | http://www.iontorrent.com/ |
| Pacific Biosciences | Random indel errors | Full-length transcriptomics, discovering large structural variants and haplotypes | http://www.pacificbiosciences.com/ |
As it is mentioned before, RNA viruses have very high mutation frequencies. The error rate of viral RNA-dependent RNA polymerases is estimated to be between 10-3 and 10-6 per nucleotide copied compared to 10-8-10-11 for DNA polymerases[72]. Consequently, an RNA virus population consists not of a single genotype but of an ensemble of closely related genotypes, termed as a quasispecies, centred on a master sequence. This genetic diversity creates a cloud of potentially beneficial mutations, which is thought to allow rapid adaptation to a constantly changing environment[52]. Quasispecies theory makes a number of predictions about the behaviour of viral populations and the consequences of altering diversity.
NGS technologies have redefined the modus operandi in virus genetics research, allowing the unprecedented generation of very large sequencing datasets on a short time scale and at affordable costs[101]. A significant technical challenge to address how we can measure viral diversity could be NGS technology. Ultra-deep sequencing has the sensitivity and quantitative nature required for this kind of investigations into viral genetic drift, natural selection and response to antiviral drugs. The analysis of viral quasispecies has been greatly enhanced by the recent emergence of these powerful technologies that allow the simultaneous sequencing of 400000-10000000 individual target sequences[104].
As previously reported, drug-resistant variants may already exist previous to a particular antiviral treatment embedded in a predominantly wild-type virus quasispecies population. Consequently, as they may be present at different low-level frequencies, this may lead to varying degrees of viral response and therefore the mutant genome will become enriched upon treatment with HCV inhibitors. Hence, determining the natural levels of low frequency resistant variants before starting a treatment might be relevant to better predict viral response to HCV inhibitors[105].
The classic method for detecting these drug-resistant variants in infected patients is population-based DNA sequencing, in other words: bacterial cloning. This method provides a good idea of the major sequences present, but unfortunately it cannot detect minor variants that are present at a frequency below 20%-25%[106]. With the development of deep sequencing technologies, detection of drug-resistant variants became more sensitive allowing the identification of variants present at very low frequencies (about 0.1%-1%)[107-109]. For HCV, the deep sequencing method was first used to detect emergence of NS3 mutants. This way it was clearly demonstrated that de novo telaprevir-resistant NS3 mutants arose in mice injected with wild-type HCV only 2 wk after the beginning of treatment[108]. Deep sequencing was also used to confirm results of naturally occurring drug-resistant HCV mutants detected by a novel mismatch amplification mutation assay polymerase chain reaction[107].
As can be noted, deep sequencing technology provided a comprehensive view of the viral population dynamics during monotherapy with NS3 protease inhibitors. It allowed the estimation of pre-treatment levels of NS3 drug resistant mutants, suggesting therefore a limitation of HCV viral suppression with NS3 protease inhibitor monotherapy caused by the pre-existence of drug resistant mutants. This finding strongly evidences the need for a combined therapy to durably treat HCV infection[105].
In addition, NGS technology has already been implemented in order to study the transmission event of HCV among injection drug users. In this case NGS was used to determine intra-host viral genetic variation by deep sequencing the HCV hypervariable region, which allowed a detailed analysis of the structure of the viral quasispecies in the patients’ population under study[110].
NGS approaches are powerful methods that allow a rather comprehensive analysis of the intra-host viral genetic variation[107]. Moreover, these technologies are becoming rapidly accessible all around the world which will likely revolutionise the field of molecular epidemiology[110].
It is worth mentioning that these technologies offer, as already discussed, several advantages over conventional methods, such as consensus sequencing, bacterial cloning, and endpoint limiting dilution[107]. Furthermore, as the development of a variety of software and algorithms capable of handling the massive amount of data generated by NGS platforms is increasing in parallel with the advances in these technologies, it will likely expedite the implementation of such approaches in a variety of settings in the near future[110]. Taking this into consideration, the use of NGS in HCV outbreak investigations will presumably improve molecular epidemiology studies as well as provide a vast amount of information that will need to be handled appropriately both for the benefit of infected patients and the management of public health systems.
In addition to mutations, it is widely accepted that recombination plays an important role in the evolution of RNA viruses by creating genetic variation through the exchange of nucleotide sequences between different genomic RNA molecules[111]. Therefore it is considered as a key mechanism for the production of new genomes with selective growth advantage[112]. Homologous recombination occurs, in some cases of RNA virus recombination, when the donor sequence neatly replaces a homologous region of the acceptor sequence leaving its structure unchanged. In these cases, not only the parental RNAs are homologous, but also crossovers occur at homologous sites[112,113]. Nevertheless, hybrid sequences may originate as a result of aberrant homologous recombination (when similar viruses exchange sequence without maintaining strict alignment) and non-homologous recombination (recombination between unrelated RNA sequences)[112,113]. RNA recombination involves replication of genomic RNA as a necessary component of the process. If in the middle of the replication process, the viral RNA-dependent RNA polymerase complex switches from one RNA parental strand to another, hybrid complementary RNA strands will be formed. Yet, if the replicase continues to copy the new strand at the same site where it left the parental one, this constitutes a homologous recombination event. On the other hand, aberrant or non-homologous recombination will occur if the copying process is not as precise[112]. This template strand exchange mechanism of recombination is known as “copy choice”. The exact mechanism of exchange of strand is not known, but could be promoted by the pause of the polymerase during chain elongation. Thus far, nearly all studies on the mechanisms of recombination in RNA viruses support a copy-choice model, originally proposed for poliovirus[114]. It is of note, that this template-switching mechanism greatly differs from the enzyme-driven breakage-rejoining mechanism of homologous recombination in DNA, mainly because it resorts to replication as an essential step of the process[112].
Recombination in RNA virus was first discovered in poliovirus infected cells in which the frequent recovery of poliovirus that results from recombination has the potential to produce “escape mutants” in nature as well as in experiments[115]. Subsequently, recombination was found to occur in other RNA viruses positive and negative-stranded and more recently, recombination between unrelated groups of RNA and DNA viruses was discovered in a novel virus genome isolated from an extreme environment[116]. The presence of recombination in several members of the family Flaviviridae such as Pestivirus, Flavivirus and Hepacivirus has been demonstrated[117-122].
Regarding HCV, recombination has been reported both inter and intragenotypic in populations in different geographic locations (Table 2). Some earlier reports described some HCV strains from Honduras in which the study of partial sequences from different regions of the viral genome resulted in HCV discordant genotype, providing first evidence for the possible existence of HCV recombination[123]. However it was not until 2002 that the first convincing report of an intergenotypic HCV recombinant strain was published by Kalinina et al[124] (2002) in Saint Petersburg (Russia)[49,124]. These authors described six different natural HCV strains that belonged to different subtypes, 2k and 1b. They found that the 5’ untranslated region and the core coding region belonged to subtype 2k, whereas the NS5B region corresponded to subtype 1b[124]. Sequencing the E2-p7-NS2 region, they were able to map the crossover point within the NS2 region, estimating it most likely between positions 3175 and 3176 (according to the numbering system for strain pj6CF). The reported recombinant was cautiously designated RF1_2k/1b, in agreement with the nomenclature used for human immunodeficiency virus (HIV) recombinants[124]. This same recombinant strain has since then been isolated in other countries, like Ireland[125], Uzbekistan[126], Cyprus[127], France[128] and Estonia[129] which would suggest that, although its generation might not be favoured by natural selection, it would also not be selected against[49]. Additionally, at least ten other different intergenotypic recombinant forms (RFs) of HCV have been described and are totally or partially characterised (Table 1). Within this group of recombinants, we can observe the presence of recombinant forms between genotypes 2 and 6 described in Vietnam[130] and Taiwan[131], between genotypes 2 and 5 described in France[132], between genotypes 3 and 1 in Taiwan[131] and China[133], and between genotypes 2 and 1 reported in Japan, the United States and the Philippines[134-137]. It is interesting to note that recombinant forms found so far have a wide geographic distribution. Besides, all HCV genotypes except for genotype 4 and 7 have been found in them. Other interesting feature is that all these recombinants but one (RF_3a/1b) originated by the combination of a 5’-end of genotype 2 and a 3’-end of a different genotype. The 3’-end of subtype 1b seems to be the only one appearing in more than one recombinant form[49]. Oddly enough, genotype 2 is present in the majority of the recombinants found to date, which might suggest a critical role in order for the process to take place[138] or even for the stability and functionality of the resulting recombinant genome. The fact that some recombinants involving genotype 2 and subtype 1b have been frequently found in older patients and in cases not usually related to the epidemic spread associated to a higher use of intravenous drugs, makes it difficult to assess whether this pattern derives from adaptive selection or is simply due to chance[1,49]. As we can observe in Table 2, another characteristic feature of intergenotypic recombination in HCV seems to be that the crossover points appear to be located within either gene NS2 or NS3[49]. An apparent hotspot has been identified between amino acids 1022 and 1042 (corresponding to the vicinity of the NS2/NS3 junction). Considering how short this region seems to be, and despite the existence of only a few reports inquiring into RNA secondary structures involved in the recombination process[130,139], all seems to indicate that a copy choice mechanism might be responsible for the generation of these recombinant forms[49].
| Strain | Genotype | Country | Recombination breakpoint(s) | Ref. |
| Intergenotype | ||||
| RF1_2k/1b | 2k/1b | Russia, Ireland, Uzbekistan, Georgia, France, Cyprus, Estonia | NS2, positions 3175-3176 | [124-129] |
| D3 | 2i/6p | Viet Nam | NS2/NS3 junction, between positions 3405 and 3464 | [130] |
| SE-03-07-1689 | RF3_2b/1b | Philippines | NS3, positions 3466-3467 | [137] |
| HC10-0804 | 2b/1b | Japan | NS2/NS3 junction, positions 3443-3444 | [135] |
| B5808, M2123, M4430, M4416, M4414, M4431, M2777, M8774 | 2b/1b | Japan | NS2, putative position 3301 | [136] |
| R1 | 2/5 | France | NS2/NS3 junction, between residues 3420 and 3440 | [132] |
| D177 | RF_2b/6w RF_3a/1b RF_2a/1a | Taiwan Taiwan, China Taiwan | NS2/NS3 junction, position 3429 Undetermined Undetermined | [131,133] |
| JF779679 | 2b/1a | United States | NS2/NS3 junction, positions 3405-3416 | [134] |
| Intersubtype | Subtypes | |||
| PE22 | RF2_1b/1a | Peru | NS5B, position 8321 | [141] |
| H23 | 1b/1a | Uruguay | Core, at position 387 | [111] |
| HC-J1 | 1a/1c | Japan | 2 sites in E1-E2, at positions 1407 and 2050 | [142] |
| Khajal | 1a/1c | India | 5 sites, from core to NS3, at positions 801 1261, 2181, 3041 and 3781 | [143] |
| R49 | 4a/4d | Portugal | Undetermined | [140] |
| EU246930 | 6a/6o | Viet Nam | NS5B, between positions 8345-9073 | [144] |
| EU246932 | 6e/6o | Viet Nam | NS5B, between positions 8358-8977 | [144] |
| EU246937 | 6n/6o | Thailand | NS5B, between positions 8372-9033 | [144] |
| EU246931 | 6e/6h | Viet Nam | NS5B, between positions 8356-9019 | [144] |
| Intrapatient | Subtype | |||
| 1b | Spain | NS5B, at residue 286 | [145] | |
| 1a, 1b, 3a | Spain | 1 or 2 sites within E1-E2 or NS5A | [146] | |
| 4a | Ireland | E2 glycoprotein, HVR1 region | [147] |
With respects to intragenotypic recombination, nine recombinant forms have been described. As we mentioned before, each of the six major genotypes of HCV (except for genotype 7 for which there is only one complete-genome sequence available) can be subdivided into closely related sub-types that differ from each other by at least 15% in nucleotide sequences[24]. Therefore, the same methodological procedures based on phylogenetic incongruence used to detect intergenotypic recombinants are also applicable to detect intragenotype/intersubtype RFs. Only one of the intragenotype recombinants described involved genotype 4[140], four involved different subtypes of genotype 1 and the remaining four involved different subtypes of genotype 6. Examples of recombinants (1a/1b) have been identified in Peru as well as in Uruguay[111,141] and (1a/1c) in Japan and in India[142,143]. Interestingly, although the recombinants reported in Uruguay and Peru are (1a/1b), their recombination breakpoints were found in different regions of the genome as shown in Table 2. The same happens in the case of the recombinant forms described in India and in Japan. Therefore, unlike what it is observed in intergenotype recombination, where all shared a common genome region in which recombination occurs, in this case these points are highly variable in their location. Only two of the intragenotype 1 RFs have been fully sequenced[142,143] and both involve the same subtypes (1a and 1c). Interestingly, they revealed the existence of more than one cross-over point, resulting in mosaic recombinant forms. In addition, the sequences showed a very dissimilar size, exhibiting relatively short segments of one subtype embedded within a genome of the other subtype[49]. The most recent description of intersubtype recombinant forms involves genotype 6, and these were identified thanks to full-genome sequence analysis[144]. The remaining three cases of intersubtypic RFs reported have only been partially characterised at the genome level[49]. One of the cases was detected by discordant phylogenetic analysis of the regions coding for E1 and NS5B isolated from an intravenous drug user from Portugal[140]. This case should be considered as a putative example since no recombination breakpoint has been mapped for this RF. The two remaining cases are different as they have been described only by sequencing one single portion of the HCV genome. Contrary to what occurred with the other example, the corresponding breakpoints for these two were identified within the core and NS5B genes each[111,141], genes that are relatively conserved and are therefore suitable for phylogenetic typing and subtyping of HCV isolates[49].
When we talk about recombination in HCV we can not forget to mention the existence of intrapatient or also called intra quasispecies recombination. With respect to this matter, three reports have been published thus far. Two of them involve intrapatient recombination in individual patients undergoing therapy[145,146]. The most recent one involves quasispecies evolution from a chronically infected, treatment naïve individual[147]. In the first report of intra-quasispecies recombination, analysed sequences were obtained from the NS5A gene of HCV quasispecies populations from six patients being treated with IFN + RBV combined therapy[145]. Only one recombinant strain was detected in all patient quasispecies populations studied and its recombination crossover point was found within the protein kinase R (PKR)-binding region of NS5A[145]. This region has a particular importance since previous work by Enomoto et al[148] (1995) suggested that the genetic heterogeneity of the interferon sensitivity determining region domain of HCV NS5A (IFN sensitivity-determining region), linked to response to therapy in Japanese patients[148]. Although there seems not to be a consensus on this issue[85], the published information supports the hypothesis that an association indeed exists between NS5A and response to therapy[149-151]. Some reports suggest that HCV NS5A protein can act in vivo repressing PKR function, and presumably allowing HCV to escape the antiviral effects of interferon[1,3,152,153]. Analysing NS5A protein sequences of both the recombinant and putative parental-like virus provided evidence in favour of the possibility that the recombinant isolate might have acquired amino acids already known for being present in HCV strains resistant to interferon treatment[145]. The results of these studies support the fact that recombination cannot be denied as an evolutionary mechanism for generating diversity in HCV in vivo in patients undergoing antiviral therapy[145]. In spite of this fact, recombination does not seem to play a major role in the evolution of HCV quasispecies populations; at least this is what can be extrapolated from the study of NS5A genes, since only one recombinant isolate was found among all HCV quasispecies populations studied. Contrary to this finding, Sentandreu et al[146] (2008) identified a high frequency of intra-patient recombination events (18.01% of the 111 analysed patients) analysing a large data set of HCV sequences (around 17700) from intra-patient viral population[146]. They retrospectively studied NS5A and E1-E2 coding regions from samples isolated from two different sets of groups: HCV mono-infected patients, both naïve and non-responders to antiviral treatment; and HCV/HIV co-infected patients, both treatment-naïve and under HAART[146]. These authors found recombination within the E1-E2 region (9.1%), and within the NS5A region (9.6%), with specific areas being proposed as the crossover points. Although no structural analyses were performed in this study, these results are consistent with the implication of RNA secondary structure in favouring the hotspots or zones where recombination can occur within the HCV genome[49]. As per these results, where recombination intra-patient was found in 18% of the HCV infected patients studied, intra-quasispecies recombination events seem relatively frequent. Moreover, this might be an underestimation of the real frequency of HCV recombination due to the difficulty in detecting recombination events if they occur between genetically very similar variants as is the case of variants within a quasispecies[49,146]. More recently, Palmer et al[147] (2012) detected putative intra-subtype recombinants, as well as the likely ancestral parental donors. By retrospective clonal analysis they explored quasispecies evolution evaluated at the HVR1 region from serum samples isolated from the same chronically infected, treatment naïve individual which were collected over 9.6 years[147]. Their detailed analysis clearly documents the emergence, maintenance and final removal of HCV variants, which in this case has been demonstrated in a patient who did not undergo antiviral therapy highlighting the importance of HCV quasispecies dynamics even in absence of a clear selective pressure.
Even though the regions apparently involved in the crossover events seem to be different when comparing intergenotype vs intraquasispecies recombination, they seem to be quite conserved within each of the different categories. This might represent another indication of the importance of RNA secondary structure for recombination events to take place and might as well hint at a possible factor determining their occurrence.
Having detected so many recombinant strains (Table 2) proves that HCV is capable of successfully completing all the steps leading to this event: simultaneous infection of the same cell by different viral strains, simultaneous replication of both viral genomes, strand shift by the viral RNA polymerase without disturbing the correct reading frame, and encapsidation and release of the recombinant genomes as viable viral particles[49]. The resulting products will then be subjected to the same population processes governing the maintenance, growth or disappearance of new variants in a heterogeneous viral population[49,146].
If we analyse in depth the recombination events, we can consider that recombination in HCV may be underestimated. Why? Three different factors might account for this: Firstly, in recombination events between subtype viral strains, there is a trade-off between the capability of homologous recombination event to occur, and the intra-patient viral diversity, since homologous recombination requires a minimum length of sequence identity. Secondly, another trade-off occurs between the intra-patient viral diversity and the discrimination power to detect recombination with the different methods available. Finally, despite recombination events between different genotypes/subtypes co-infecting the same patient are probably easier to detect, they are less likely to occur, since the strains of different subtypes differ more between them than between those from the same subtype; this would imply a lower probability of template switching and moreover, if a recombination event does indeed happen, it will likely generate recombinant sequences less viable than the parental ones. Some or even all these factors acting in concert might explain why the frequency of recombinant HCV sequences reported to date is so low[49,111,124-137,140-147].
How relevant recombination for HCV long term evolution and its incidence in HCV infection is, has not been thoroughly investigated yet, but these findings support a potentially significant role for recombination by creating genetic variation through the reshuffling of independent variants[146]. Recombination may serve two opposite purposes: simply to explore a new genomic combination or to rescue viable genomes from debilitated parental ones[29,154]. Considering that recombination may influence vaccine development, virus control programs, patient management as well as antiviral therapies, it is clearly important to determine the extent to which this mechanism plays a role in HCV evolution[111].
RNA viruses exist as complex mutant distributions commonly known as viral quasispecies. This is the result of high mutation rates due to the lack of proofreading activities in their RNA-dependent RNA polymerases[87]. The evolution of the HCV quasispecies is a highly dynamic process that has therapeutic implications due to the continuous generation and selection of fitter variants within the quasispecies spectrum which might allow viruses to escape control by antiviral drugs and treatment[15,74]. Further studies on HCV quasispecies is needed in order to develop appropriate strategies for effective antiviral control[55]. HCV utilizes all known genetic mechanisms, including mutation and recombination. NGS technologies may represent an important improvement to identify key parameters in our understanding of HCV evolution in relation to current and new therapies against HCV.
P- Reviewer: Bare P, Sayiner AA S- Editor: Ji FF L- Editor: A E- Editor: Liu SQ
| 1. | Simmonds P. Genetic diversity and evolution of hepatitis C virus--15 years on. J Gen Virol. 2004;85:3173-3188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 622] [Cited by in RCA: 632] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 2. | Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21-S29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 354] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 3. | Pawlotsky JM. The nature of interferon-alpha resistance in hepatitis C virus infection. Curr Opin Infect Dis. 2003;16:587-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 4. | Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962-973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1070] [Cited by in RCA: 1083] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 5. | Rosen HR, Gretch DR. Hepatitis C virus: current understanding and prospects for future therapies. Mol Med Today. 1999;5:393-399. [PubMed] |
| 6. | Keyvani H, Fazlalipour M, Monavari SH, Mollaie HR. Hepatitis C virus--proteins, diagnosis, treatment and new approaches for vaccine development. Asian Pac J Cancer Prev. 2012;13:5931-5949. [PubMed] |
| 7. | Hu KQ, Vierling JM, Redeker AG. Viral, host and interferon-related factors modulating the effect of interferon therapy for hepatitis C virus infection. J Viral Hepat. 2001;8:1-18. [PubMed] |
| 8. | Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335-1374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2320] [Cited by in RCA: 2242] [Article Influence: 140.1] [Reference Citation Analysis (1)] |
| 9. | Kanda T, Imazeki F, Yokosuka O. New antiviral therapies for chronic hepatitis C. Hepatol Int. 2010;4:548-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 10. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol. 2011;55:245-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 889] [Cited by in RCA: 919] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 11. | Editors of the Drafting Committee for Hepatitis Management Guidelines: The Japan Society of Hepatology. Guidelines for the Management of Hepatitis C Virus Infection: First edition, May 2012, The Japan Society of Hepatology. Hepatol Res. 2013;43:1-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 12. | Available from: http: //www.who.int. |
| 13. | Lawitz EJ. Diagnosis and management of telaprevir-associated rash. Gastroenterol Hepatol (N Y). 2011;7:469-471. [PubMed] |
| 14. | Bartenschlager R, Penin F, Lohmann V, André P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011;19:95-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 337] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 15. | Cristina J, del Pilar Moreno M, Moratorio G. Hepatitis C virus genetic variability in patients undergoing antiviral therapy. Virus Res. 2007;127:185-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 16. | Varaklioti A, Vassilaki N, Georgopoulou U, Mavromara P. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J Biol Chem. 2002;277:17713-17721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 113] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 17. | Xu Z, Choi J, Lu W, Ou JH. Hepatitis C virus f protein is a short-lived protein associated with the endoplasmic reticulum. J Virol. 2003;77:1578-1583. [PubMed] |
| 18. | Ma HC, Lin TW, Li H, Iguchi-Ariga SM, Ariga H, Chuang YL, Ou JH, Lo SY. Hepatitis C virus ARFP/F protein interacts with cellular MM-1 protein and enhances the gene trans-activation activity of c-Myc. J Biomed Sci. 2008;15:417-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 19. | Shao SW, Wu WB, Bian ZQ, Yu JG, Zhao P, Zhao LJ, Zhu SY, Qi ZT. Hepatitis C virus F protein inhibits cell apoptosis by activation of intracellular NF-kappaB pathway. Hepatol Res. 2009;39:282-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Wu WB, Shao SW, Zhao LJ, Luan J, Cao J, Gao J, Zhu SY, Qi ZT. Hepatitis C virus F protein up-regulates c-myc and down-regulates p53 in human hepatoma HepG2 cells. Intervirology. 2007;50:341-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 21. | Friebe P, Bartenschlager R. Genetic analysis of sequences in the 3’ nontranslated region of hepatitis C virus that are important for RNA replication. J Virol. 2002;76:5326-5338. [PubMed] |
| 22. | Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194-206. [PubMed] |
| 23. | Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67-83. [PubMed] |
| 24. | Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 965] [Cited by in RCA: 982] [Article Influence: 89.3] [Reference Citation Analysis (1)] |
| 25. | Bukh J, Purcell RH, Miller RH. Sequence analysis of the 5’ noncoding region of hepatitis C virus. Proc Natl Acad Sci USA. 1992;89:4942-4946. [PubMed] |
| 26. | Piñeiro D, Martinez-Salas E. RNA structural elements of hepatitis C virus controlling viral RNA translation and the implications for viral pathogenesis. Viruses. 2012;4:2233-2250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Argentini C, Genovese D, Dettori S, Rapicetta M. HCV genetic variability: from quasispecies evolution to genotype classification. Future Microbiol. 2009;4:359-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 28. | Le Guillou-Guillemette H, Vallet S, Gaudy-Graffin C, Payan C, Pivert A, Goudeau A, Lunel-Fabiani F. Genetic diversity of the hepatitis C virus: impact and issues in the antiviral therapy. World J Gastroenterol. 2007;13:2416-2426. [PubMed] |
| 29. | Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1104] [Cited by in RCA: 1044] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 30. | Davis GL, Esteban-Mur R, Rustgi V, Hoefs J, Gordon SC, Trepo C, Shiffman ML, Zeuzem S, Craxi A, Ling MH. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. International Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1493-1499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 927] [Cited by in RCA: 895] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 31. | McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485-1492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2509] [Cited by in RCA: 2435] [Article Influence: 90.2] [Reference Citation Analysis (0)] |
| 32. | Poynard T, Marcellin P, Lee SS, Niederau C, Minuk GS, Ideo G, Bain V, Heathcote J, Zeuzem S, Trepo C. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group (IHIT). Lancet. 1998;352:1426-1432. [PubMed] |
| 33. | Ogata N, Alter HJ, Miller RH, Purcell RH. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:3392-3396. [PubMed] |
| 34. | Zeuzem S. Heterogeneous virologic response rates to interferon-based therapy in patients with chronic hepatitis C: who responds less well? Ann Intern Med. 2004;140:370-381. [PubMed] |
| 35. | Fried MW, Hadziyannis SJ. Treatment of chronic hepatitis C infection with peginterferons plus ribavirin. Semin Liver Dis. 2004;24 Suppl 2:47-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 54] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 36. | Ghany MG, Nelson DR, Strader DB, Thomas DL, Seeff LB. An update on treatment of genotype 1 chronic hepatitis C virus infection: 2011 practice guideline by the American Association for the Study of Liver Diseases. Hepatology. 2011;54:1433-1444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 803] [Cited by in RCA: 844] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 37. | Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, Poordad F, Goodman ZD, Sings HL, Boparai N. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1207-1217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1306] [Cited by in RCA: 1308] [Article Influence: 93.4] [Reference Citation Analysis (0)] |
| 38. | Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med. 2011;364:2405-2416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1905] [Cited by in RCA: 1862] [Article Influence: 133.0] [Reference Citation Analysis (0)] |
| 39. | Poordad F, McCone J, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1996] [Cited by in RCA: 1981] [Article Influence: 141.5] [Reference Citation Analysis (0)] |
| 40. | Zeuzem S, Andreone P, Pol S, Lawitz E, Diago M, Roberts S, Focaccia R, Younossi Z, Foster GR, Horban A. Telaprevir for retreatment of HCV infection. N Engl J Med. 2011;364:2417-2428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1230] [Cited by in RCA: 1214] [Article Influence: 86.7] [Reference Citation Analysis (0)] |
| 41. | Rosenquist Å, Samuelsson B, Johansson PO, Cummings MD, Lenz O, Raboisson P, Simmen K, Vendeville S, de Kock H, Nilsson M. Discovery and development of simeprevir (TMC435), a HCV NS3/4A protease inhibitor. J Med Chem. 2014;57:1673-1693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 158] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 42. | Sofia MJ, Bao D, Chang W, Du J, Nagarathnam D, Rachakonda S, Reddy PG, Ross BS, Wang P, Zhang HR. Discovery of a β-d-2’-deoxy-2’-α-fluoro-2’-β-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J Med Chem. 2010;53:7202-7218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 494] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 43. | Everson GT, Sims KD, Rodriguez-Torres M, Hézode C, Lawitz E, Bourlière M, Loustaud-Ratti V, Rustgi V, Schwartz H, Tatum H. Efficacy of an interferon- and ribavirin-free regimen of daclatasvir, asunaprevir, and BMS-791325 in treatment-naive patients with HCV genotype 1 infection. Gastroenterology. 2014;146:420-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 168] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 44. | Fontana RJ, Hughes EA, Bifano M, Appelman H, Dimitrova D, Hindes R, Symonds WT. Sofosbuvir and daclatasvir combination therapy in a liver transplant recipient with severe recurrent cholestatic hepatitis C. Am J Transplant. 2013;13:1601-1605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 114] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 45. | Jacobson IM, Gordon SC, Kowdley KV, Yoshida EM, Rodriguez-Torres M, Sulkowski MS, Shiffman ML, Lawitz E, Everson G, Bennett M. Sofosbuvir for hepatitis C genotype 2 or 3 in patients without treatment options. N Engl J Med. 2013;368:1867-1877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 846] [Cited by in RCA: 838] [Article Influence: 69.8] [Reference Citation Analysis (0)] |
| 46. | Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, Davis MN, Kayali Z, Reddy KR. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;368:1878-1887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1322] [Cited by in RCA: 1325] [Article Influence: 110.4] [Reference Citation Analysis (0)] |
| 47. | Sulkowski MS, Gardiner DF, Rodriguez-Torres M, Reddy KR, Hassanein T, Jacobson I, Lawitz E, Lok AS, Hinestrosa F, Thuluvath PJ. Daclatasvir plus sofosbuvir for previously treated or untreated chronic HCV infection. N Engl J Med. 2014;370:211-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 933] [Cited by in RCA: 911] [Article Influence: 82.8] [Reference Citation Analysis (0)] |
| 48. | Cristina J. Genetic diversity and evolution of hepatitis C virus in the Latin American region. J Clin Virol. 2005;34 Suppl 2:S1-S7. [PubMed] |
| 49. | González-Candelas F, López-Labrador FX, Bracho MA. Recombination in hepatitis C virus. Viruses. 2011;3:2006-2024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 50. | Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631-1648. [PubMed] |
| 51. | Kim T, Mudry RA, Rexrode CA, Pathak VK. Retroviral mutation rates and A-to-G hypermutations during different stages of retroviral replication. J Virol. 1996;70:7594-7602. [PubMed] |
| 52. | Biebricher CK, Eigen M. The error threshold. Virus Res. 2005;107:117-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 131] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 53. | Chambers TJ, Fan X, Droll DA, Hembrador E, Slater T, Nickells MW, Dustin LB, Dibisceglie AM. Quasispecies heterogeneity within the E1/E2 region as a pretreatment variable during pegylated interferon therapy of chronic hepatitis C virus infection. J Virol. 2005;79:3071-3083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 54. | Domingo E, Baranowski E, Ruiz-Jarabo CM, Martín-Hernández AM, Sáiz JC, Escarmís C. Quasispecies structure and persistence of RNA viruses. Emerg Infect Dis. 1998;4:521-527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 140] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 55. | Domingo E, Escarmís C, Lázaro E, Manrubia SC. Quasispecies dynamics and RNA virus extinction. Virus Res. 2005;107:129-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 85] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 56. | Feliu A, Gay E, García-Retortillo M, Saiz JC, Forns X. Evolution of hepatitis C virus quasispecies immediately following liver transplantation. Liver Transpl. 2004;10:1131-1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 47] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 57. | Laskus T, Wilkinson J, Gallegos-Orozco JF, Radkowski M, Adair DM, Nowicki M, Operskalski E, Buskell Z, Seeff LB, Vargas H. Analysis of hepatitis C virus quasispecies transmission and evolution in patients infected through blood transfusion. Gastroenterology. 2004;127:764-776. [PubMed] |
| 58. | Martell M, Esteban JI, Quer J, Genescà J, Weiner A, Esteban R, Guardia J, Gómez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225-3229. [PubMed] |
| 59. | Eigen M, Gardiner W, Schuster P, Winkler-Oswatitsch R. The origin of genetic information. Sci Am. 1981;244:88-92, 96, et passim. [PubMed] |
| 60. | Eigen M, Schuster P. The hypercycle. A principle of natural self-organization. Part A: Emergence of the hypercycle. Naturwissenschaften. 1977;64:541-565. [PubMed] |
| 61. | Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465-523. [PubMed] |
| 62. | Domingo E, Martin V, Perales C, Grande-Pérez A, García-Arriaza J, Arias A. Viruses as quasispecies: biological implications. Curr Top Microbiol Immunol. 2006;299:51-82. [PubMed] |
| 63. | Pellerin M, Lopez-Aguirre Y, Penin F, Dhumeaux D, Pawlotsky JM. Hepatitis C virus quasispecies variability modulates nonstructural protein 5A transcriptional activation, pointing to cellular compartmentalization of virus-host interactions. J Virol. 2004;78:4617-4627. [PubMed] |
| 64. | Domingo E, Gomez J. Quasispecies and its impact on viral hepatitis. Virus Res. 2007;127:131-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 98] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 65. | Martín V, Domingo E. Influence of the mutant spectrum in viral evolution: focused selection of antigenic variants in a reconstructed viral quasispecies. Mol Biol Evol. 2008;25:1544-1554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 66. | Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344-348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 954] [Cited by in RCA: 831] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 67. | Wilke CO. Quasispecies theory in the context of population genetics. BMC Evol Biol. 2005;5:44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 184] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 68. | Elena SF, Sanjuán R. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J Virol. 2005;79:11555-11558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 220] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 69. | Holmes EC. Error thresholds and the constraints to RNA virus evolution. Trends Microbiol. 2003;11:543-546. [PubMed] |
| 70. | Arnold JJ, Vignuzzi M, Stone JK, Andino R, Cameron CE. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J Biol Chem. 2005;280:25706-25716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 134] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 71. | Pfeiffer JK, Kirkegaard K. Ribavirin resistance in hepatitis C virus replicon-containing cell lines conferred by changes in the cell line or mutations in the replicon RNA. J Virol. 2005;79:2346-2355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 67] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 72. | Vignuzzi M, Stone JK, Andino R. Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications. Virus Res. 2005;107:173-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 121] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 73. | Fint SJEL, Racaniello VR, Skalka AM. Principles of virology. Third ed. Washington, DC, United States of America: ASM Press 2009; 569. |
| 74. | Pawlotsky JM. Hepatitis C virus population dynamics during infection. Curr Top Microbiol Immunol. 2006;299:261-284. [PubMed] |
| 75. | Baccam P, Thompson RJ, Fedrigo O, Carpenter S, Cornette JL. PAQ: Partition Analysis of Quasispecies. Bioinformatics. 2001;17:16-22. [PubMed] |
| 76. | Cody SH, Nainan OV, Garfein RS, Meyers H, Bell BP, Shapiro CN, Meeks EL, Pitt H, Mouzin E, Alter MJ. Hepatitis C virus transmission from an anesthesiologist to a patient. Arch Intern Med. 2002;162:345-350. [PubMed] |
| 77. | Gretch DR, Polyak SJ, Wilson JJ, Carithers RL, Perkins JD, Corey L. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J Virol. 1996;70:7622-7631. [PubMed] |
| 78. | Lin HJ, Seeff LB, Barbosa L, Hollinger FB. Occurrence of identical hypervariable region 1 sequences of hepatitis C virus in transfusion recipients and their respective blood donors: divergence over time. Hepatology. 2001;34:424-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 79. | Manzin A, Solforosi L, Debiaggi M, Zara F, Tanzi E, Romanò L, Zanetti AR, Clementi M. Dominant role of host selective pressure in driving hepatitis C virus evolution in perinatal infection. J Virol. 2000;74:4327-4334. [PubMed] |
| 80. | Weiner AJ, Thaler MM, Crawford K, Ching K, Kansopon J, Chien DY, Hall JE, Hu F, Houghton M. A unique, predominant hepatitis C virus variant found in an infant born to a mother with multiple variants. J Virol. 1993;67:4365-4368. [PubMed] |
| 81. | Hijikata M, Mizuno K, Rikihisa T, Shimizu YK, Iwamoto A, Nakajima N, Yoshikura H. Selective transmission of hepatitis C virus in vivo and in vitro. Arch Virol. 1995;140:1623-1628. [PubMed] |
| 82. | Kojima M, Osuga T, Tsuda F, Tanaka T, Okamoto H. Influence of antibodies to the hypervariable region of E2/NS1 glycoprotein on the selective replication of hepatitis C virus in chimpanzees. Virology. 1994;204:665-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 49] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 83. | Cuevas JM, Torres-Puente M, Jiménez-Hernández N, Bracho MA, García-Robles I, Carnicer F, Olmo JD, Ortega E, González-Candelas F, Moya A. Combined therapy of interferon plus ribavirin promotes multiple adaptive solutions in hepatitis C virus. J Med Virol. 2009;81:650-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 84. | Cuevas JM, Gonzalez M, Torres-Puente M, Jiménez-Hernández N, Bracho MA, García-Robles I, González-Candelas F, Moya A. The role of positive selection in hepatitis C virus. Infect Genet Evol. 2009;9:860-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 85. | Puig-Basagoiti F, Forns X, Furcić I, Ampurdanés S, Giménez-Barcons M, Franco S, Sánchez-Tapias JM, Saiz JC. Dynamics of hepatitis C virus NS5A quasispecies during interferon and ribavirin therapy in responder and non-responder patients with genotype 1b chronic hepatitis C. J Gen Virol. 2005;86:1067-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 86. | Ueda E, Enomoto N, Sakamoto N, Hamano K, Sato C, Izumi N, Watanabe M. Changes of HCV quasispecies during combination therapy with interferon and ribavirin. Hepatol Res. 2004;29:89-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 87. | Ojosnegros S, Agudo R, Sierra M, Briones C, Sierra S, González-López C, Domingo E, Cristina J. Topology of evolving, mutagenized viral populations: quasispecies expansion, compression, and operation of negative selection. BMC Evol Biol. 2008;8:207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 88. | Torres-Puente M, Cuevas JM, Jiménez-Hernández N, Bracho MA, García-Robles I, Wrobel B, Carnicer F, Del Olmo J, Ortega E, Moya A. Genetic variability in hepatitis C virus and its role in antiviral treatment response. J Viral Hepat. 2008;15:188-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 89. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2776] [Cited by in RCA: 2723] [Article Influence: 170.2] [Reference Citation Analysis (0)] |
| 90. | Rallón NI, Naggie S, Benito JM, Medrano J, Restrepo C, Goldstein D, Shianna KV, Vispo E, Thompson A, McHutchison J. Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS. 2010;24:F23-F29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 91. | Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100-1104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1505] [Cited by in RCA: 1504] [Article Influence: 94.0] [Reference Citation Analysis (0)] |
| 92. | Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1779] [Cited by in RCA: 1775] [Article Influence: 110.9] [Reference Citation Analysis (0)] |
| 93. | Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, Hong L, McKenzie A, Patel K, Shianna KV. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888-1896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 303] [Cited by in RCA: 314] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 94. | Yuan H, Adams-Huet B, Petersen T, Attar N, Lee WM, Jain MK. A single nucleotide polymorphism in IL28B affects viral evolution of hepatitis C quasispecies after pegylated interferon and ribavirin therapy. J Med Virol. 2012;84:1913-1919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 95. | Aparicio E, Franco S, Parera M, Andrés C, Tural C, Clotet B, Martínez MA. Complexity and catalytic efficiency of hepatitis C virus (HCV) NS3 and NS4A protease quasispecies influence responsiveness to treatment with pegylated interferon plus ribavirin in HCV/HIV-coinfected patients. J Virol. 2011;85:5961-5969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 96. | Miura M, Maekawa S, Kadokura M, Sueki R, Komase K, Shindo H, Ohmori T, Kanayama A, Shindo K, Amemiya F. Analysis of viral amino acids sequences and the IL28B SNP influencing the development of hepatocellular carcinoma in chronic hepatitis C. Hepatol Int. 2011;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 97. | Valenti L, Pulixi E, La Spina S. IL28B, HCV core mutations, and hepatocellular carcinoma: does host genetic make-up shape viral evolution in response to immunity? Hepa Inter. 2012;6:356-359. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 98. | Bushman FD, Hoffmann C, Ronen K, Malani N, Minkah N, Rose HM, Tebas P, Wang GP. Massively parallel pyrosequencing in HIV research. AIDS. 2008;22:1411-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 99. | Eriksson N, Pachter L, Mitsuya Y, Rhee SY, Wang C, Gharizadeh B, Ronaghi M, Shafer RW, Beerenwinkel N. Viral population estimation using pyrosequencing. PLoS Comput Biol. 2008;4:e1000074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 153] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 100. | Margeridon-Thermet S, Shulman NS, Ahmed A, Shahriar R, Liu T, Wang C, Holmes SP, Babrzadeh F, Gharizadeh B, Hanczaruk B. Ultra-deep pyrosequencing of hepatitis B virus quasispecies from nucleoside and nucleotide reverse-transcriptase inhibitor (NRTI)-treated patients and NRTI-naive patients. J Infect Dis. 2009;199:1275-1285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 170] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 101. | Luciani F, Bull RA, Lloyd AR. Next generation deep sequencing and vaccine design: today and tomorrow. Trends Biotechnol. 2012;30:443-452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 102. | Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4795] [Cited by in RCA: 4100] [Article Influence: 256.3] [Reference Citation Analysis (0)] |
| 103. | Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet. 2010;19:R227-R240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 611] [Cited by in RCA: 636] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 104. | Kelleher RJ, Geigenmüller U, Hovhannisyan H, Trautman E, Pinard R, Rathmell B, Carpenter R, Margulies D. High-throughput sequencing of mGluR signaling pathway genes reveals enrichment of rare variants in autism. PLoS One. 2012;7:e35003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 105. | Svarovskaia ES, Martin R, McHutchison JG, Miller MD, Mo H. Abundant drug-resistant NS3 mutants detected by deep sequencing in hepatitis C virus-infected patients undergoing NS3 protease inhibitor monotherapy. J Clin Microbiol. 2012;50:3267-3274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 106. | Leitner T, Halapi E, Scarlatti G, Rossi P, Albert J, Fenyö EM, Uhlén M. Analysis of heterogeneous viral populations by direct DNA sequencing. Biotechniques. 1993;15:120-127. [PubMed] |
| 107. | Fonseca-Coronado S, Escobar-Gutiérrez A, Ruiz-Tovar K, Cruz-Rivera MY, Rivera-Osorio P, Vazquez-Pichardo M, Carpio-Pedroza JC, Ruíz-Pacheco JA, Cazares F, Vaughan G. Specific detection of naturally occurring hepatitis C virus mutants with resistance to telaprevir and boceprevir (protease inhibitors) among treatment-naïve infected individuals. J Clin Microbiol. 2012;50:281-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 108. | Hiraga N, Abe H, Imamura M, Tsuge M, Takahashi S, Hayes CN, Ochi H, Tateno C, Yoshizato K, Nakamura Y. Impact of viral amino acid substitutions and host interleukin-28b polymorphism on replication and susceptibility to interferon of hepatitis C virus. Hepatology. 2011;54:764-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 109. | Ninomiya M, Ueno Y, Funayama R, Nagashima T, Nishida Y, Kondo Y, Inoue J, Kakazu E, Kimura O, Nakayama K. Use of illumina deep sequencing technology to differentiate hepatitis C virus variants. J Clin Microbiol. 2012;50:857-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 110. | Escobar-Gutiérrez A, Vazquez-Pichardo M, Cruz-Rivera M, Rivera-Osorio P, Carpio-Pedroza JC, Ruíz-Pacheco JA, Ruiz-Tovar K, Vaughan G. Identification of hepatitis C virus transmission using a next-generation sequencing approach. J Clin Microbiol. 2012;50:1461-1463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 111. | Moreno P, Alvarez M, López L, Moratorio G, Casane D, Castells M, Castro S, Cristina J, Colina R. Evidence of recombination in Hepatitis C Virus populations infecting a hemophiliac patient. Virol J. 2009;6:203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 112. | Worobey M, Holmes EC. Evolutionary aspects of recombination in RNA viruses. J Gen Virol. 1999;80:2535-2543. [PubMed] |
| 113. | Lai MM. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61-79. [PubMed] |
| 114. | Copper PD, Steiner-Pryor A, Scotti PD, Delong D. On the nature of poliovirus genetic recombinants. J Gen Virol. 1974;23:41-49. [PubMed] |
| 115. | Kew O, Nottay B. Evolution of the oral polio vaccine strains in humans occurs by both mutation and intra-molecular recombination. Modern approaches to vaccines. Molecular and chemical basis of virus virulence and immunogenicity. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory 1984; 457. |
| 116. | Diemer GS, Stedman KM. A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol Direct. 2012;7:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 124] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 117. | Becher P, Orlich M, Thiel HJ. RNA recombination between persisting pestivirus and a vaccine strain: generation of cytopathogenic virus and induction of lethal disease. J Virol. 2001;75:6256-6264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 92] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 118. | Holmes EC, Worobey M, Rambaut A. Phylogenetic evidence for recombination in dengue virus. Mol Biol Evol. 1999;16:405-409. [PubMed] |
| 119. | Tolou HJ, Couissinier-Paris P, Durand JP, Mercier V, de Pina JJ, de Micco P, Billoir F, Charrel RN, de Lamballerie X. Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J Gen Virol. 2001;82:1283-1290. [PubMed] |
| 120. | Twiddy SS, Holmes EC. The extent of homologous recombination in members of the genus Flavivirus. J Gen Virol. 2003;84:429-440. [PubMed] |
| 121. | Uzcategui NY, Camacho D, Comach G, Cuello de Uzcategui R, Holmes EC, Gould EA. Molecular epidemiology of dengue type 2 virus in Venezuela: evidence for in situ virus evolution and recombination. J Gen Virol. 2001;82:2945-2953. [PubMed] |
| 122. | Worobey M, Holmes EC. Homologous recombination in GB virus C/hepatitis G virus. Mol Biol Evol. 2001;18:254-261. [PubMed] |
| 123. | Yun Z, Lara C, Johansson B, Lorenzana de Rivera I, Sönnerborg A. Discrepancy of hepatitis C virus genotypes as determined by phylogenetic analysis of partial NS5 and core sequences. J Med Virol. 1996;49:155-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 124. | Kalinina O, Norder H, Mukomolov S, Magnius LO. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J Virol. 2002;76:4034-4043. [PubMed] |
| 125. | Moreau I, Hegarty S, Levis J, Sheehy P, Crosbie O, Kenny-Walsh E, Fanning LJ. Serendipitous identification of natural intergenotypic recombinants of hepatitis C in Ireland. Virol J. 2006;3:95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 126. | Kurbanov F, Tanaka Y, Avazova D, Khan A, Sugauchi F, Kan N, Kurbanova-Khudayberganova D, Khikmatullaeva A, Musabaev E, Mizokami M. Detection of hepatitis C virus natural recombinant RF1_2k/1b strain among intravenous drug users in Uzbekistan. Hepatol Res. 2008;38:457-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 127. | Demetriou VL, Kyriakou E, Kostrikis LG. Near-full genome characterisation of two natural intergenotypic 2k/1b recombinant hepatitis C virus isolates. Adv Virol. 2011;2011:710438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 128. | Morel V, Descamps V, François C, Fournier C, Brochot E, Capron D, Duverlie G, Castelain S. Emergence of a genomic variant of the recombinant 2k/1b strain during a mixed Hepatitis C infection: a case report. J Clin Virol. 2010;47:382-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 129. | Tallo T, Norder H, Tefanova V, Krispin T, Schmidt J, Ilmoja M, Orgulas K, Pruunsild K, Priimägi L, Magnius LO. Genetic characterization of hepatitis C virus strains in Estonia: fluctuations in the predominating subtype with time. J Med Virol. 2007;79:374-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 130. | Noppornpanth S, Lien TX, Poovorawan Y, Smits SL, Osterhaus AD, Haagmans BL. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J Virol. 2006;80:7569-7577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 119] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 131. | Lee YM, Lin HJ, Chen YJ, Lee CM, Wang SF, Chang KY, Chen TL, Liu HF, Chen YM. Molecular epidemiology of HCV genotypes among injection drug users in Taiwan: Full-length sequences of two new subtype 6w strains and a recombinant form_2b6w. J Med Virol. 2010;82:57-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 132. | Legrand-Abravanel F, Claudinon J, Nicot F, Dubois M, Chapuy-Regaud S, Sandres-Saune K, Pasquier C, Izopet J. New natural intergenotypic (2/5) recombinant of hepatitis C virus. J Virol. 2007;81:4357-4362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 94] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 133. | Du H, Qi Y, Hao F, Huang Y, Mao L, Ji S, Huang M, Qin C, Yan R, Zhu X. Complex patterns of HCV epidemic in Suzhou: evidence for dual infection and HCV recombination in East China. J Clin Virol. 2012;54:207-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 134. | Bhattacharya D, Accola MA, Ansari IH, Striker R, Rehrauer WM. Naturally occurring genotype 2b/1a hepatitis C virus in the United States. Virol J. 2011;8:458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 135. | Yokoyama K, Takahashi M, Nishizawa T, Nagashima S, Jirintai S, Yotsumoto S, Okamoto H, Momoi MY. Identification and characterization of a natural inter-genotypic (2b/1b) recombinant hepatitis C virus in Japan. Arch Virol. 2011;156:1591-1601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 136. | Hoshino H, Hino K, Miyakawa H, Takahashi K, Akbar SM, Mishiro S. Inter-genotypic recombinant hepatitis C virus strains in Japan noted by discrepancies between immunoassay and sequencing. J Med Virol. 2012;84:1018-1024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 137. | Kageyama S, Agdamag DM, Alesna ET, Leaño PS, Heredia AM, Abellanosa-Tac-An IP, Jereza LD, Tanimoto T, Yamamura J, Ichimura H. A natural inter-genotypic (2b/1b) recombinant of hepatitis C virus in the Philippines. J Med Virol. 2006;78:1423-1428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 73] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 138. | Morel V, Fournier C, François C, Brochot E, Helle F, Duverlie G, Castelain S. Genetic recombination of the hepatitis C virus: clinical implications. J Viral Hepat. 2011;18:77-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 139. | Kalinina O, Norder H, Magnius LO. Full-length open reading frame of a recombinant hepatitis C virus strain from St Petersburg: proposed mechanism for its formation. J Gen Virol. 2004;85:1853-1857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 60] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 140. | Calado RA, Rocha MR, Parreira R, Piedade J, Venenno T, Esteves A. Hepatitis C virus subtypes circulating among intravenous drug users in Lisbon, Portugal. J Med Virol. 2011;83:608-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 141. | Colina R, Casane D, Vasquez S, García-Aguirre L, Chunga A, Romero H, Khan B, Cristina J. Evidence of intratypic recombination in natural populations of hepatitis C virus. J Gen Virol. 2004;85:31-37. [PubMed] |
| 142. | Cristina J, Colina R. Evidence of structural genomic region recombination in Hepatitis C virus. Virol J. 2006;3:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 143. | Ross RS, Verbeeck J, Viazov S, Lemey P, Van Ranst M, Roggendorf M. Evidence for a complex mosaic genome pattern in a full-length hepatitis C virus sequence. Evol Bioinform Online. 2008;4:249-254. [PubMed] |
| 144. | Shi W, Freitas IT, Zhu C, Zheng W, Hall WW, Higgins DG. Recombination in hepatitis C virus: identification of four novel naturally occurring inter-subtype recombinants. PLoS One. 2012;7:e41997. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 145. | Moreno MP, Casane D, López L, Cristina J. Evidence of recombination in quasispecies populations of a Hepatitis C Virus patient undergoing anti-viral therapy. Virol J. 2006;3:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 146. | Sentandreu V, Jiménez-Hernández N, Torres-Puente M, Bracho MA, Valero A, Gosalbes MJ, Ortega E, Moya A, González-Candelas F. Evidence of recombination in intrapatient populations of hepatitis C virus. PLoS One. 2008;3:e3239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 54] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 147. | Palmer BA, Moreau I, Levis J, Harty C, Crosbie O, Kenny-Walsh E, Fanning LJ. Insertion and recombination events at hypervariable region 1 over 9.6 years of hepatitis C virus chronic infection. J Gen Virol. 2012;93:2614-2624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 148. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest. 1995;96:224-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 452] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 149. | Gale MJ, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 631] [Cited by in RCA: 608] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 150. | Giménez-Barcons M, Franco S, Suárez Y, Forns X, Ampurdanès S, Puig-Basagoiti F, Sánchez-Fueyo A, Barrera JM, Llovet JM, Bruix J. High amino acid variability within the NS5A of hepatitis C virus (HCV) is associated with hepatocellular carcinoma in patients with HCV-1b-related cirrhosis. Hepatology. 2001;34:158-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 151. | Witherell GW, Beineke P. Statistical analysis of combined substitutions in nonstructural 5A region of hepatitis C virus and interferon response. J Med Virol. 2001;63:8-16. [PubMed] |
| 152. | Gale M, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208-5218. [PubMed] |
| 153. | Pawlotsky JM, Germanidis G, Neumann AU, Pellerin M, Frainais PO, Dhumeaux D. Interferon resistance of hepatitis C virus genotype 1b: relationship to nonstructural 5A gene quasispecies mutations. J Virol. 1998;72:2795-2805. [PubMed] |
| 154. | Costa-Mattioli M, Ferré V, Casane D, Perez-Bercoff R, Coste-Burel M, Imbert-Marcille BM, Andre EC, Bressollette-Bodin C, Billaudel S, Cristina J. Evidence of recombination in natural populations of hepatitis A virus. Virology. 2003;311:51-59. [PubMed] |
| 155. | Yusim K, Richardson R, Tao N, Dalwani A, Agrawal A, Szinger J, Funkhouser R, Korber B, Kuiken C. Los alamos hepatitis C immunology database. Appl Bioinformatics. 2005;4:217-225. [PubMed] |