Published online Feb 27, 2015. doi: 10.4254/wjh.v7.i2.139
Peer-review started: September 20, 2014
First decision: November 1, 2014
Revised: November 21, 2014
Accepted: December 3, 2014
Article in press: December 10, 2014
Published online: February 27, 2015
Processing time: 147 Days and 10.1 Hours
Biomarkers for surveillance, diagnosis and prediction of prognosis in patients with hepatocellular carcinoma (HCC) are currently not ready for introduction into clinical practice because of limited sensitivity and specificity. Especially for the early detection of small HCC novel biomarkers are needed to improve the current effectiveness of screening performed by ultrasound. The use of high-throughput technologies in hepatocellular research allows to identify molecules involved in the complex pathways in hepatocarcinogenesis. Several invasive and non-invasive biomarkers have been identified already and have been evaluated in different clinical settings. Gene signatures with prognostic potential have been identified by gene expression profiling from tumor tissue. However, a single “all-in-one” biomarker that fits all-surveillance, diagnosis, prediction of prognosis-has not been found so far. The future of biomarkers most probably lies in a combination of non-invasive biomarkers, imaging and clinical parameters in a surveillance setting. Molecular profiling of tumorous and non-tumorous liver tissue may allow a prediction of prognosis for the individual patient and hopefully clear the way for individual treatment approaches. This article gives an overview on current developments in biomarker research in HCC with a focus on currently available and novel biomarkers, in particular on microRNA.
Core tip: The aim of this review is to provide an overview on current invasive and non-invasive biomarkers in hepatocellular carcinoma (HCC) with respect to their use in surveillance, diagnosis and prediction of prognosis. We also give an outlook on the future development of HCC biomarker research with a focus on microRNA.
- Citation: Schütte K, Schulz C, Link A, Malfertheiner P. Current biomarkers for hepatocellular carcinoma: Surveillance, diagnosis and prediction of prognosis. World J Hepatol 2015; 7(2): 139-149
- URL: https://www.wjgnet.com/1948-5182/full/v7/i2/139.htm
- DOI: https://dx.doi.org/10.4254/wjh.v7.i2.139
The incidence of hepatocellular carcinoma (HCC) is rising throughout the world as a consequence of a rising prevalence of hepatitis B virus (HBV) and hepatitis C virus (HCV) infection and an increase in prevalence of (non-alcoholic) fatty liver disease due to the metabolic syndrome[1-4]. The number of patients that are at risk to develop HCC and need to undergo structured surveillance is therefore constantly rising in parallel.
Transabdominal ultrasound currently is the only recommended tool for surveillance in the Western world which has been shown to be cost-effective. Its sensitivity is dependent on many factors, including the quality of the ultrasound machine, the experience of the examiner and also the patient. Especially in obese patients with NASH cirrhosis liver ultrasonography may be difficult and therefore not always appropriate to rule out HCC. In patients with liver cirrhosis regenerative nodules may be hard to distinguish from HCC on ultrasound, and the sensitivity of ultrasound to detect early HCC lies in a range of 32% to 65%[5,6]. On the other hand, surveillance by contrast-enhanced computed tomography or contrast-enhanced magnetic resonance tomography is rather expensive and not cost-effective. In addition, they are associated with the additional exposure to radiation with an increased risk of tumor development and/or contrast-media related deterioration of kidney function. In developing countries, where even the availability of ultrasound surveillance is quite low, serological markers for surveillance are of special interest[7].
Biomarkers in blood, other body fluids or tissue for screening, prediction of prognosis and monitoring of response to a therapy would be an important contribution to the management of patients with HCC.
Early detection of HCC is the most important factor to offer the patient the chance of cure. α-fetoprotein (AFP) is the most widely used and broadly known biomarker for HCC, but the measurement of serum AFP levels has been dropped from current surveillance guidelines in Europe and the United States because of low sensitivity and specificity. This is based on the knowledge that almost 80% of small HCCs do not show increased levels of AFP, and the sensitivity decreases to 25% in tumors smaller than 3 cm[8]. Nonetheless, serum AFP measurement is still combined with ultrasound by many physicians worldwide to reduce the risk of missing small lesions in the cirrhotic liver that have not been detected by ultrasound. Alternative or additional biomarkers may be useful tools for surveillance or as a decisional tool in clinical practice to identify patients that will benefit from advanced imaging methods in a surveillance setting to augment the proportion of patients with HCC diagnosed in an early tumor stage.
Apart from their role as a surveillance tool, biomarkers may play a role as diagnostic tool once a suspicious lesion in a patient with liver cirrhosis has been detected. In the past, a significant concentration of AFP in the serum of a patient with liver cirrhosis and a suspicious mass in the liver larger than 2 cm was sufficient to diagnose HCC[9]. However, diagnostic algorithms endorsed by the American Association for the Study of Liver Diseases (AASLD) and the European Association for the Study of the Liver (EASL) nowadays strictly rely on typical radiological hallmarks in dynamic contrast-enhanced imaging apart from biomarkers[9-11].
Once the diagnosis of HCC is confirmed, molecular biomarkers could potentially be used for prediction of prognosis of the individual patient and also for the guidance of therapeutic decisions. Currently, early studies on predictive biomarkers are on their way to make a step towards a personalized and individualized therapy of patients with HCC.
This review gives a concise overview on current clinical-translational knowledge on biomarkers in surveillance, diagnosis and prediction of prognosis with a focus on miRNA.
Geographical differences in tumor prevalence, tumor biology and resources have resulted in differences in current guidelines with respect to screening recommendations. The main pillar of surveillance in high risk populations is repeated transabdominal ultrasound with small differences with respect to the definition of target populations in various guidelines. Throughout the world three serum biomarkers are suggested as tools to determine the risk of liver cancer in high risk populations: AFP, the proportion of the fucosylated isoform of AFP, AFP-L3, and des-gamma-carboxy-prothrombin (DCP). These markers are FDA approved for this indication, but not a part of the surveillance guidelines published by the AASLD and the EASL[10,11]. Current expert opinion from Western countries has been rather critical on these biomarkers regarding their clinical value[12]. On the other hand, the Asia Pacific guideline recommends to combine ultrasound with the measurement of AFP levels in the serum and the Japanese society even recommends to apply all three mentioned biomarkers[13,14] for surveillance.
However, most studies on the performance of biomarkers in HCC detection have not been performed in a surveillance setting but compared levels of predefined biomarkers in patients with HCC with a comparator group, in most cases in patients with chronic liver disease.
A randomized controlled study performed in a high-risk population in China showed that screening by AFP measurement led to earlier diagnosis of HCC but had no impact on mortality[15]. On the other hand, semiannual screening for HCC by AFP measurement in a population-based study in Alaska was effective in detecting HCC at early stages and significantly prolonged survival rates[16].
As the discussion on the rise and fall of AFP as biomarker in HCC surveillance and diagnosis has been intense and sometimes even emotional during the last decade[17-19], data on the most important diagnostic studies referred to in this discussion are summarized in Table 1.
| Ref. | Year | n | Comparator | Cut-off-level | Sensitivity | Specificity | AUC |
| Marrero et al[20] | 2009 | 836 total (419 HCC) | Liver cirrhosis | 20 ng/mL | 59% | 90% | 0.8 |
| Mao et al[21] | 2010 | 4217 total (789 HCC) | Amongst others healthy controls, HBV carriers, liver cirrhosis | 35 ng/mL | 58.20% | 85.30% | |
| Farinati et al[22] | 2006 | 1158 HCC | No control | 400 ng/mL | 18% | 0.59 | |
| Lok et al[23] | 2010 | 39 HCC, 77 matched controls | Hepatitis C | 20 ng/mL | 61% | 81% | 0.79 |
A recent meta-analysis on the performance of AFP in diagnosis of HCC included seven studies and revealed a pooled sensitivity of 66% with a specificity of 86% and an area under the cure (AUC) of 0.87[24]. In a further meta-analysis including ten studies the pooled sensitivity of AFP for the diagnosis of HCC was 51.9% at a specificity of 94% (AUC = 0.81)[25]. It is a major drawback of AFP as surveillance tool that its serum levels are influenced by the activity of the underlying liver disease and therefore increased in patients with elevated ALT levels even in the absence of HCC as shown in the HALT-C trial[26]. Additionally, only a proportion of patients with HCC exerts elevated AFP serum levels leading to low sensitivity of the marker. The heterogeneity of tumor biology in HCC therefore results in a necessity to find better or complementary markers to close this diagnostic gap.
The clinical utility of high-sensitivity AFP-L3 (hs-AFP-L3) in early prediction of HCC development in patients with chronic HBV or HCV infection was recently evaluated in a large Japanese study. Even at low AFP levels and in absence of suspicious ultrasound findings an elevation of hs-AFP-L3 was an early predictor of HCC development with an elevation in 34.3% of patients one year prior to diagnosis of HCC[27]. In patients with low AFP levels (< 20 ng/mL), the diagnostic sensitivity for hs-AFP-L 3% at a cut-off of 5% was 41.5% with a specificity of 85.1%[28].
Numerous studies have investigated the performance of alternative markers or combinations of already established markers. New candidate markers include squamous cell carcinoma antigen-immunoglobulin M complex (SCCA-IGM), α-l-fucosidase[29], glypican-3 (GPC-3), insulin-like growth factor (IGF)[30], vascular endothelial growth factor (VEGF), or Dickkopf-1 (DKK1)[31].
Three further biomarkers have intensively been studied for their potential use in screening for HCC, namely Golgi protein 73 (GP73), interleukin-6 (IL-6) and squamous cell carcinoma antigen (SCCA) and were addressed in a recent meta-analysis[32]. The transmembrane glycoprotein GP73 has a sensitivity of 62% with a specificity of 88% at a cut-off of 10 relative units in a study comparing 144 patients with HCC to 152 patients with cirrhosis and 56 healthy controls[33]. A further study including 4217 subjects of whom 789 were patients with HCC revealed a sensitivity of 74.6% with a specificity of 97.4% at a cut-off of 8.5 relative units[21]. Two smaller studies were identified in the meta-analysis studying the cytokine IL-6. Using different cut-off-values, sensitivity for HCC ranged from 46% to 73% with a specificity of 87% to 95%[32,34,35]. The largest study on the role of the serine protease inhibitor SCCA included 961 patients and resulted in a sensitivity of 42% at specificity of 83% using a cut-off of 3.8 ng/mL[32,36].
Seven well-designed studies on the diagnostic performance of osteopontin, an integrin-binding glycophosphoprotein, were published and recently summarized in a meta-analysis[24]. Osteopontin is expressed by transformed malignant cells and has been evaluated also in colon and pancreatic cancer. All of the reported studies were retrospective in design and included a range of 30 to 179 patients with HCC. The pooled sensitivity of osteopontin for HCC was 86% with a specificity of 86% resulting in a diagnostic accuracy comparable to that of AFP in the included studies. The authors of the meta-analysis conclude that further validation studies are needed before the marker could be suggested for the use in daily clinical routine.
By combining two or more biomarkers the diagnostic performance of a single non-invasive test can be optimized. This has been investigated for the three best established non-invasive biomarkers in HCC, AFP, AFP-L3 and DCP.
When comparing 164 European patients with HCC to 422 controls with chronic liver disease a significant increase in AFP serum levels was mainly shown in patients with advanced stages of HCC and in patients suffering from viral hepatitis while DCP was more frequently elevated in patients with early-stage and NASH associated HCC. Taken alone, neither of the two parameters could detect more than one third of HCC patients independently of stage or etiology but by combination of AFP with DCP a sensitivity of 55% for early stage HCC and 78% for all stages (cut-off for AFP 10 ng/mL and for DCP 5 ng/mL) was reached[37]. The addition of AFP-L3% to this combination, led to a further gain in sensitivity (84%) in another European study[38].
The incorporation of clinical variables like age and gender into models based on a combination of biomarkers for HCC detection further improve the predictive performance of these models[39]. A model using a combination of age, gender, AFP, AFP-L3 and DCP estimates the probability to suffer from HCC in an individual patient with chronic liver disease with a sensitivity of 86% for HCC in BCLC stage 0 or A and a sensitivity of 94% for later tumor stages[40]. The diagnostic performance of novel circulating biomarkers and scores is summarized in Table 2.
| Ref. | Year | Marker | n | Comparator | Cut-off-level | Sensitivity | Specifity | AUC |
| Toyoda et al[31] | 2011 | hs-AFP-L3% | 666 | Chronic liver disease and AFP < 20 ng/mL | 5% | 41.50% | 85.10% | 0.707 |
| Ertle et al[37] | 2013 | DCP | 586 | Chronic liver disease | 5 ng/mL | 45.80% | 95% | 0.87 |
| Wan et al[24] | 2014 | Osteopontin | Meta-analysis (7 studies) | mixed | Pooled: 86% | Pooled: 86% | 0.92 | |
| Hsia et al[35] | 2007 | IL-6 | 128 | Mixed, including chronic liver disease and healthy controls | 3 pg/mL | 46% | 95% | |
| Mao et al[21] | 2010 | GP73 | 4217 | Mixed, including chronic liver disease and healthy controls | 8.5 rel. units | 74.60% | 97.40% | 0.94 |
| Giannelli et al[36] | 2007 | SCCA | 961 | Liver cirrhosis | 3.8 ng/mL | 41.90% | 82.60% | 0.656 |
| Ertle et al[37] | 2013 | AFP combined with DCP | 586 | Chronic liver disease | DCP 5 ng/mL AFP 10 ng/mL | 78% | 89.30% | 0.91 |
| Johnson et al[40] | 2014 | GALAD-score | 670 | Chronic liver disease | 93% | 89% |
Although the complex process of hepatocarcinogenesis is still not fully understood, several signal transduction pathways have been identified as critical players in the pathophysiology of HCC, including the Wnt/β-Catenin pathway, the p53 pathway, the tumor suppressor retinoblastoma protein pRb1 pathway, the mitogen-activated protein kinase pathway, the Ras pathway, JAK/STAT signaling, mechanisms of cellular stress response like heat shock proteins and epidermal growth factor receptor and transforming growth factor-β signaling[41]. As a consequence of different risk factors causing HCC in the individual patient, the alterations in these pathways differ in different settings which is probably the cause of insufficient sensitivity of single biomarkers. Genetic and epigenetic alterations occur in these pathways and mediate cell proliferation. The possibility to perform proteomic profiling and whole genome sequencing in combination with systems biology has led to a new era in biomarker development that will hopefully help to understand the complex interactions in hepatocarcinogenesis of multiple proteins, genes and transcription factors. First examples of this approach have successfully been evaluated in clinical studies, but none of the signatures has been validated in large prospective studies.
In patients with chronic HBV infection and liver cirrhosis, proteomic analyses in the plasma identified a cluster of 11 proteins that is able to identify patients at high risk for HCC development (OR = 4.83, 95%CI: 1.26-18.56)[42].
Gene expression profiling of peripheral blood mononuclear cells in HCC patients using microarrays and bioinformatics-driven analysis of the data has identified a blood-based signature of three genes, namely Chemokine (C-X-C motif) receptor 2 (CXCR2), C-C chemokine receptor type 2 (CCR2) and E1A-Binding Protein P400 (EP400), that predicts HCC with an AUC of 0.96 yielding at a sensitivity of 93% with a specificity of 89%[43].
High-throughput metabolomics technologies with the comprehensive analysis of small molecular metabolites may additionally identify serum metabolic profiles that can be used as diagnostic biomarkers. First steps into this direction have also already been taken[44,45].
To distinguish dysplastic nodules from well-differentiated HCC is a challenge, not only for the radiologist on imaging, but also for the pathologist on tissue samples.
Molecular signatures derived from gene expression profiling have been identified that are helpful to answer this critical question that is decisive for the further management of the patient.
Characteristic genomic changes during hepatocarcinogenesis have been identified. Specific gene signatures accurately reflect the pathological progression of disease from cirrhosis to dysplasia to early and advanced HCC in patients with HCV infection in Asian and Western patients[46,47].
A three gene set in the tissue including glypican 3 (GPC3; 18-fold increase in HCC, P = 0.01), LYVE1 (12-fold decrease in HCC, P = 0.0001), and survivin (2.2-fold increase in HCC, P = 0.02) has an accuracy of 94% to discriminate dysplastic nodules from early HCC in HCV cirrhosis. Especially immunostaining for GPC3 is highly discriminative[48].
Heat shock protein 70 and cyclase-associated protein 2 are further examples for tissue biomarkers identified in comprehensive approaches that found their way into clinical testing and application[49,50].
With respect to novel potential biomarkers, non-coding RNA and specifically microRNA (miRNA) have received the greatest attention over the past years[51]. MiRNAs are small non-coding and evolutionary conserved RNA molecules that serve as posttranscriptional regulators of mRNA expression and interfere with translation to protein. Following several common modifications steps, miRNA become a part of the so called RISC (RNA silencing complex) to be functionally active[52]. MiRNAs can either preserve their function intracellularly by regulating the expression of a target population of molecules, or can be released from the cell bound to other proteins and also as a free molecule[53,54]. As part of the released vesicles, specific miRNAs can further preserve their functional activity locally or be transported in blood or probably other specimens to other tissues or organs[55].
The most exiting advantages of miRNA over various other molecules are their stability against degradation, cell-type specific miRNA expression patterns and detectability in all types of human specimens such as blood, feces, saliva, etc.[56-59]. While mRNA or various proteins are relatively sensitive to extracellular enzymes, miRNA expression levels remain, as long as they are preserved in a natural milieu, relatively resistant to RNA digestions, heating, storage, drying, formalin fixation, etc. For detailed information regarding the biogenesis of miRNA as well as regarding the current knowledge on molecular function we refer to the several excellent reviews from the field[51,60,61].
Shortly after definite recognition of miRNA, several groups have provided seminal evidence for differences in miRNA expression patterns between different tissues and malignant conditions including HCC[62,63]. High quality analyses using deep sequencing have recently provided an important view in microRNAome in liver tissue and HCC[64]. Interestingly, about 86% of the miRNA were expressed in very low concentrations and only about 1% were expressed abundantly. Three of those miRNAs, namely miR-122, miR-192 and miR-199a/b-3p, were responsible for 74% of all miRNA in normal liver tissue with miR-122 accounting for almost 52% suggesting that those miRNAs are the most important ones in liver biology[64]. Many recent reports have shown a broad spectrum of changes in microRNAoma in HCC[64-67]. Therefore, miRNAs may have the potential to become valid biomarkers in HCC.
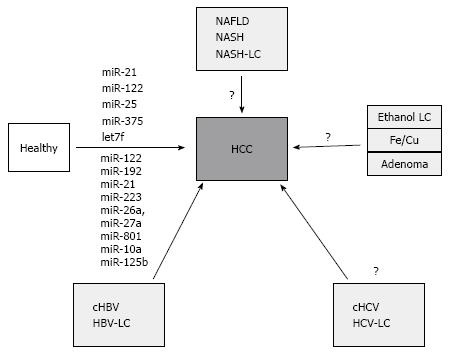
The biggest effort from miRNA-based biomarker research has been made to improve the diagnostic utility in HCC. In parallel with the dominant etiological factors in HCC development, the largest body of data comes from Asian populations and virus-related HCC cohorts. In one of the first profiling studies, Li et al[68] performed deep sequencing in pooled samples from chronic HBV virus patients, HCC patients and controls with and without cancer. They identified a pattern of 21 miRNA that show differential expression in cHBV patients and 6 miRNA differentially expressed in HCC patients. Following subsequent testing and validation, 13 miRNA, including miR-122, miR-375, miR-92a, miR-10a and let-7c, were identified as a biomarker for patients with HBV (acute and chronic) and HCV virus infection. Furthermore, using only 3 miRNAs (miR-25, miR-375, let7f) the authors could reach an AUC of 99.7% with a 97.9% sensitivity and a 99.1% specificity to discriminate controls from HCC patients. Most interestingly for HCC diagnosis, the comparison of the two cohorts with chronic HBV and HBV-associated HCC lead to identification of two miRNAs (miR-10a and miR-125b) that could separate the HCC cohort with an AUC of 99.2% (sensitivity 98.5% and specificity 98.5)[68]. Recently, another large scale study studied plasma samples from 934 patients with various conditions including healthy subjects, patients with chronic HBV, liver cirrhosis and HBV-related HCC[69]. Following discovery and training phases, the authors identified a panel of 7 microRNAs (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a and miR-801) that provided the highest diagnostic accuracy for the identification of HBV-related HCC. Using an independent validation cohort of 390 samples, the area under the curve value was comparable with the training data and reached 0.888 with a sensitivity of 81.8% and a specificity of 83.5%. Interestingly, the diagnostic accuracy was independent of disease stage and was comparable to healthy subjects, patients with chronic hepatitis or livers cirrhosis. At present, this study is one of the largest to evaluate the biomarker potential of miRNAs in cancer. Notably, the expression of selected miRNA was analyzed using RT-PCR which may be critical for clinical translation of the results. Whether miR-1228 is the optimal normalizer needs further evaluation[69].
Besides profiling studies, a candidate-based approach has been also used to evaluate the expression differences of liver and tumor-related miRNA. In particular miR-21, which is most frequently deregulated miRNA in cancer, was found at higher level both in sera and plasma from HCC patients[70,71], while other showed no significant expression differences[72,73]. In similar fashion, miR-122, the most abundant miRNA of the liver, was also found at high level in sera from HCC patients[71,72]. However, the level of circulating miR-122 may be strongly influenced by inflammation or apoptosis of hepatocytes in such conditions as acute or chronic hepatitis or nonalcoholic fatty liver disease, suggesting that background condition of the liver inflammation may strongly influence the miR-122 level[74]. Nevertheless, the data from candidate-based studies correlate with the data from Link et al[54] at least for both miRNAs miR-21 and miR-122[69]. There are several other studies that have identified additional miRNA, however, at present no independent validation has been performed (Figure 1). It is further more important to mention that the potential of miRNA as biomarker has not been equally analyzed in all HCC-related risk conditions. Systemic analyses for alcohol, NASH or HCV-related conditions are pending.
The molecular heterogeneity of HCC results in differences in outcome of affected patients. Clinically, main factors that have an impact on patient survival have been identified. These include tumor related factors like number and size of nodules, vascular invasion, existence of extrahepatic metastases, liver function and patient related factors. The Barcelona Classification summarizes these factors in a comprehensive algorithm and is endorsed by current guidelines[10,11]. However, within the defined tumor stages the survival of patients is still heterogeneous, and some patients that are treated in curative intent or even undergo liver transplantation show early recurrence of disease. Knowledge on high-risk profiles would therefore be important to guide individualized treatment.
Several of the non-invasive biomarkers that have been evaluated for their diagnostic power in HCC have also been studied for their prognostic significance.
High expression of AFP in serum correlates with high cell proliferation, high angiogenesis and low apoptosis and is associated with poor prognosis[75,76]. The fraction of AFP-L3 is another prognostic biomarker for survival after resection of HCC[77,78]. Patients that have undergone resection of HCC and had elevated levels of AFP, AFP-L3 and DCP at baseline had a worse prognosis than those patients that are positive for just one or two of the markers before surgery[79]. The combination of AFP, the percentage of AFP-L3 and DCP combined with the concentrations of bilirubin and albumin, summarized in the BALAD score, is prognostic for survival of patients with HCC in an Asian population[80]. Recently, a modification of this model, the BALAD-2 score, was validated in an international setting and confirmed to reliably predict the prognosis of patients with HCC[81].
Other circulating biomarkers that mirror current knowledge on pathways involved in hepatocarcinogenesis and shown to be of prognostic value are, amongst others, IGF1[82], DKK1[83,84], GPC-3 and HSP 70[85] although prospective validation studies are still to come. In patients with advanced HCC, baseline angiopoietin 2 (Ang2), and VEGF concentration in the plasma also independently predict survival[76].
In addition to their diagnostic potential, miRNAs may be helpful in prediction of the prognosis of HCC. Li et al[86] studied the expression of several miRNAs in sera from 46 HCC patients and 20 controls. Specifically, miR-221 was fond in high concentration in HCC sera samples, which correlated with tumor size, cirrhosis and tumor stage. Kaplan-Meier survival analyses revealed an inverse correlation between miR-221 expression and survival rates. In another study, Tomimaru et al[70] analyzed miR-21 expression in plasma from 126 HCC patients. MiR-21 expression was high in HCC and diminished after surgical treatment. Most importantly, high miR-21 expression level in plasma correlated with shorter cumulative survival following treatment. Köberle et al[87] analyzed the performance of miR-1 and miR-122 in European HCC patients. Higher miR-1 and miR-122 serum levels were associated with longer overall survival compared to low expression of those miRNAs. However, miR-122, but not miR-1, showed a correlation with hepatic inflammation, liver function and synthetic capacity. The authors conclude that miR-1 may be a liver function independent predictive biomarker of HCC. There is also growing evidence that miRNA signature profiling can be useful in prognostic stratification[88]. A signature of 31-miRNA correlates with stage of disease[89]. A distinct 20-miRNA signature associated with metastases of HCC has also been identified[90].
Despite of the promising potential, there are several pitfalls in utility and implementation of miRNA-based biomarkers in clinical practice. First, the majority of data comes from Asian populations with predominantly virus-related HCC (Figure 1)[54]. However, in European or American populations the incidence of virus-related HCC is dropping and increases for NAFLD-related conditions therefore the data in these patients may probably be different. Second, the complexity of the miRNA alterations in the background of liver pathology (ex. chronic hepatitis with early fibrosis or cirrhosis) may impact the pattern of miRNA expression with increasing expression in one of the conditions and decreasing level in another. Furthermore, the ideal biomarker is probably the one that is expressed in HCC tissue with increasing concentration during progression of the disease. A combination of miRNA with the established-although not ideal-biomarker AFP may probably be beneficial[70].
In the above section, we provided a brief insight into the growing field of miRNA-based biomarker research for HCC. Before this approach may be further utilized for clinical testing there are also critical technical questions that need to be answered. What is the best non-invasive specimen for the early diagnosis of HCC: plasma or serum? What normalizer is the best for the analyses? What is the best method for translational testing? Indeed, array-based analyses may be probably too expensive to apply, therefore, a candidate-based approach will need to be standardized for effective implementation. Those are only few reasons why the currently available data are so heterogeneous[54,67]. Nevertheless, this miRNA-based approach may provide an additional value in personal-based management by prediction and application of new therapeutic targets[91].
After curative treatment of HCC the prognosis of the patient depends on the characteristics of the resected cancer but in addition on the risk of carcinogenesis due to the underlying etiology and inflammatory activity of chronic liver disease which persist after surgical resection or ablation. In a landmark study Hoshida et al[92] demonstrated that gene-expression profiling can be performed in frozen as well as in formalin-fixed paraffin-embedded tissues and identified a gene-expression signature in liver tissue adjacent to tumor in patients who underwent resection of HCC that correlated with survival.
Since then, a large number of gene-expression profile studies has been performed in HCC with the aim to distinguish molecular subtypes. A validated and commonly accepted molecular classification has not been identified so far. Based on a meta-analysis of gene expression profiles from eight European cohorts of patients with HCC, a classification framework for HCC based on gene expression profiles was proposed that distinguishes three HCC subclasses, each correlated with clinical parameters such as tumor size, extent of cellular differentiation, and serum α-fetoprotein levels[50,93]. The results of a selection of recent studies with respect to prognostic gene expression profiles are summarized in Table 3. In a large validation study gene expression profiles of tumor and adjacent tissue were evaluated for their prognostic significance and a composite prognostic model was developed[94]. A further validation of this model is pending before it may be considered for clinical use.
| Ref. | Year | Correlation with | No. of genes in signature | AUC | P-value |
| Nault et al[95] | 2013 | Disease-free survival after resection | 5 (tumor) | 0.8 | < 0.0001 |
| Lim et al[96] | 2013 | Disease-free survival after resection | 25 (tumor) | 0.002 | |
| Kurokawa et al[97] | 2004 | Tumor recurrence after resection | 20 (tumor) | 0.001 | |
| Yoshioka et al[98] | 2009 | Tumor recurrence after resection | 172 (tumor) | < 0.0001 | |
| Woo et al[99] | 2008 | Recurrence free survival | 628 (tumor) | < 0.01 |
High-throughput technologies allow the identification of new molecules involved in complex pathways and their interaction in hepatocarcinogenesis. A single perfect biomarker has not been found so far to accomplish with the clinical demand for optimal HCC patient care. A combination of serological biomarkers may offer a better risk stratification of patients belonging to high-risk populations in the future. The combination of clinical characteristics and morphological signatures of tumor and the surrounding tissue will most likely be the best option for risk stratification and prediction of prognosis in patients with HCC in the future. Individualized treatment approaches that take into account the patient’s own cancer genetic profile need to be addressed in further research[12].
However, the critical step for translational research is to move the identified candidate signatures or single biomarkers from bench to bedside. There is a great hope that new molecular biomarkers can support clinicians in their daily routine and improve the care of patients with HCC. However, analyses tools need to be standardized and simplified in order to be useful, reliable and widely available.
P- Reviewer: Kapoor S, Sazci A, Zhang X S- Editor: Tian YL L- Editor: A E- Editor: Lu YJ
| 1. | Njei B, Rotman Y, Ditah I, Lim JK. Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology. 2014;61:191-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 433] [Article Influence: 43.3] [Reference Citation Analysis (1)] |
| 2. | El-Serag HB, Kanwal F. Epidemiology of hepatocellular carcinoma in the United States: Where are we? Where do we go? Hepatology. 2014;60:1767-1775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 492] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 3. | Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47 Suppl:S2-S6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 770] [Cited by in RCA: 880] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 4. | Schütte K, Kipper M, Kahl S, Bornschein J, Götze T, Adolf D, Arend J, Seidensticker R, Lippert H, Ricke J. Clinical characteristics and time trends in etiology of hepatocellular cancer in Germany. Digestion. 2013;87:147-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Singal A, Volk ML, Waljee A, Salgia R, Higgins P, Rogers MA, Marrero JA. Meta-analysis: surveillance with ultrasound for early-stage hepatocellular carcinoma in patients with cirrhosis. Aliment Pharmacol Ther. 2009;30:37-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 573] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 6. | Singal AG, Conjeevaram HS, Volk ML, Fu S, Fontana RJ, Askari F, Su GL, Lok AS, Marrero JA. Effectiveness of hepatocellular carcinoma surveillance in patients with cirrhosis. Cancer Epidemiol Biomarkers Prev. 2012;21:793-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 202] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 7. | Kim do Y, Han KH. Epidemiology and surveillance of hepatocellular carcinoma. Liver Cancer. 2012;1:2-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 137] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 8. | Saffroy R, Pham P, Reffas M, Takka M, Lemoine A, Debuire B. New perspectives and strategy research biomarkers for hepatocellular carcinoma. Clin Chem Lab Med. 2007;45:1169-1179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 89] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 9. | Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4333] [Cited by in RCA: 4508] [Article Influence: 225.4] [Reference Citation Analysis (0)] |
| 10. | Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5972] [Cited by in RCA: 6573] [Article Influence: 469.5] [Reference Citation Analysis (1)] |
| 11. | European Association For The Study Of The Liver, European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4059] [Cited by in RCA: 4521] [Article Influence: 347.8] [Reference Citation Analysis (2)] |
| 12. | Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 929] [Cited by in RCA: 1108] [Article Influence: 100.7] [Reference Citation Analysis (1)] |
| 13. | Omata M, Lesmana LA, Tateishi R, Chen PJ, Lin SM, Yoshida H, Kudo M, Lee JM, Choi BI, Poon RT. Asian Pacific Association for the Study of the Liver consensus recommendations on hepatocellular carcinoma. Hepatol Int. 2010;4:439-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 797] [Cited by in RCA: 840] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 14. | Makuuchi M, Kokudo N, Arii S, Futagawa S, Kaneko S, Kawasaki S, Matsuyama Y, Okazaki M, Okita K, Omata M. Development of evidence-based clinical guidelines for the diagnosis and treatment of hepatocellular carcinoma in Japan. Hepatol Res. 2008;38:37-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 232] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 15. | Chen JG, Parkin DM, Chen QG, Lu JH, Shen QJ, Zhang BC, Zhu YR. Screening for liver cancer: results of a randomised controlled trial in Qidong, China. J Med Screen. 2003;10:204-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 223] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 16. | McMahon BJ, Bulkow L, Harpster A, Snowball M, Lanier A, Sacco F, Dunaway E, Williams J. Screening for hepatocellular carcinoma in Alaska natives infected with chronic hepatitis B: a 16-year population-based study. Hepatology. 2000;32:842-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 242] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 17. | Sherman M. Serological surveillance for hepatocellular carcinoma: time to quit. J Hepatol. 2010;52:614-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 18. | Forner A, Reig M, Bruix J. Alpha-fetoprotein for hepatocellular carcinoma diagnosis: the demise of a brilliant star. Gastroenterology. 2009;137:26-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 75] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 19. | Giannini EG, Trevisani F. Surveillance for hepatocellular carcinoma: just do it! Am J Gastroenterol. 2013;108:1013-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 20. | Marrero JA, Feng Z, Wang Y, Nguyen MH, Befeler AS, Roberts LR, Reddy KR, Harnois D, Llovet JM, Normolle D. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology. 2009;137:110-118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 583] [Cited by in RCA: 571] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 21. | Mao Y, Yang H, Xu H, Lu X, Sang X, Du S, Zhao H, Chen W, Xu Y, Chi T. Golgi protein 73 (GOLPH2) is a valuable serum marker for hepatocellular carcinoma. Gut. 2010;59:1687-1693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 197] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 22. | Farinati F, Marino D, De Giorgio M, Baldan A, Cantarini M, Cursaro C, Rapaccini G, Del Poggio P, Di Nolfo MA, Benvegnù L. Diagnostic and prognostic role of alpha-fetoprotein in hepatocellular carcinoma: both or neither? Am J Gastroenterol. 2006;101:524-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 312] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 23. | Lok AS, Sterling RK, Everhart JE, Wright EC, Hoefs JC, Di Bisceglie AM, Morgan TR, Kim HY, Lee WM, Bonkovsky HL. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology. 2010;138:493-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 451] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 24. | Wan H, Xu H, Gu Y, Wang H, Xu W, Zu M. Comparison osteopontin vs AFP for the diagnosis of HCC: A meta-analysis. Clin Res Hepatol Gastroenterol. 2014;38:706-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 25. | Xu C, Yan Z, Zhou L, Wang Y. A comparison of glypican-3 with alpha-fetoprotein as a serum marker for hepatocellular carcinoma: a meta-analysis. J Cancer Res Clin Oncol. 2013;139:1417-1424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 26. | Richardson P, Duan Z, Kramer J, Davila JA, Tyson GL, El-Serag HB. Determinants of serum alpha-fetoprotein levels in hepatitis C-infected patients. Clin Gastroenterol Hepatol. 2012;10:428-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 27. | Mossad NA, Mahmoud EH, Osman EA, Mahmoud SH, Shousha HI. Evaluation of squamous cell carcinoma antigen-immunoglobulin M complex (SCCA-IGM) and alpha-L-fucosidase (AFU) as novel diagnostic biomarkers for hepatocellular carcinoma. Tumour Biol. 2014;35:11559-11564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 28. | Bertino G, Ardiri A, Malaguarnera M, Malaguarnera G, Bertino N, Calvagno GS. Hepatocellualar carcinoma serum markers. Semin Oncol. 2012;39:410-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 29. | Zhang J, Zhao Y, Yang Q. Sensitivity and specificity of Dickkopf-1 protein in serum for diagnosing hepatocellular carcinoma: a meta-analysis. Int J Biol Markers. 2014;29:e403-e410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 30. | Kumada T, Toyoda H, Tada T, Kiriyama S, Tanikawa M, Hisanaga Y, Kanamori A, Tanaka J, Kagebayashi C, Satomura S. High-sensitivity Lens culinaris agglutinin-reactive alpha-fetoprotein assay predicts early detection of hepatocellular carcinoma. J Gastroenterol. 2014;49:555-563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 31. | Toyoda H, Kumada T, Tada T, Kaneoka Y, Maeda A, Kanke F, Satomura S. Clinical utility of highly sensitive Lens culinaris agglutinin-reactive alpha-fetoprotein in hepatocellular carcinoma patients with alpha-fetoprotein & lt; 20 ng/mL. Cancer Sci. 2011;102:1025-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 32. | Witjes CD, van Aalten SM, Steyerberg EW, Borsboom GJ, de Man RA, Verhoef C, Ijzermans JN. Recently introduced biomarkers for screening of hepatocellular carcinoma: a systematic review and meta-analysis. Hepatol Int. 2013;7:59-64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 33. | Marrero JA, Romano PR, Nikolaeva O, Steel L, Mehta A, Fimmel CJ, Comunale MA, D’Amelio A, Lok AS, Block TM. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J Hepatol. 2005;43:1007-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 285] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 34. | Porta C, De Amici M, Quaglini S, Paglino C, Tagliani F, Boncimino A, Moratti R, Corazza GR. Circulating interleukin-6 as a tumor marker for hepatocellular carcinoma. Ann Oncol. 2008;19:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 125] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 35. | Hsia CY, Huo TI, Chiang SY, Lu MF, Sun CL, Wu JC, Lee PC, Chi CW, Lui WY, Lee SD. Evaluation of interleukin-6, interleukin-10 and human hepatocyte growth factor as tumor markers for hepatocellular carcinoma. Eur J Surg Oncol. 2007;33:208-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 95] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 36. | Giannelli G, Fransvea E, Trerotoli P, Beaugrand M, Marinosci F, Lupo L, Nkontchou G, Dentico P, Antonaci S. Clinical validation of combined serological biomarkers for improved hepatocellular carcinoma diagnosis in 961 patients. Clin Chim Acta. 2007;383:147-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 37. | Ertle JM, Heider D, Wichert M, Keller B, Kueper R, Hilgard P, Gerken G, Schlaak JF. A combination of α-fetoprotein and des-γ-carboxy prothrombin is superior in detection of hepatocellular carcinoma. Digestion. 2013;87:121-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 83] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 38. | Hadziyannis E, Sialevris K, Georgiou A, Koskinas J. Analysis of serum α-fetoprotein-L3% and des-γ carboxyprothrombin markers in cases with misleading hepatocellular carcinoma total α-fetoprotein levels. Oncol Rep. 2013;29:835-839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 39. | Wang M, Mehta A, Block TM, Marrero J, Di Bisceglie AM, Devarajan K. A comparison of statistical methods for the detection of hepatocellular carcinoma based on serum biomarkers and clinical variables. BMC Med Genomics. 2013;6 Suppl 3:S9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 40. | Johnson PJ, Pirrie SJ, Cox TF, Berhane S, Teng M, Palmer D, Morse J, Hull D, Patman G, Kagebayashi C. The detection of hepatocellular carcinoma using a prospectively developed and validated model based on serological biomarkers. Cancer Epidemiol Biomarkers Prev. 2014;23:144-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 229] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 41. | Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047-2063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 512] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 42. | Liu CC, Wang YH, Chuang EY, Tsai MH, Chuang YH, Lin CL, Liu CJ, Hsiao BY, Lin SM, Liu LY. Identification of a liver cirrhosis signature in plasma for predicting hepatocellular carcinoma risk in a population-based cohort of hepatitis B carriers. Mol Carcinog. 2014;53:58-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 43. | Shi M, Chen MS, Sekar K, Tan CK, Ooi LL, Hui KM. A blood-based three-gene signature for the non-invasive detection of early human hepatocellular carcinoma. Eur J Cancer. 2014;50:928-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 44. | Wang X, Zhang A, Sun H. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology. 2013;57:2072-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 175] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 45. | Chen F, Xue J, Zhou L, Wu S, Chen Z. Identification of serum biomarkers of hepatocarcinoma through liquid chromatography/mass spectrometry-based metabonomic method. Anal Bioanal Chem. 2011;401:1899-1904. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 78] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 46. | Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 506] [Cited by in RCA: 558] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 47. | Nam SW, Park JY, Ramasamy A, Shevade S, Islam A, Long PM, Park CK, Park SE, Kim SY, Lee SH. Molecular changes from dysplastic nodule to hepatocellular carcinoma through gene expression profiling. Hepatology. 2005;42:809-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 125] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 48. | Llovet JM, Chen Y, Wurmbach E, Roayaie S, Fiel MI, Schwartz M, Thung SN, Khitrov G, Zhang W, Villanueva A. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology. 2006;131:1758-1767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 277] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 49. | Sakamoto M, Mori T, Masugi Y, Effendi K, Rie I, Du W. Candidate molecular markers for histological diagnosis of early hepatocellular carcinoma. Intervirology. 2008;51 Suppl 1:42-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 50. | Mínguez B, Lachenmayer A. Diagnostic and prognostic molecular markers in hepatocellular carcinoma. Dis Markers. 2011;31:181-190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 44] [Reference Citation Analysis (0)] |
| 51. | Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 406] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 52. | Link A, Kupcinskas J, Wex T, Malfertheiner P. Macro-role of microRNA in gastric cancer. Dig Dis. 2012;30:255-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 53. | Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003-5008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2345] [Cited by in RCA: 2643] [Article Influence: 188.8] [Reference Citation Analysis (0)] |
| 54. | Link A, Goel A. MicroRNA in gastrointestinal cancer: a step closer to reality. Adv Clin Chem. 2013;62:221-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 55. | Redis RS, Calin S, Yang Y, You MJ, Calin GA. Cell-to-cell miRNA transfer: from body homeostasis to therapy. Pharmacol Ther. 2012;136:169-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 142] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 56. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5636] [Cited by in RCA: 6315] [Article Influence: 371.5] [Reference Citation Analysis (0)] |
| 57. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3218] [Cited by in RCA: 3554] [Article Influence: 209.1] [Reference Citation Analysis (0)] |
| 58. | Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733-1741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 2087] [Article Influence: 139.1] [Reference Citation Analysis (0)] |
| 59. | Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, Goel A. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766-1774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 251] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 60. | Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143:1442-1460.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 201] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 61. | Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3368] [Cited by in RCA: 4121] [Article Influence: 374.6] [Reference Citation Analysis (1)] |
| 62. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7370] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 63. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4471] [Cited by in RCA: 4523] [Article Influence: 238.1] [Reference Citation Analysis (0)] |
| 64. | Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 588] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 65. | Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537-2545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 876] [Cited by in RCA: 896] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 66. | Toffanin S, Hoshida Y, Lachenmayer A, Villanueva A, Cabellos L, Minguez B, Savic R, Ward SC, Thung S, Chiang DY. MicroRNA-based classification of hepatocellular carcinoma and oncogenic role of miR-517a. Gastroenterology. 2011;140:1618-1628.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 179] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 67. | Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 297] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 68. | Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, Shen HB, Zhang CY, Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798-9807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 378] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 69. | Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang JF, Zhang Z, Lu S, Huang X. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781-4788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 498] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 70. | Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I, Umeshita K. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 285] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 71. | Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 450] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 72. | Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One. 2011;6:e28486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 241] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 73. | Bihrer V, Waidmann O, Friedrich-Rust M, Forestier N, Susser S, Haupenthal J, Welker M, Shi Y, Peveling-Oberhag J, Polta A. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PLoS One. 2011;6:e26971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 116] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 74. | Tan Y, Ge G, Pan T, Wen D, Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS One. 2014;9:e105192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 141] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 75. | Mitsuhashi N, Kobayashi S, Doki T, Kimura F, Shimizu H, Yoshidome H, Ohtsuka M, Kato A, Yoshitomi H, Nozawa S. Clinical significance of alpha-fetoprotein: involvement in proliferation, angiogenesis, and apoptosis of hepatocellular carcinoma. J Gastroenterol Hepatol. 2008;23:e189-e197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 76. | Llovet JM, Peña CE, Lathia CD, Shan M, Meinhardt G, Bruix J. Plasma biomarkers as predictors of outcome in patients with advanced hepatocellular carcinoma. Clin Cancer Res. 2012;18:2290-2300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 464] [Cited by in RCA: 455] [Article Influence: 35.0] [Reference Citation Analysis (2)] |
| 77. | Matsuda M, Asakawa M, Amemiya H, Fujii H. Lens culinaris agglutinin-reactive fraction of AFP is a useful prognostic biomarker for survival after repeat hepatic resection for HCC. J Gastroenterol Hepatol. 2011;26:731-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 78. | Saito Y, Shimada M, Utsunomiya T, Morine Y, Imura S, Ikemoto T, Mori H, Hanaoka J, Yamada S, Asanoma M. Prediction of recurrence of hepatocellular carcinoma after curative hepatectomy using preoperative Lens culinaris agglutinin-reactive fraction of alpha-fetoprotein. Hepatol Res. 2012;42:887-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 79. | Kiriyama S, Uchiyama K, Ueno M, Ozawa S, Hayami S, Tani M, Yamaue H. Triple positive tumor markers for hepatocellular carcinoma are useful predictors of poor survival. Ann Surg. 2011;254:984-991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 80. | Toyoda H, Kumada T, Osaki Y, Oka H, Urano F, Kudo M, Matsunaga T. Staging hepatocellular carcinoma by a novel scoring system (BALAD score) based on serum markers. Clin Gastroenterol Hepatol. 2006;4:1528-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 82] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 81. | Fox R, Berhane S, Teng M, Cox T, Tada T, Toyoda H, Kumada T, Kagebayashi C, Satomura S, Johnson PJ. Biomarker-based prognosis in hepatocellular carcinoma: validation and extension of the BALAD model. Br J Cancer. 2014;110:2090-2098. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 82. | Kaseb AO, Morris JS, Hassan MM, Siddiqui AM, Lin E, Xiao L, Abdalla EK, Vauthey JN, Aloia TA, Krishnan S. Clinical and prognostic implications of plasma insulin-like growth factor-1 and vascular endothelial growth factor in patients with hepatocellular carcinoma. J Clin Oncol. 2011;29:3892-3899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 83. | Huang Y, Yang X, Zhao F, Shen Q, Wang Z, Lv X, Hu B, Yu B, Fan J, Qin W. Overexpression of Dickkopf-1 predicts poor prognosis for patients with hepatocellular carcinoma after orthotopic liver transplantation by promoting cancer metastasis and recurrence. Med Oncol. 2014;31:966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 84. | Tung EK, Mak CK, Fatima S, Lo RC, Zhao H, Zhang C, Dai H, Poon RT, Yuen MF, Lai CL. Clinicopathological and prognostic significance of serum and tissue Dickkopf-1 levels in human hepatocellular carcinoma. Liver Int. 2011;31:1494-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 109] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 85. | Wang L, Yao M, Dong Z, Zhang Y, Yao D. Circulating specific biomarkers in diagnosis of hepatocellular carcinoma and its metastasis monitoring. Tumour Biol. 2014;35:9-20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 86. | Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;406:70-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 168] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 87. | Köberle V, Kronenberger B, Pleli T, Trojan J, Imelmann E, Peveling-Oberhag J, Welker MW, Elhendawy M, Zeuzem S, Piiper A. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur J Cancer. 2013;49:3442-3449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 148] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 88. | Li X, Yang W, Lou L, Chen Y, Wu S, Ding G. microRNA: a promising diagnostic biomarker and therapeutic target for hepatocellular carcinoma. Dig Dis Sci. 2014;59:1099-1107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 89. | Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098-1112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 306] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 90. | Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 568] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 91. | Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685-1694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1665] [Cited by in RCA: 1702] [Article Influence: 141.8] [Reference Citation Analysis (0)] |
| 92. | Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995-2004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1044] [Cited by in RCA: 1006] [Article Influence: 59.2] [Reference Citation Analysis (0)] |
| 93. | Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385-7392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 956] [Cited by in RCA: 945] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 94. | Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, Cornella H, Liberzon A, Kobayashi M, Kumada H. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501-15122.e2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 334] [Cited by in RCA: 334] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 95. | Nault JC, De Reyniès A, Villanueva A, Calderaro J, Rebouissou S, Couchy G, Decaens T, Franco D, Imbeaud S, Rousseau F. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 266] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 96. | Lim HY, Sohn I, Deng S, Lee J, Jung SH, Mao M, Xu J, Wang K, Shi S, Joh JW. Prediction of disease-free survival in hepatocellular carcinoma by gene expression profiling. Ann Surg Oncol. 2013;20:3747-3753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 97. | Kurokawa Y, Matoba R, Takemasa I, Nagano H, Dono K, Nakamori S, Umeshita K, Sakon M, Ueno N, Oba S. Molecular-based prediction of early recurrence in hepatocellular carcinoma. J Hepatol. 2004;41:284-291. [PubMed] |
| 98. | Yoshioka S, Takemasa I, Nagano H, Kittaka N, Noda T, Wada H, Kobayashi S, Marubashi S, Takeda Y, Umeshita K. Molecular prediction of early recurrence after resection of hepatocellular carcinoma. Eur J Cancer. 2009;45:881-889. [PubMed] |
| 99. | Woo HG, Park ES, Cheon JH, Kim JH, Lee JS, Park BJ, Kim W, Park SC, Chung YJ, Kim BG. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res. 2008;14:2056-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 135] [Article Influence: 7.9] [Reference Citation Analysis (0)] |