Published online Oct 18, 2015. doi: 10.4254/wjh.v7.i23.2482
Peer-review started: May 6, 2015
First decision: July 29, 2015
Revised: August 18, 2015
Accepted: September 29, 2015
Article in press: September 30, 2015
Published online: October 18, 2015
Processing time: 173 Days and 20.5 Hours
AIM: To review published methods for detection of hepatitis B virus (HBV) infection.
METHODS: A thorough search on Medline database was conducted to find original articles describing different methods or techniques of detection of HBV, which are published in English in last 10 years. Articles outlining methods of detection of mutants or drug resistance were excluded. Full texts and abstracts (if full text not available) were reviewed thoroughly. Manual search of references of retrieved articles were also done. We extracted data on different samples and techniques of detection of HBV, their sensitivity (Sn), specificity (Sp) and applicability.
RESULTS: A total of 72 studies were reviewed. HBV was detected from dried blood/plasma spots, hepatocytes, ovarian tissue, cerumen, saliva, parotid tissue, renal tissue, oocytes and embryos, cholangiocarcinoma tissue, etc. Sensitivity of dried blood spot for detecting HBV was > 90% in all the studies. In case of seronegative patients, HBV DNA or serological markers have been detected from hepatocytes or renal tissue in many instances. Enzyme linked immunosorbent assay and Chemiluminescent immunoassay (CLIA) are most commonly used serological tests for detection. CLIA systems are also used for quantitation. Molecular techniques are used qualitatively as well as for quantitative detection. Among the molecular techniques version 2.0 of the CobasAmpliprep/CobasTaqMan assay and Abbott’s real time polymerase chain reaction kit were found to be most sensitive with a lower detection limit of only 6.25 IU/mL and 1.48 IU/mL respectively.
CONCLUSION: Serological and molecular assays are predominant and reliable methods for HBV detection. Automated systems are highly sensitive and quantify HBV DNA and serological markers for monitoring.
Core tip: The article was aimed to review published methods of detection of hepatitis B virus (HBV) infection. A thorough search on medline database was conducted and 72 studies were included. It was observed that HBV can be detected reliably from dried blood spot (sensitivity > 90%). Serological and Molecular assays are predominant and reliable methods. Chemiluminescent immunoassay is more sensitive than Enzyme linked immunosorbent assay. Rapid tests are useful for screening. Real time polymerase chain reaction (PCR), branched DNA probe assays are principal methods for quantitation. Automated systems are more sensitive compared to in house assays. Abbott real time PCR was found to be most sensitive with a lower detection limit of only 1.48 IU/mL.
- Citation: Ghosh M, Nandi S, Dutta S, Saha MK. Detection of hepatitis B virus infection: A systematic review. World J Hepatol 2015; 7(23): 2482-2491
- URL: https://www.wjgnet.com/1948-5182/full/v7/i23/2482.htm
- DOI: https://dx.doi.org/10.4254/wjh.v7.i23.2482
The enigma of hepatitis started long back in 3rd millennium B.C. in Sumeria with the first description of jaundice. Epidemic icterus was reported initially by Hippocrates (460 to 375 B.C.) followed by various vague descriptions by Greeks and Romans. But the perception of transmissibility came into acceptance with the spread of syphilis by Columbus and crew in 1494[1]. Further innumerable epidemics occurred in recipients of vaccines containing human serum or lymph. The largest was in 1942 among United States Army personnel, who received yellow fever vaccine containing human serum[2]. In 1940’s several experiments in human volunteers by Cameron (1943)[3], Mac Callam (1944)[2,3], Paul et al[4] (1945) confirmed the viral etiology of hepatitis. 2 distinct clinicoepidemiological forms of viral hepatitis: Serum hepatitis and infectious hepatitis was evidenced by the study of Krugman[3] in the late 1950’s and 1960’s at WillowBrook State Schools, NewYork[4]. But the most important exploration in the history of viral hepatitis was of Sir B. Blumberg in the year 1960’s. He observed an unusual reaction between the serum of hemophiliac patient and that of Australian aborigine in immunodiffusion gel and named this unusual protein Australia Antigen (Au Ag) which was further linked to viral hepatitis[5]. In 1968 Alfred Prince also described a serum antigen (SH Ag) in the serum of post transfusion patients[6]. These Au Antigen and SH Ag were soon found to be identical[1]. In the year 1970, Dane et al[7] discovered 42 nm sized virus like particles while observing Au Ag immune complexes under Electron Microscope. It was obvious that Au Ag was the surface antigen, whereas the Dane particles were actual virus. Hence the Au Ag was named hepatitis B surface antigen (HBsAg). By treating these “Dane particles” with mild detergents core particles were released by Almeida et al[8]. Antibody present in post hepatitis serum reacted with these inner/core particles. Researchers could comprehend soon that to assess the infectivity of the disease mere presence of HBsAg is not sufficient. In 1972 hepatitis B e antigen (HBeAg) was identified by Magnius et al[9] which helped to differentiate between highly infectious and less infectious forms. Simultaneously hepatitis B virus (HBV) DNA was identified by Robinson et al[10]. In earlier days infection with HBV was detected by demonstration of antibody titer by Complement Fixation Test[2]. The first solid phase sandwich radio immunoassay named Ausria 125 was developed by Ling et al[11] at Abbott Laboratories (North Chicago). This highly sensitive detection method became a major discovery in the diagnosis of viral transfusion hepatitis and screening of blood donors[2]. Since then innumerable serological and molecular methods have been developed for diagnosing HBV. This article provides an overview of detection of HBV infection employing different techniques.
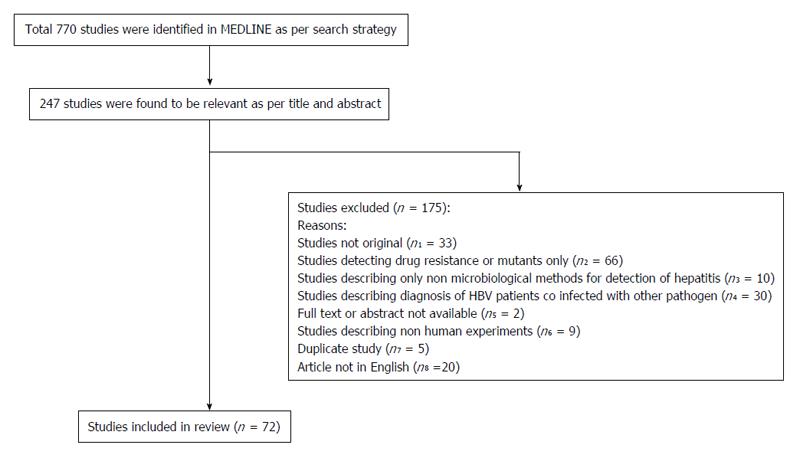
The review followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines[12]. A protocol was developed and pertinent studies were identified as per inclusion and exclusion criteria (Figure 1). A thorough search on Medline database was conducted for articles related to diagnosis of HBV infection. The search was based on the following keywords or medical subject heading terms in the database: (detection[All Fields] AND (“hepatitis B virus”[MeSH Terms] OR “hepatitis B virus”[All Fields]) AND (“infection”[MeSH Terms] OR “infection”[All Fields])) AND (“2005/04/02”[PDat] : “2015/03/30”[PDat]).
The inclusion criteria were (1) articles describing methods or techniques of diagnosis of HBV; (2) published in English language; and (3) published in last 10 years. Articles were excluded if (1) study not original (review or editorial or case report); (2) studies describing methods of detection of drug resistance or mutants; (3) studies describing non microbiological serum biomarkers for diagnosing hepatitis only; (4) studies describing diagnosis of patients coinfected with other viruses [hepatitis C virus (HCV), human immunodeficiency virus, etc.] or bacteria (Mycobacterium tuberculosis); and (5) full text or abstract not available in Medline.
HBV is most commonly detected in serum or whole blood. But we retrieved total 17 studies, which have been published in MEDLINE in last 10 years, discussing about detection of HBV from samples other than serum or whole blood. Researchers have detected HBV from dried blood/plasma spots[13-16], hepatocytes[17-20], ovarian tissue[21], cerumen[22,23], saliva[24], parotid tissue[25], renal tissue[26], oocytes and embryos[27,28], cholangiocarcinoma tissue[29], etc. (Table 1).
| Ref. | Year of publication | Sample used | Method used | Comments |
| Mendy et al[13] | 2005 | Dried blood spots along with serum | HBsAg detected by determine (TM) HBsAg | Comparison of DBS results with serum testing results: Sn 96%, Sp 100% |
| Chen et al[21] | 2005 | Ovarian tissue | HBsAg and HBcAg detected by immunocytochemistry and HBV DNA by PCR | Positivity rate of HBV DNA was 58.3% |
| van der Laan et al[17] | 2007 | Hepatocytes | Flow cytometric quantitation | A significant correlation was found between the percentage of infected hepatocytes and the intracellular expression level of HBsAg (R = 0.841, P < 0.001) |
| Goh et al[22] | 2008 | Cerumen and otorrhoea samples along with serum | HBsAg and HBeAg were detected by Enzyme Immunoassay and HBV DNA was detected by quantitative PCR | HBV DNA was detected in 66.7% of cerumen samples and 100% of otorrhoea samples |
| Chen et al[25] | 2009 | Parotid tissue | Serological markers by immunocytrochemistry and HBV DNA by PCR | Overall positivity rate was 54.5% to 58.3% |
| Nuriya et al[18] | 2010 | Hepatocyres | PCR - In situ hybridisation | All hepatocytes were infected with HBV in chronic liver disease |
| Villar et al[14] | 2011 | Dried blood spots | HBsAg, anti-HBc, and anti-HBs were detected by ELISA | Sn was 90.5%, 97.6%, and 78% for anti-HBc, HBsAg, anti-HBs assays, and Sp was 92.6%, 96.7%, and 97.3% for anti-HBc, HBsAg, and anti-HBs assays, respectively |
| Wu et al[29] | 2012 | Paraffin embedded intrahepatic and extrahepaticcholangiocarcinoma tissue | HBV DNA by nested PCR and HBV related antigens by immunohistochemistry method | HBV DNA and HBV antigens were detected significantly in cases of intrahepatic cholangiocarcinoma |
| Arora et al[24] | 2012 | Saliva | HBsAg was detected by ELISA | Sn 74.29% and Sp 100% |
| Cobo et al[27] | 2012 | Spent culture media and liquid nitrogen samples of oocytes and embryos | Reverse transcriptase PCR | Viral sequences were not detected in these samples from seropositive patients |
| Ye et al[28] | 2013 | Discarded test tube embryos from mothers with chronic HBV infection undergoing in vitro fertilization treatment | Single cell reverse transcriptase PCR | Detection rate was 13.2% |
| Eftekharian et al[23] | 2013 | Cerumen along with serum | HBV DNA was detected by PCR | HBV DNA was detected in 6.6% of HBsAg positive patients |
| Kong et al[26] | 2013 | Frozen renal tissue | HBsAg and HBcAg detected by immunohistochemistry | Found positive in 9 out of 500 patients of glomerulonephritis without serological evidence |
| Ross et al[15] | 2013 | Dried blood spots | HBsAG, anti HBcAg, anti HBsAg detected by Abbott Architect and HBV DNA by artus HBV LC PCR | Sensitivity was 98.6%, 97.1%, 97.5%, 93% |
| Alidjinou et al[16] | 2014 | Dried plasma spots | HBsAg and HBV DNA detected by ELISA and PCR | Sn and Sp 100% for serological markers and Sn 96%, Sp 100% for HBV DNA |
| Zhong et al[19] | 2014 | Hepatocytes | Covalently closed circular HBV DNA detected by in situ PCR | Helps to detect recurrence of HBV |
| Huang et al[20] | 2015 | Formalin fixed paraffin embedded hepatocellular carcinoma tissue | Droplet digital PCR to detect HBV copy number | Highly sensitive method |
Dried blood spots were first used in medical diagnostics by Guthrie et al[30] to detect Phenylketonuria. Dried blood spot (DBS) collection is much easier than taking venous blood. More over different antibodies, medications, metabolites, and nucleic acids remain stable for a longer period in these samples[15]. As researchers have validated this sample in diagnosis of HBV, it has been used much conveniently in field settings or resource poor settings. This review highlights that serological markers and nucleic acid of HBV can be detected from this sample by Point Of Care Tests (POCT), Enzyme Linked Immunosorbent Assay (ELISA) or Nucleic Acid Amplification Techniques with high sensitivity (Table 1). The combination of DBS and POCT is even more advantageous to use in resource poor settings. Sn of detection of HBsAg from saliva was 74.29% in the study of Arora et al[24] Presence of viral antigen in saliva makes dentistry personnel more vulnerable. Saliva can also be collected very easily without technical expertise and with the help of POCTs diagnosis can be made in resource poor settings rapidly.
In certain cases of chronic infection with low level viremia or seronegative patients, HBV DNA has been detected from hepatocytes by polymerase chain reaction (PCR) - In situ hybridization, while couldn’t be detected from blood[18]. Other novel and highly sensitive techniques like flowcytometric quantitation, droplet digital PCR has increased the sensitivity of HBV detection from hepatocytes even more. This is especially important in diagnosing the etiology of chronic hepatitis/hepatocellular carcinoma in seronegative or low viremic patients. Again persistent detection of covalently closed circular DNA (cccDNA) helps to predict recurrence of the disease[19]. Detection of serological markers and HBV DNA from ovarian tissue, oocytes or embryo becomes important in case of in vitro fertilization[21,28]. Though in one study nucleic acid couldn’t be detected after culture and vitrification of oocytes or embryos from seropositive mothers during the procedure[27]. In the study of Kong et al[26], HBsAg and HBcAg were detected in frozen renal tissue by immunohistochemistry in 1.9% of seronegative patients with glomerulonephritis. As it is a common extrahepatic manifestation of viral hepatitis, in occult infections renal tissues can be used to detect the presence of virus.
The detection of HBV is very important in controlling its spread. After the discovery of Ausria 125 various serological, molecular and automated detection methods have been introduced and validated by different researchers. While searching Medline database in last 10 years total 55 studies were found describing different methods of detection.
Serological methods are most common, rapid and cost effective methods to detect different markers like HBsAg, anti-HBsAg, anti-HBcAg, HBeAg, anti-HBeAg, etc.
ELISA: ELISA is a type of solid phase immunoassay in which antigens or antibodies are covalently bound with suitable enzymes that can catalyze the conversion of a substrate into colored products. It is a validated method to detect different serological markers. Various ELISA kits are commercially available. Maity et al[31], 2012 evaluated 3 ELISA kits (Span diagnostics Ltd., J. Mitra and Co. Pvt. Ltd., and Transasia Biomedicals Ltd.) in 300 samples. All the kits were found to be good at screening having higher specificity. Positive predictive value (PPV) and negative predictive value (NPV) were 100% when panels were tested by kits of J. Mitra and Co. Pvt Ltd. and Transasia Biomedicals Ltd, though little less in case of kit of Span Diagnostics Ltd. Though in most of the cases kits are evaluated against a pretested panel, when the results are projected to a population, PPV and NPVs depend widely on the prevalence of that infection. Different researchers have modified this method even. Yazdani et al[32], 2010 used novel monoclonal antibodies as capture layer and a polyclonal biotinylated antibody as detector phase to develop one new ELISA system. Sensitivity and specificity of the assay were 98.98% and 99.6%, respectively when compared to established commercial kit. The performance of ELISA depends on concentration of coating antibody, conjugates and sera. Using different concentrations by checkerboard titration method Fatema et al[33], found that, optimal concentration of coating antibody to be 0.25 ng/mL and 1 in 9 diution of both conjugate and sera. Poly L lysine coated magnetic beads were used to concentrate the virus by Satoh et al[34]. HBsAg and Anti HBc were tested by Enzyme Immuno Assay (AxSYM, Abbott), and haemaglutination inhibition test. By HBsAg EIA they were able to detect 27 out of 40 occult HBV infection. Antigen/ antibody quality is very important for diagnostic accuracy. Recombinant HBcAg is expressed in Escherichia coli and Pichia pastoris (P. pastoris) by Li et al[35], 2007 and used in ELISA for detection of anti HBcAg. P. pastoris derived antigen was more specific and sensitive in detection than the other counterpart.
Chemiluminescent enzyme immunoassay and its modifications: This rapid immunoassay method uses antigen or antibodies labeled with luminescent molecules. This is more sensitive than ELISA. In comparative studies with PCR the sensitivity of chemiluminescent enzyme immunoassay (CLEIA/ CLIA) is 96%[36]. Its sensitivity is even more enhanced by different modifications by researchers. Matsubara et al[37], 2009 developed a highly sensitive CLEIA method for quantitative detection of HBsAg by a combination of monoclonal antibodies each specific for epitopes of HBsAg. This method was 230 fold more sensitive than existing CLIA methods. Incorporating firefly luciferase as labelling enzyme a bioluminescent enzyme immunoassay was developed by Minekawa et al[38]. This became 50 fold more sensitive than conventional CLIAs. Liu et al[39], 2013 developed an amplified luminescent proximity homogeneous assay (AlphaLISA) for HBsAg. The detection sensitivity was as low as 0.01 IU/mL, when compared with the commercial light-initiated chemiluminescence assay. The correlation coefficient of this assay was 0.921.
Automated systems: AxSYM (Abbott) is the first automated third generation immunoassay system. Abbott PRISM HBsAg assay is an in vitro chemiluminescent immunoassay. A new prototype assay based on magnetic micro particle was developed in this sysyem to increase its sensitivity and ability to detect mutants. Lou et al[40] demonstrated that it can detect more commercially available seroconversion panel members (185 of 384) than PRISM (181). Researchers have evaluated different automated CLIA systems across the world. Elecsys (Roche) and Architect (Abbott) gave comparable results for quantitation of HBsAg when assessed by Gupta et al[41]. Beckman Coulter’s anti-HBs chemiluminescence immunoassay (Access AbHBsII) was evaluated in 1207 routine samples prescreened with AxSYM (Abbott) for detection of anti HBsAg by Motte et al[42]. Sn, Sp, PPV and NPV were 97.8%, 98.1%, 96%, and 99%, respectively. ADVIA centaur CP Immunoassay System is based on chemiluminescent with advanced acridinium ester technology. van Helden et al[43] compared its performance with AxSYM, Abbott. It’s Sn and Sp was 100% and 99.5%. The automated chemiluminescent micro particle immunoassay of Abbott (Architect) detects anti-HBc. Borderline reactivity in this system was reassessed by 2 other tests: Microparticle enzyme immunoassay (MEIA, AxSYM, Abbott), and enzyme linked fluorescent assay (ELFA, VIDAS Anti-HBc Total II, bioMérieux) by Ollier et al[44]. 42.99% of borderline reactive samples were found to be positive by MEIA, ELFA. So, other confirmatory tests should be done in this scenario. This commonly used Abbott’s Architect system was also compared with another fully automated and closed DiaSorinLIAISON(®)XL by Krawczyk et al[45] and Kinn et al[46]. The two tests were in > 95% agreement in both the studies. In a multicentre study, automated VIDAS HBsAg Ultra [long (L) and short (S)] incubation protocol (Biomérieux) was compared to AxSYM (Abbott) by Weber et al[47]. Sn of the VIDAS HBsAg Ultra (L), (S) and the AxSYM HBsAg v2 were 99.07%, 97.87% and 94.14% respectively. Sp was 100% for VIDAS. The mean time of the diagnostic window was shortened with the VIDAS HBsAg Ultra (L) and (S) when compared with the AxSYM HBsAg v2 by 1.06 and 0.66 d, respectively. Sn for the VIDAS HBsAg Ultra (L), (S) and AxSYM HBsAg v2 were 99.07%, 97.87% and 94.14%. The Sp were 100% (VIDAS HBsAg Ultra L and S) and 99.6% (AxSYM HBsAg v2)[47].
Other methods: A biosensor based imaging ellipsometry was developed and validated for 169 patients by Qi et al[48]. They concluded that this method could detect 5 markers within 1 h with acceptable agreement when compared to ELISA. Another novel assay based on magnetic beads and time resolved fluroimmunoassay (TR FIA) was developed by Ren et al[49], 2014. The detection antibodies were europium labeled and capturing monoclonal antibodies were immobilized on magnetic beads. The test results had correlation with CLIA (Y = 1.182X - 0.017, R = 0.989). The same TRFIA method was also used to detect HBV Pre S1 antigen by Hu et al[50] and HBsAg by Myyryläinen et al[51]. Burbelo et al[52] used Luciferase Immmunoprecipitation system to detect HBV infection. This could correctly predict the HBV status in all but 2 of 99 assays. Fletcher et al[53] standardised an in house neutralization test for confirmation of HBsAg. Six hundred and fifteen HBsAg samples were subjected to the test. 100% of high reactive samples and 93% of low reactive samples were neutralized by this method, whereas 100% of grey zone reactive samples were negative.
POCT: POCTs are developed to make diagnosis more rapid and accessible to patients. Njai et al[54] validated 3 POCTs (Determine, Vikia and Espline) for detecting HBsAg in field or laboratory setting in Gambia, Western Africa. All the 3 tests gave acceptable result when compared to AxSYM HBsAg ELISA as reference test. Rapid kits (J. Mitra and Co. Pvt. Ltd., Span diagnostic Ltd., Standard Diag. Inc.) were also evaluated by Maity et al[31], 2012. Sn, Sp, PPV and NPV of all the kits were 100%.
Liu et al[55] compared test results of 4 different types of serological tests in 116455 samples. Chemiluminescent microparticle immunoassay (CMIA), electrochemiluminescent immunoassay (ECLIA), ELISA and golden immunochromato-graphic assay (GICA) were used to test the HBsAg level. For qualitative results GICA was significantly less specific than the other 3 tests. Compared to CMIA the false negativity rate of ECLIA, ELISA and GICA were 0.2%, 1.3%, 12.3%.
Molecular methods: Molecular methods used in diagnosis can be categorized as nucleic acid hybridization, nucleic acid amplification, sequencing and enzymatic digestion of nucleic acids.
Hybridization technique: Conventional hybridization technique, though highly specific, lacks sensitivity. Yao et al[56] constructed a peptide nucleic acid probe which combined with target DNA sequences more efficiently than DNA probes. The detection limit was 8.6 pg/L and Sp was 94.4%.
Nucleic acid amplification technique: Amplification techniques can be: (1) target amplification: PCR, nucleic acid sequence based amplification, transcription mediated amplification, Strand Displacement amplification, etc.; (2) Signal amplification: Branched DNA probe (bDNA); and (3) probe amplification: Ligase chain reaction. These techniques can qualitatively or quantitatively detect minute amount of HBV DNA present in the sample. Some researchers have even combined 2 different methods to increase Sn. Combination of bDNA and HBV PCR helped in detection of HBe Ag positive chronic HBV patients by Ozdarendeli et al[57].
Quantitative detection is very important for monitoring of HBV infection. Molecular methods have been used for quantitation by different researchers (Table 2). In this review, of the entire in house and automated molecular techniques version 2.0 (v2.0) of the CobasAmpliPrep/CobasTaqMan (CAP/CTM) assay was found to be most sensitive, with a lower detection limit of only 6.25 IU/mL[63]. Commercial assays were more sensitive than in house assays.
| Ref. | Year of publication | Method of quantitation | Detection limit |
| Garson et al[58] | 2005 | FRET based real time PCR assay | Sn at 95% detection level was 24.2 IU/mL |
| Welzel et al[59] | 2006 | Novel real time PCR | Sn at 95% detection level was 56 IU/mL |
| Mazet-Wagner et al[60] | 2006 | Real time PCR assay to detect total HBV DNA and cccDNA from serum and peripheral blood mononuclear cells | 27 IU/mL |
| McCormick et al[61] | 2006 | Procleix Ultrio Assay (Multiplex PCR) to detect HIV 1, HCV RNA and HBV DNA simultaneously | Sp ≥ 99.5%, Sn is > 95% with detection limit for HBV DNA of 15 IU/mL |
| Cai et al[62] | 2008 | Real time fluorogenic Loop Mediated Isothermal Amplification (RtF-LAMP) | At 95% detection level 210 copies/mL |
| Paraskevis et al[63] | 2010 | New ultrasensitive in house real time PCR assay | Sn at 95% and 50% detection level: 22.2 IU/mL and 8.4 IU/mL |
| Chevaliez et al[64] | 2010 | v2.0 of the CAP/CTM assay | Highly Sn, could even detect 6.25 IU/mL HBV DNA; Sp is 99%, Intra-assay and interassay coefficients of variation ranged from 0.21% to 2.67% and from 0.65% to 2.25%, respectively |
| Sun et al[65] | 2011 | Duplex real-time PCR assay using two sets of primers/probes and a specific armored DNA as internal control | Detection limit 29.5 IU/mL; Sp 100% |
| Cha et al[66] | 2013 | ExiStation HBV diagnostic system | 9.55 IU/mL |
| Yang et al[67] | 2014 | Colorimetric PCR with DNA zyme containing probe | Broad range of linearity and high sensitivity |
| Kania et al[68] | 2014 | 2 in house real time PCR targeting X (qPCR1) or S (qPCR2) genes | qPCR1: 104 IU/mL; qPCR2: 91 IU/mL |
Park et al[69] evaluated Magicplex™ HepaTrio Real-time Detection test, a multiplex PCR assay for the detection of hepatitis A virus, HBV and HCV. Sn and Sp was 93.8% and 98.2%. Monjezi et al[70] developed a Taq Man real time detection assay based on the concept of phage display mediated immune PCR for the detection of HBcAg. This method was able to detect about 10 ng of HBcAg.
A rapid real time micro scale chip based PCR system consisting of 6 individual thermal cycling modules was developed by Cho et al[71]. It took less than 20 min to complete 40 thermal cycles. They conducted large clinical evaluation study to detect HBV infection. The sn and sp was 94% and 93% respectively.
The persistence of HBV can be detected by demonstration of covalently cccDNA. Takkenberg et al[72] developed a sensitive, specific and reproducible Real Time PCR to detect and quantitate cccDNA in chronic HBV patients. The lower limit of detection was 15 copies/PCR. cccDNA is detected by Southern blot analysis in cell cultures by Cai et al[73]. Guo et al[74] developed magnetic capture hybridization and quantitative PCR assay to detect cccDNA with a detection limit of 90 IU/mL.
Studies have been conducted to compare different methods (Table 3). Abbott’s real time PCR kit was most sensitive with lower limit of detection of only 1.48 IU/mL. In comparison most of the automated systems had good agreement.
| Ref. | Year of publication | Comparison between | Remarks |
| Hochberger et al[75] | 2006 | Automated COBAS AmpliPrep/COBAS TaqMan system (real time PCR) and Versant HBV 3.0 | Good correlation between two |
| Juman Awadh et al[76] | 2008 | Versant HBV 3.0 (Bayer, branched DNA mediated assay) and Biotitre B (real time PCR variant) | Both were highly specific, though reproducibility of Versant HBV 3.0 was higher |
| Yang et al[77] | 2009 | Real Art HBV PCR Kit (Abbott, real time PCR) and Versant bDNA 3.0 | Abbott's kit was more sn, detection limit 27 IU/mL |
| Louisirirotchanakul et al[78] | 2010 | fully automated ElecsysHBsAg II assay, Architect, AxSYM and Advia Centaur HBsAg assays | The later 2 tests appeared less sensitive in detecting early HBV infection |
| Berger et al[79] | 2010 | CAP/CTM assay v2.0 and v1.0 | Comparable results for all 278 tested samples |
| Lunel-Fabiani et al[80] | 2010 | Access immunoassay system from Beckman coulter with Abbott AxSYM and PRISM HBsAg assays; VIDAS was used to conclude discrepant results | Sn: 100%, Sp: 99.96% |
| Caliendo et al[81] | 2011 | Abbott RealTime HBV IUO, the Roche CAP/CTM HBV test, the Roche CobasTaqMan HBV test with HighPure system, and the Qiagen artus HBV TM ASR | Limit of detection of artus 1.5 log(10) IU/mL, of other 3 tests 1.0 log(10) IU/mL |
| Ismail et al[82] | 2011 | Abbott HBV real-time PCR (Abbott PCR), artus HBV real-time PCR with QIAamp DNA blood kit purification (artus-DB), and artus HBV real-time PCR with the QIAamp DSP virus kit purification (artus-DSP) | Lower limit of detection against WHO standards were 1.43, 82 and 9 IU/mL respectively |
| Yeh et al[83] | 2014 | Abbott real time HBV (RealTime assay) and CAP/CTM HBV assays 2.0 (TaqMan assay) | Real time assay's Sn: 98.2%, Sp: 100%. Good level of agreement between the two |
HBV can be detected reliably from DBS (Sn > 90% in all cases). In certain cases of occult infections or seronegative patients, HBV have been detected from hepatocytes or renal tissues also. Serological and Molecular assays are predominant and reliable methods for HBV detection. CLIA is more sensitive than ELISA. Rapid tests are also dependable and useful for screening purpose, especially in resource poor settings. Quantitation is important for monitoring. Real time PCR, bDNA assays are principal methods used for this purpose. Automated systems are more sensitive when compared to in house assays. Among the molecular techniques v2.0 of the CAP/CTM assay and Abbott real time PCR were found to be most sensitive with a lower detection limit of only 6.25 IU/mL and 1.48 IU/mL respectively.
We acknowledge staff of Virology Division, National Institute of Cholera and Enteric Diseases for providing support.
In earlier days infection with hepatitis B virus (HBV) was detected by demonstration of antibody titer by Complement Fixation Test. The first solid phase sandwich radio immunoassay named Ausria 125 was developed by Ling et al at Abbott Laboratories (North Chicago). This highly sensitive detection method became a major discovery in the diagnosis of viral transfusion hepatitis and screening of blood donors. Since then innumerable serological and molecular methods have been developed for diagnosing HBV.
This article provides an overview of detection of HBV infection employing different techniques.
Beside serum/plasma, HBV can be detected reliably from dried blood spots (DBS) (Sn > 90% in all cases). In occult infections or seronegative patients, HBV was detected from hepatocytes or renal tissues. Serological and Molecular assays are predominant and reliable methods. Chemiluminescent immunoassay is more sensitive than enzyme Linked Immunosorbent Assay. Rapid tests are useful for screening. Real time polymerase chain reaction (PCR), branched DNA assays are principal methods for quantitation. Automated systems are more sensitive compared to in house assays. CobasAmpliprep/CobasTaqMan version 2.0 assay and Abbott real time PCR were found to be most sensitive with a lower detection limit of only 6.25 IU/mL and 1.48 IU/mL respectively. Rapid tests are also highly sensitive and specific as evaluated by different researchers.
Use of DBS and validated rapid tests can aid in initial diagnosis in resource poor settings. Quantitation is important for monitoring and prognostic evaluation and automated systems are highly sensitive and efficient for this purpose.
The authors have performed a good study, the manuscript is interesting.
P- Reviewer: Changotra H S- Editor: Tian YL L- Editor: A E- Editor: Liu SQ
| 1. | Trepo C. A brief history of hepatitis milestones. Liver Int. 2014;34 Suppl 1:29-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 2. | Gerlich WH. Medical virology of hepatitis B: how it began and where we are now. Virol J. 2013;10:239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 220] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 3. | Krugman S. Viral hepatitis: overview and historical perspectives. Yale J Biol Med. 1976;49:199-203. [PubMed] |
| 4. | Paul JR, Havens WP, Sabin AB, Philip CB. Transmission experiments in serum jaundice and infectious hepatitis. J Am Med Assoc. 1945;128:911. [RCA] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | Blumberg BS, Alter HJ, Visnich S. A “new” antigen in leukemia sera. JAMA. 1965;191:541-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 933] [Cited by in RCA: 732] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 6. | Prince AM. An antigen detected in the blood during the incubation period of serum hepatitis. Proc Natl Acad Sci USA. 1968;60:814-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 501] [Cited by in RCA: 464] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 7. | Dane DS, Cameron CH, Briggs M. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet. 1970;1:695-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 648] [Cited by in RCA: 560] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 8. | Almeida JD, Rubenstein D, Stott EJ. New antigen-antibody system in Australia-antigen-positive hepatitis. Lancet. 1971;2:1225-1227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 178] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Magnius LO, Espmark A. A new antigen complex co-occurring with Australia antigen. Acta Pathol Microbiol Scand B Microbiol Immunol. 1972;80:335-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 10. | Robinson WS, Clayton DA, Greenman RL. DNA of a human hepatitis B virus candidate. J Virol. 1974;14:384-391. [PubMed] |
| 11. | Ling CM, Overby LR. Prevalence of hepatitis B virus antigen as revealed by direct radioimmune assay with 125 I-antibody. J Immunol. 1972;109:834-841. [PubMed] |
| 12. | PRISMA 2009 checklist [Internet]. [Accessed 2015 Mar 25]. Available from: http://www.prismastatement.org/2.1.2PRISMA 2009 Checklist.pdf. |
| 13. | Mendy M, Kirk GD, van der Sande M, Jeng-Barry A, Lesi OA, Hainaut P, Sam O, McConkey S, Whittle H. Hepatitis B surface antigenaemia and alpha-foetoprotein detection from dried blood spots: applications to field-based studies and to clinical care in hepatitis B virus endemic areas. J Viral Hepat. 2005;12:642-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | Villar LM, de Oliveira JC, Cruz HM, Yoshida CF, Lampe E, Lewis-Ximenez LL. Assessment of dried blood spot samples as a simple method for detection of hepatitis B virus markers. J Med Virol. 2011;83:1522-1529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 15. | Ross RS, Stambouli O, Grüner N, Marcus U, Cai W, Zhang W, Zimmermann R, Roggendorf M. Detection of infections with hepatitis B virus, hepatitis C virus, and human immunodeficiency virus by analyses of dried blood spots--performance characteristics of the ARCHITECT system and two commercial assays for nucleic acid amplification. Virol J. 2013;10:72. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 16. | Alidjinou EK, Moukassa D, Sané F, Twagirimana Nyenyeli S, Akoko EC, Mountou MV, Bocket L, Ibara JR, Hober D. Detection of hepatitis B virus infection markers in dried plasma spots among patients in Congo-Brazzaville. Diagn Microbiol Infect Dis. 2014;78:229-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 17. | van der Laan LJ, Taimr P, Kok A, Sprengers D, Zondervan PE, Tilanus HW, Janssen HL. Flowcytometric quantitation of hepatitis B viral antigens in hepatocytes from regular and fine-needle biopsies. J Virol Methods. 2007;142:189-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 18. | Nuriya H, Inoue K, Tanaka T, Hayashi Y, Hishima T, Funata N, Kaji K, Hayashi S, Kaneko S, Kohara M. Detection of hepatitis B and C viruses in almost all hepatocytes by modified PCR-based in situ hybridization. J Clin Microbiol. 2010;48:3843-3851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 19. | Zhong Y, Hu S, Xu C, Zhao Y, Xu D, Zhao Y, Zhao J, Li Z, Zhang X, Zhang H. A novel method for detection of HBVcccDNA in hepatocytes using rolling circle amplification combined with in situ PCR. BMC Infect Dis. 2014;14:608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Huang JT, Liu YJ, Wang J, Xu ZG, Yang Y, Shen F, Liu XH, Zhou X, Liu SM. Next generation digital PCR measurement of hepatitis B virus copy number in formalin-fixed paraffin-embedded hepatocellular carcinoma tissue. Clin Chem. 2015;61:290-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 21. | Chen LZ, Fan XG, Gao JM. Detection of HBsAg, HBcAg, and HBV DNA in ovarian tissues from patients with HBV infection. World J Gastroenterol. 2005;11:5565-5567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 22. | Goh EK, Son BH, Kong SK, Chon KM, Cho KS. Analysis of hepatitis B virus in the cerumen and otorrhea of chronic HBV-infected patients: is there a hepatitis B virus infectivity? Otol Neurotol. 2008;29:929-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 23. | Eftekharian A, Moghaddasi H, Gachkar L, Amlashi SS. Detection of hepatitis B virus in the cerumen of patients with chronic hepatitis B infection. J Laryngol Otol. 2013;127:1065-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 24. | Arora G, Sheikh S, Pallagatti S, Singh B, Singh VA, Singh R. Saliva as a tool in the detection of hepatitis B surface antigen in patients. Compend Contin Educ Dent. 2012;33:174-176, 178; quiz 180, 182. [PubMed] |
| 25. | Chen L, Liu F, Fan X, Gao J, Chen N, Wong T, Wu J, Wen SW. Detection of hepatitis B surface antigen, hepatitis B core antigen, and hepatitis B virus DNA in parotid tissues. Int J Infect Dis. 2009;13:20-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | Kong D, Wu D, Wang T, Li T, Xu S, Chen F, Jin X, Lou G. Detection of viral antigens in renal tissue of glomerulonephritis patients without serological evidence of hepatitis B virus and hepatitis C virus infection. Int J Infect Dis. 2013;17:e535-e538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 27. | Cobo A, Bellver J, de los Santos MJ, Remohí J. Viral screening of spent culture media and liquid nitrogen samples of oocytes and embryos from hepatitis B, hepatitis C, and human immunodeficiency virus chronically infected women undergoing in vitro fertilization cycles. Fertil Steril. 2012;97:74-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 28. | Ye F, Jin Y, Kong Y, Shi JZ, Qiu HT, Zhang X, Zhang SL, Lin SM. The presence of HBV mRNA in the fertilized in vitro embryo of HBV patients confirms vertical transmission of HBV via the ovum. Epidemiol Infect. 2013;141:926-930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 29. | Wu Y, Wang T, Ye S, Zhao R, Bai X, Wu Y, Abe K, Jin X. Detection of hepatitis B virus DNA in paraffin-embedded intrahepatic and extrahepatic cholangiocarcinoma tissue in the northern Chinese population. Hum Pathol. 2012;43:56-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 30. | Guthrie R, Susi A. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics. 1963;32:338-343. [PubMed] |
| 31. | Maity S, Nandi S, Biswas S, Sadhukhan SK, Saha MK. Performance and diagnostic usefulness of commercially available enzyme linked immunosorbent assay and rapid kits for detection of HIV, HBV and HCV in India. Virol J. 2012;9:290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | Yazdani Y, Roohi A, Khoshnoodi J, Shokri F. Development of a sensitive enzyme-linked immunosorbent assay for detection of hepatitis B surface antigen using novel monoclonal antibodies. Avicenna J Med Biotechnol. 2010;2:207-214. [PubMed] |
| 33. | Fatema K, Tabassum S, Nessa A, Jahan M. Development and evaluation of an in-house ELISA to detect hepatitis B virus surface antigen in resource-limited settings. Bangladesh Med Res Counc Bull. 2013;39:65-68. [PubMed] |
| 34. | Satoh K, Iwata-Takakura A, Yoshikawa A, Gotanda Y, Tanaka T, Yamaguchi T, Mizoguchi H. A new method of concentrating hepatitis B virus (HBV) DNA and HBV surface antigen: an application of the method to the detection of occult HBV infection. Vox Sang. 2008;95:174-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 35. | Li ZX, Hong GQ, Hu B, Liang MJ, Xu J, Li L. Suitability of yeast- and Escherichia coli-expressed hepatitis B virus core antigen derivatives for detection of anti-HBc antibodies in human sera. Protein Expr Purif. 2007;56:293-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 36. | Khadem-Ansari MH, Omrani MD, Rasmi Y, Ghavam A. Diagnostic validity of the chemiluminescent method compared to polymerase chain reaction for hepatitis B virus detection in the routine clinical diagnostic laboratory. Adv Biomed Res. 2014;3:116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 37. | Matsubara N, Kusano O, Sugamata Y, Itoh T, Mizuii M, Tanaka J, Yoshizawa H. A novel hepatitis B virus surface antigen immunoassay as sensitive as hepatitis B virus nucleic acid testing in detecting early infection. Transfusion. 2009;49:585-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 38. | Minekawa T, Ohkuma H, Abe K, Maekawa H, Arakawa H. Development of ultra-high sensitivity bioluminescent enzyme immunoassay for hepatitis B virus surface antigen using firefly luciferase. Luminescence. 2009;24:394-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 39. | Liu TC, Huang H, Dong ZN, He A, Li M, Wu YS, Xu WW. Development of an amplified luminescent proximity homogeneous assay for quantitative determination of hepatitis B surface antigen in human serum. Clin Chim Acta. 2013;426:139-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 40. | Lou SC, Pearce SK, Lukaszewska TX, Taylor RE, Williams GT, Leary TP. An improved Abbott ARCHITECT assay for the detection of hepatitis B virus surface antigen (HBsAg). J Clin Virol. 2011;51:59-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 41. | Gupta E, Pandey P, Kumar A, Sharma MK, Sarin SK. Correlation between two chemiluminescence based assays for quantification of hepatitis B surface antigen in patients with chronic hepatitis B infection. Indian J Med Microbiol. 2015;33:96-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 42. | Motte A, Colson P, Tamalet C. Evaluation of the clinical performance of the Beckman Coulter Access AbHBsII immunoassay for the detection of hepatitis B surface antibodies. J Clin Virol. 2006;37:213-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 43. | van Helden J, Cornely C, Dati F, Levy HR, Bal T, Seeger M, Wright T, Baker L. Performance evaluation of the ADVIA Centaur anti-HBe and HBeAg assays. J Clin Virol. 2008;43:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 44. | Ollier L, Laffont C, Kechkekian A, Doglio A, Giordanengo V. Detection of antibodies to hepatitis B core antigen using the Abbott ARCHITECT anti-HBc assay: analysis of borderline reactive sera. J Virol Methods. 2008;154:206-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 45. | Krawczyk A, Hintze C, Ackermann J, Goitowski B, Trippler M, Grüner N, Neumann-Fraune M, Verheyen J, Fiedler M. Clinical performance of the novel DiaSorin LIAISON(®) XL murex: HBsAg Quant, HCV-Ab, HIV-Ab/Ag assays. J Clin Virol. 2014;59:44-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 46. | Kinn S, Akhavan S, Agut H, Thibault V. Performance of the DiaSorin LIAISON(®) anti-HBs II for the detection of hepatitis B surface antibodies: comparison with the Abbott Architect anti-HBs assay. J Clin Virol. 2011;50:297-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 47. | Weber B, Van der Taelem-Brulé N, Berger A, Simon F, Geudin M, Ritter J. Evaluation of a new automated assay for hepatitis B surface antigen (HBsAg) detection VIDAS HBsAg Ultra. J Virol Methods. 2006;135:109-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 48. | Qi C, Zhu W, Niu Y, Zhang HG, Zhu GY, Meng YH, Chen S, Jin G. Detection of hepatitis B virus markers using a biosensor based on imaging ellipsometry. J Viral Hepat. 2009;16:822-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 49. | Ren ZQ, Liu TC, Hou JY, Chen MJ, Chen ZH, Lin GF, Wu YS. A rapid and sensitive method based on magnetic beads for the detection of hepatitis B virus surface antigen in human serum. Luminescence. 2014;29:591-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 50. | Hu Z, Li M, Huang B, Liu J, Yu L, Chen G. Detection of hepatitis B virus PreS1 antigen using a time-resolved fluoroimmunoassay. J Immunoassay Immunochem. 2012;33:156-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 51. | Myyryläinen T, Talha SM, Swaminathan S, Vainionpää R, Soukka T, Khanna N, Pettersson K. Simultaneous detection of Human Immunodeficiency Virus 1 and Hepatitis B virus infections using a dual-label time-resolved fluorometric assay. J Nanobiotechnology. 2010;8:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 52. | Burbelo PD, Ching KH, Mattson TL, Light JS, Bishop LR, Kovacs JA. Rapid antibody quantification and generation of whole proteome antibody response profiles using LIPS (luciferase immunoprecipitation systems). Biochem Biophys Res Commun. 2007;352:889-895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 53. | Fletcher GJ, Gnanamony M, David J, Ismail AM, Subramani T, Abraham P. Do we need an ‘in-house’ neutralization assay for confirmation of hepatitis B surface antigen? Answers from a tertiary care hospital in India. J Gastroenterol Hepatol. 2010;25:942-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 54. | Njai HF, Shimakawa Y, Sanneh B, Ferguson L, Ndow G, Mendy M, Sow A, Lo G, Toure-Kane C, Tanaka J. Validation of rapid point-of-care (POC) tests for detection of hepatitis B surface antigen in field and laboratory settings in the Gambia, Western Africa. J Clin Microbiol. 2015;53:1156-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 55. | Liu C, Chen T, Lin J, Chen H, Chen J, Lin S, Yang B, Shang H, Ou Q. Evaluation of the performance of four methods for detection of hepatitis B surface antigen and their application for testing 116,455 specimens. J Virol Methods. 2014;196:174-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (1)] |
| 56. | Yao C, Zhu T, Tang J, Wu R, Chen Q, Chen M, Zhang B, Huang J, Fu W. Hybridization assay of hepatitis B virus by QCM peptide nucleic acid biosensor. Biosens Bioelectron. 2008;23:879-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 64] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 57. | Ozdarendeli A, Toroman ZA, Bulut Y, Demirbag K, Kalkan A, Ozden M, Kilic SS. Combined branched-DNA and conventional HBV PCR assays for detection of serum HBV-DNA in hepatitis B e antigen-positive chronic hepatitis B patients. Hepatogastroenterology. 2006;53:106-109. [PubMed] |
| 58. | Garson JA, Grant PR, Ayliffe U, Ferns RB, Tedder RS. Real-time PCR quantitation of hepatitis B virus DNA using automated sample preparation and murine cytomegalovirus internal control. J Virol Methods. 2005;126:207-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 125] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 59. | Welzel TM, Miley WJ, Parks TL, Goedert JJ, Whitby D, Ortiz-Conde BA. Real-time PCR assay for detection and quantification of hepatitis B virus genotypes A to G. J Clin Microbiol. 2006;44:3325-3333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 60. | Mazet-Wagner AA, Baclet MC, Loustaud-Ratti V, Denis F, Alain S. Real-time PCR quantitation of hepatitis B virus total DNA and covalently closed circular DNA in peripheral blood mononuclear cells from hepatitis B virus-infected patients. J Virol Methods. 2006;138:70-79. [PubMed] |
| 61. | McCormick MK, Dockter J, Linnen JM, Kolk D, Wu Y, Giachetti C. Evaluation of a new molecular assay for detection of human immunodeficiency virus type 1 RNA, hepatitis C virus RNA, and hepatitis B virus DNA. J Clin Virol. 2006;36:166-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 62. | Cai T, Lou G, Yang J, Xu D, Meng Z. Development and evaluation of real-time loop-mediated isothermal amplification for hepatitis B virus DNA quantification: a new tool for HBV management. J Clin Virol. 2008;41:270-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 63. | Paraskevis D, Beloukas A, Haida C, Katsoulidou A, Moschidis Z, Hatzitheodorou H, Varaklioti A, Sypsa V, Hatzakis A. Development of a new ultra sensitive real-time PCR assay (ultra sensitive RTQ-PCR) for the quantification of HBV-DNA. Virol J. 2010;7:57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 64. | Chevaliez S, Bouvier-Alias M, Laperche S, Hézode C, Pawlotsky JM. Performance of version 2.0 of the Cobas AmpliPrep/Cobas TaqMan real-time PCR assay for hepatitis B virus DNA quantification. J Clin Microbiol. 2010;48:3641-3647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 65. | Sun S, Meng S, Zhang R, Zhang K, Wang L, Li J. Development of a new duplex real-time polymerase chain reaction assay for hepatitis B viral DNA detection. Virol J. 2011;8:227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 66. | Cha YJ, Yoo SJ, Sohn YH, Kim HS. Performance evaluation of ExiStation HBV diagnostic system for hepatitis B virus DNA quantitation. J Virol Methods. 2013;193:492-497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 67. | Yang L, Du F, Chen G, Yasmeen A, Tang Z. A novel colorimetric PCR-based biosensor for detection and quantification of hepatitis B virus. Anal Chim Acta. 2014;840:75-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 68. | Kania D, Ottomani L, Meda N, Peries M, Dujols P, Bolloré K, Rénier W, Viljoen J, Ducos J, Van de Perre P. Performance of two real-time PCR assays for hepatitis B virus DNA detection and quantitation. J Virol Methods. 2014;201:24-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 69. | Park Y, Kim BS, Choi KH, Shin DH, Lee MJ, Cho Y, Kim HS. A novel multiplex real-time PCR assay for the concurrent detection of hepatitis A, B and C viruses in patients with acute hepatitis. PLoS One. 2012;7:e49106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 70. | Monjezi R, Tan SW, Tey BT, Sieo CC, Tan WS. Detection of hepatitis B virus core antigen by phage display mediated TaqMan real-time immuno-PCR. J Virol Methods. 2013;187:121-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 71. | Cho YK, Kim J, Lee Y, Kim YA, Namkoong K, Lim H, Oh KW, Kim S, Han J, Park C. Clinical evaluation of micro-scale chip-based PCR system for rapid detection of hepatitis B virus. Biosens Bioelectron. 2006;21:2161-2169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 44] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 72. | Takkenberg RB, Zaaijer HL, Molenkamp R, Menting S, Terpstra V, Weegink CJ, Dijkgraaf MG, Jansen PL, Reesink HW, Beld MG. Validation of a sensitive and specific real-time PCR for detection and quantitation of hepatitis B virus covalently closed circular DNA in plasma of chronic hepatitis B patients. J Med Virol. 2009;81:988-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 73. | Cai D, Nie H, Yan R, Guo JT, Block TM, Guo H. A southern blot assay for detection of hepatitis B virus covalently closed circular DNA from cell cultures. Methods Mol Biol. 2013;1030:151-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 118] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 74. | Guo Y, Sheng S, Nie B, Tu Z. Development of magnetic capture hybridization and quantitative polymerase chain reaction for hepatitis B virus covalently closed circular DNA. Hepat Mon. 2015;15:e23729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 75. | Hochberger S, Althof D, Gallegos de Schrott R, Nachbaur N, Röck H, Leying H. Fully automated quantitation of hepatitis B virus (HBV) DNA in human plasma by the COBAS AmpliPrep/COBAS TaqMan system. J Clin Virol. 2006;35:373-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 54] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 76. | Juman Awadh A, Kyuregyan KK, Isaeva OV, Mikhailov MI. Comparative characterization of two tests for measurement of hepatitis B virus DNA in the blood serum and plasma, based on the use of two different detection methods. Bull Exp Biol Med. 2008;146:246-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 77. | Yang JF, Lin YY, Huang JF, Liu SF, Chu PY, Hsieh MY, Lin ZY, Chen SC, Wang LY, Dai CY. Comparison of clinical application of the Abbott HBV PCR kit and the VERSANT HBV DNA 3.0 test to measure serum hepatitis B virus DNA in Taiwanese patients. Kaohsiung J Med Sci. 2009;25:413-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 78. | Louisirirotchanakul S, Khupulsup K, Akraekthalin S, Chan KP, Saw S, Aw TC, Cho DH, Shin MG, Lim J. Comparison of the technical and clinical performance of the Elecsys HBsAg II assay with the Architect, AxSym, and Advia Centaur HBsAg screening assays. J Med Virol. 2010;82:755-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 79. | Berger A, Gohl P, Stürmer M, Rabenau HF, Nauck M, Doerr HW. Detection and quantitation of HBV DNA in miniaturized samples: multi centre study to evaluate the performance of the COBAS ® AmpliPrep/COBAS ® TaqMan ® hepatitis B virus (HBV) test v2.0 by the use of plasma or serum specimens. J Virol Methods. 2010;169:404-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 80. | Lunel-Fabiani F, Duburcq X, Levayer T, Descamps F, Maniez-Montreuil M, Pivert A, Ducancelle A, Pouzet A, Marant L, Clèment A. Multi-center evaluation of the hepatitis B surface antigen (HBsAg) assay and HbsAg confirmatory assay for the family of Access immunoassay systems. Clin Lab. 2010;56:281-290. [PubMed] |
| 81. | Caliendo AM, Valsamakis A, Bremer JW, Ferreira-Gonzalez A, Granger S, Sabatini L, Tsongalis GJ, Wang YF, Yen-Lieberman B, Young S. Multilaboratory evaluation of real-time PCR tests for hepatitis B virus DNA quantification. J Clin Microbiol. 2011;49:2854-2858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 82. | Ismail AM, Sivakumar J, Anantharam R, Dayalan S, Samuel P, Fletcher GJ, Gnanamony M, Abraham P. Performance characteristics and comparison of Abbott and artus real-time systems for hepatitis B virus DNA quantification. J Clin Microbiol. 2011;49:3215-3221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 83. | Yeh ML, Huang CF, Hsieh MY, Huang JF, Dai CY, Yu ML, Chuang WL. Comparison of the Abbott RealTime HBV assay with the Roche Cobas AmpliPrep/Cobas TaqMan HBV assay for HBV DNA detection and quantification. J Clin Virol. 2014;60:206-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |