Published online Aug 27, 2014. doi: 10.4254/wjh.v6.i8.613
Revised: March 13, 2014
Accepted: July 12, 2014
Published online: August 27, 2014
Processing time: 212 Days and 9.9 Hours
AIM: To identify novel non-invasive biomarkers for non-alcoholic fatty liver disease (NAFLD).
METHODS: Twenty patients with histologically proven NAFLD and 20 controls were included. All NAFLD cases were scored using the NAFLD activity score. The relative expressions of miR-197, miR-146b, miR-10b, miR-181d, miR-34a, miR-122, miR-99a and miR-29a were analyzed using real-time polymerase chain reaction.
RESULTS: Serum levels of miR-181d, miR-99a, miR-197 and miR-146b were significantly lower in biopsy-proven NAFLD patients than in the healthy controls. Serum levels of miR-197 and miR-10b were inversely correlated with degree of inflammation and miR-181d and miR-99a were inversely correlated with serum gamma glutamyl transferase levels in non-alcoholic steatohepatitis patients.
CONCLUSION: NAFLD is associated with altered serum miRNA expression pattern. This study provides clues for defining the non-invasive diagnosis of NAFLD.
Core tip: Due to the limitations of liver biopsy, the use of non-invasive markers has emerged in recent years. MicroRNAs (miRNAs) are a class of naturally occurring small noncoding RNAs that regulate gene expression. Altered miRNA expression has been reported in animal and human liver samples in non-alcoholic fatty liver disease (NAFLD). There is, however, only limited information on their detection in blood and their correlation with histological disease severity in patients with NAFLD. For this reason, we measured the serum levels of some miRNAs in non-alcoholic steatohepatitis (NASH) patients. Of these microRNAs, miR-181d, miR-99a, miR-197 and miR-146b were expressed at lower levels in NASH patients than in controls. Serum levels of miR-197 and miR-10b were inversely correlated with degree of inflammation and miR-181d and miR-99a were inversely correlated with serum gamma glutamyl transferase levels in NASH patients.
- Citation: Celikbilek M, Baskol M, Taheri S, Deniz K, Dogan S, Zararsiz G, Gursoy S, Guven K, Ozbakır O, Dundar M, Yucesoy M. Circulating microRNAs in patients with non-alcoholic fatty liver disease. World J Hepatol 2014; 6(8): 613-620
- URL: https://www.wjgnet.com/1948-5182/full/v6/i8/613.htm
- DOI: https://dx.doi.org/10.4254/wjh.v6.i8.613
Non-alcoholic fatty liver disease (NAFLD) is the most common cause of chronic liver disease in Western populations. It comprises a disease spectrum which includes variable degrees of simple steatosis (fatty liver), non-alcoholic steatohepatitis (NASH) and cirrhosis which are likely to be characterized by different pathogenesis and clinical history[1]. Liver biopsy is used in daily practice for the diagnosis of NASH and histologic severity. The identification of novel non-invasive biomarkers for NAFLD is needed due to the invasive nature of liver biopsy.
MicroRNAs (miRNAs) are a class of naturally occurring small noncoding RNAs of approximately 22 nucleotides in length that regulate gene expression either by promoting mRNA degradation or by attenuating protein translation[2,3]. miRNAs can influence NAFLD through pathways involving inflammation, fibrosis, insulin resistance, lipid metabolism and the metabolic syndrome. Recently, altered miRNA expression has been reported in animal and human liver samples in NAFLD[4-6]. Serum levels of miR-29a were significantly down-regulated in fibrotic/cirrhotic livers compared with nonfibrotic livers[7]. miRNA-10b was proven to regulate the level of steatosis in human hepatocyte cell culture[8]. miR-122 was significantly underexpressed in liver samples of NASH subjects compared to controls and miR-34a was significantly overexpressed[5]. miR-181d may play a role in regulating the lipid content of hepatocytes[9]. miR-197 and miR-99, in the visceral adipose tissue, were significantly associated with pericellular fibrosis in NAFLD[10]. Serum free fatty acid (FFA) levels negatively correlated with adipose tissue level of miR-99a[11]. miR-146b was up-regulated in adipose tissue after long-term high-fat diet-induced obesity in mice[12].
There is insufficient data in the literature regarding serum miRNA expression patterns in NAFLD. In the present study, we analyzed the serum expression profile of these eight miRNAs (miR-29a, miR-10b, miR-122, miR-34a, miR-181d, miR-197, miR-99a, miR-146b) which are related to NAFLD progression and pathogenesis, and are related to serum FFAs, insulin resistance, and adipose tissue.
A total of 40 patients were enrolled in this study between September 2010 and January 2012 at Erciyes University Department of Gastroenterology. Twenty patients with histologically proven NAFLD were included. The inclusion criteria were as follows: (1) 18 years or older; (2) persistently elevated (for at least 6 mo) aminotransferases; (3) ultrasonographic presence of hyperechogenic liver and (4) liver histology with a diagnosis of non-alcoholic steatohepatitis (NASH) without cirrhosis obtained no more than 6 mo before the study. The exclusion criteria were as follows: (1) a history of any level of alcohol consumption; (2) hypertension (≥ 135 systolic, ≥ 85 diastolic or antihypertensive use); (3) any other form of chronic liver disease; (4) use of any medications thought to cause or affect NAFLD; (5) abnormal thyroid function tests; (6) plasma fasting glucose ≥ 126 mg/dL or antidiabetic drug use; (7) chronic obstructive pulmonary disease; (8) peripheral and cerebral vascular disease; (9) hematologic disorders; (10) acute or chronic infection; (11) history of cancer; (12) chronic kidney diseases and (13) documented coronary artery disease.
The control group consisted of 20 healthy age-matched subjects from our institution staff with normal liver enzymes and abdominal ultrasonography findings. All subjects underwent a clinical examination and were questioned regarding their medical history. BMI was calculated as body weight/height2.
The study was performed in accordance with the principles of the Helsinki Declaration of 1975, as revised in 2008. This study was approved by the ethics committee of the Medical School of Erciyes University and informed consent was obtained from all participants.
This work was supported by a grant, number TSD-10-3368, from the Erciyes University as well as by a grant from the Turkish Association for the Study of the Liver. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Blood samples were drawn after an overnight fast from an antecubital vein; fasting plasma glucose, serum basal insulin level, high density lipoprotein cholesterol, triglycerides, low density lipoprotein cholesterol, creatinine, alanine aminotransferase, aspartate aminotransferase, serum total bilirubin, serum indirect bilirubin, alkaline phosphatase, and gamma glutamyl transferase (GGT) were determined by standard methods.
The estimate of IR by the homeostasis model assessment insulin resistance index (HOMA-IR) was calculated using the formula: fasting serum insulin (mIU/L) × fasting plasma glucose (mmol/L)/22.5.
The liver tissue was stained with hematoxylin-eosin and Masson’s trichrome stains. The review of the specimens was carried out by a single experienced liver pathologist. NASH was diagnosed according to the consensus arrived at a meeting of the American Association for the Study of Liver Diseases[13]. All cases were scored using NAS to compare miRNA expression and laboratory parameters[14]. A 4-point scale for steatosis [(0) < 5%, (1) 5%-33%, (2) > 33%-66%, and (3) > 66%], lobular inflammation [(0) no foci, (1) < 2 foci, (2) 2-4 foci, and (3) > 4 foci] and a 3-point scale for ballooning [(0) none, (1) mild, and (2) moderate-marked] resulted in a maximal sum score of 8. A NAS score of 5 and over correlated with salient NASH, patients with scores < 3 were diagnosed as not having NASH, and patients with scores of 3 and 4 were diagnosed as having borderline NASH. Fibrosis was scored by Masson’s trichrome stain using the NASH scoring system [(0): none, (1) perisinusoidal or periportal fibrosis, (2) perisinusoidal and periportal fibrosis, (3) bridging fibrosis, and (4) cirrhosis]. Steatosis was coded as 0 = mild (steatosis grade 1) or 1 = moderate to severe (steatosis grade 2-3). Ballooning was coded as 0 = mild (ballooning grade 1), or 1 = moderate-severe (ballooning grade 2). Lobular inflammation was coded as 0 = absent-mild (lobular inflammation 0-1) or 1 = moderate-severe (lobular inflammation 2-3). Fibrosis was coded as 0 = no significant fibrosis (F0-F1) or 1 = significant fibrosis (F2-F4).
miRNA was isolated from serum using a miRNeasy Mini Kit (Qiagen, Cat; 217004) according to the manufacturer’s protocol. The yield and purity of RNA were measured using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies). cDNA was isolated from obtained miRNA samples using a miScript Reverse Transcription Kit (Qiagen, Cat; 218060). We performed quantitative Real-Time PCR (qPCR) according to the manufacturer’s instructions (miScript SYBR Green PCR Kit Qiagen Cat. No: 218073) to profile the distribution of eight miRNAs in serum samples and RUN1A1 was used as an internal control. The qPCR assays were performed in duplicate. The relative expressions of miR-197, miR-146b, miR-10b, miR-181d, miR-34a, miR-122, miR-99a and miR-29a were analyzed using the standard 2-ΔΔCT method[15]. These miRNAs were selected due to their pathophysiological relation with NAFLD.
For statistical analysis of clinical data, the Shapiro-Wilk’s test was used, and histograms and q-q plots were constructed to check data normality. Either a two-sided independent samples t- test or Mann-Whitney U test was used for the comparison of continuous variables. χ2 analysis was used for the comparison of categorical variables. Values are expressed as frequencies and percentages, mean ± SD deviation or median and 25th-75th percentiles.
miRNA expression data analysis was performed according to the 2-ΔΔCT method. For additional pre-processing we applied a logarithmic transformation to the data, then centered genes as follows: [(value)-mean(gene)/standard deviation(gene)]. A t test was used to find the differentially expressed (DE) miRNAs between the NAFLD and control groups. Significant analysis of microarrays was also performed using 100 permutations to assess if the significance of the differential expression was by chance. The agglomerative hierarchical clustering method with average linkage and the Pearson correlation distance metric were used for DE miRNAs and a cluster heat-map was constructed to visualize the data values of the samples and DE miRNAs simultaneously in a hierarchical cluster structure. Receiver operating characteristic (ROC) curves were plotted for DE miRNAs, and the area under the ROC curve values were calculated with 95% intervals and compared with each other. A series of cut-off values were applied, sensitivity and specificity statistical measures were computed for each cut-off value. Next, a ROC curve was generated and the area under this curve was calculated for each miRNA using the trapezoidal rule. Also, Pearson and point-biserial correlation coefficients were calculated to identify the relationship between laboratory parameters, NAS-scale parameters and miRNAs.
All P values obtained from clinical data and miRNA expression data were adjusted with the false discovery rate to control the multiple testing problem. Analyses were performed using several packages of R 2.14.0 software.
The clinical and laboratory data of age- and sex-matched patients and controls are summarized in Table 1. To determine whether serum levels of miR-181d, miR-10b, miR-122, miR-34a, miR-146b, miR-197, miR-99a and miR-29a change in patients with NAFLD, we measured these eight miRNAs in sera collected from 20 patients diagnosed with definite steatohepatitis according to Sanyal et al[13] and compared them with those of 20 controls.
| Variable | NAFLD (n = 20) | Control (n = 20) | P |
| Age (yr) | 42.75 ± 8.17 | 44.50 ± 9.31 | 0.761 |
| Sex (male/female) | 9 (45.0)/11(55.0) | 9 (45.0)/11 (55.0) | 0.761 |
| BMI (kg/m2) | 31.86 ± 4.76 | 27.26 ± 3.07 | 0.006 |
| Waist circumference (cm) | 103.25 ± 13.65 | 90.75 ± 11.02 | 0.014 |
| Systolic blood pressure (mmHg) | 120.25 ± 8.66 | 120.25 ± 7.69 | 0.999 |
| Diastolic blood pressure (mmHg) | 76.50 ± 6.09 | 77.50 ± 8.96 | 0.761 |
| Fasting glucose (mg/dL) | 97.85 ± 13.88 | 86.80 ± 11.92 | 0.034 |
| Triglycerides (mg/dL) | 151.10 ± 65.06 | 167.70 ± 51.46 | 0.645 |
| HDL-C (mg/dL) | 43.00 (36.50-48.50) | 45.00 (38.45-49.95) | 0.761 |
| LDL-C (mg/dL) | 125.50 (100.00-139.00) | 108.00 (89.25-121.25) | 0.511 |
| Insulin (mIU/mL) | 14.81 (11.88-22.75) | 11.76 (8.61-15.70) | 0.229 |
| HOMA-IR | 4.05 (2.71-5.51) | 2.21 (1.77-3.12) | 0.034 |
| Total biluribin (mg/dL) | 0.65 (0.50-0.80) | 0.60 (0.55-0.80) | 0.761 |
| Direct biluribin (mg/dL) | 0.20 (0.12-0.25) | 0.20 (0.10-0.20) | 0.761 |
| AST (IU/L) | 45.00 (34.50-59.00) | 21.00 (18.00-27.00) | < 0.001 |
| ALT (IU/L) | 68.50 (52.50-85.50) | 18.50 (14.00-29.00) | < 0.001 |
| AP (IU/L) | 76.00 (55.50-96.00) | 69.00 (57.00-78.50) | 0.761 |
| GGT (IU/L) | 48.00 (33.00-73.00) | 20.00 (14.50-33.00) | < 0.001 |
| Creatinine (mg/dL) | 0.87 ± 0.15 | 0.80 ± 0.18 | 0.401 |
| WBC (μL) | 6709.50 ± 1709.47 | 6618.00 ± 1425.37 | 0.892 |
| Trombocyte (103/μL) | 252.10 ± 64.85 | 275.90 ± 69.82 | 0.511 |
| Hct | 43.37 ± 3.53 | 44.25 ± 4.19 | 0.761 |
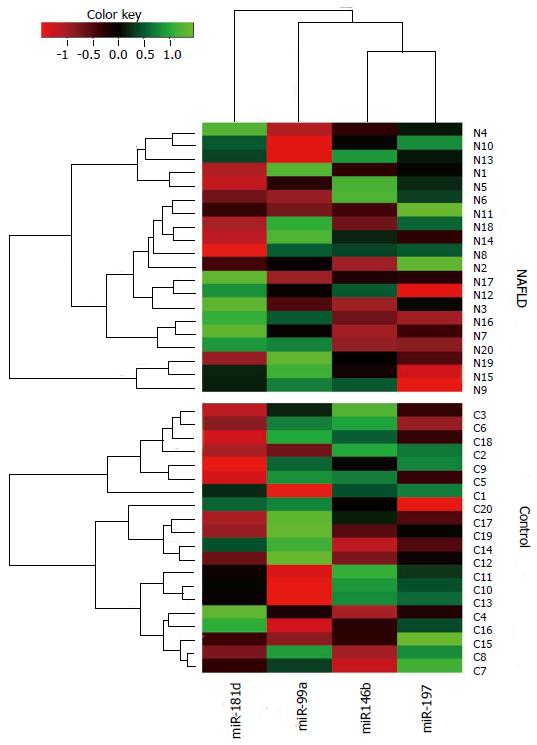
miR-181d expression was significantly lower in the serum of NASH patients compared to healthy controls (P < 0.0001). miR-99a, miR-197 and miR-146b serum levels were also lower in NASH patients (P < 0.05). Serum levels of miR-10b, miR-122, miR-34a and miR-29a did not differ between the control and patient groups (Figure 1). miR-181d levels were decreased by 2.49-fold in NASH patients compared to healthy controls (q < 5%). miR-99a levels were decreased by 1.92-fold, miR-197 levels were decreased by 1.61-fold and miR-146b levels were decreased by 1.52-fold in NASH patients compared to healthy controls (q < 5%) (Table 2). A cluster heat-map which displays the data values of samples and differentially expressed miRNAs simultaneously in a hierarchical cluster structure was produced and is shown in Figure 2.
| miRNAs | log2 (Fold change) | q value (%) | |
| Mean | SEM | ||
| Over-expressed | |||
| - | - | - | - |
| Under-expressed | |||
| miR-197 | -1.61 | 0.54 | 0 |
| miR-146b | -1.52 | 0.55 | 0 |
| miR-181d | -2.49 | 0.50 | 0 |
| miR-99a | -1.92 | 0.53 | 0 |
In our study, serum levels of miR-197 and miR-10b were negatively correlated with degree of inflammation (P < 0.05) (Table 3). Serum levels of these two miRNAs were decreased, while the degree of inflammation increased from absent-mild to moderate-severe. miRNA levels were similar in both groups according to NAS, steatosis, ballooning and fibrosis. Because both miR-197 and miR-10b levels correlated with degree of inflammation, we investigated the relationship between their serum levels and laboratory parameters in NASH patients (Table 4). Based on the correlation results between laboratory parameters and miRNAs (Table 4), a negative moderate and significant correlation was detected between GGT and miR-181d (r = -0.464, P < 0.05), and between GGT and miR-99a (r = -0.479, P < 0.05). Other correlation coefficients between laboratory parameters and miRNAs were not statistically significant (P > 0.05).
| miR-197 | miR-146b | miR-10b | miR-181d | miR-34a | miR-122 | miR-99a | miR-29a | |
| Steatosis | 0.229 | 0.191 | 0.179 | 0.179 | 0.245 | 0.067 | 0.311 | 0.256 |
| Inflammation | -0.4571 | -0.396 | -0.4921 | -0.402 | -0.302 | -0.23 | -0.431 | -0.315 |
| Ballooning | 0.154 | 0.114 | 0.175 | 0.254 | 0.165 | -0.037 | -0.029 | 0.096 |
| Fibrosis | -0.112 | -0.171 | -0.213 | -0.098 | 0.033 | -0.137 | -0.197 | -0.103 |
| NAS | 0.051 | 0.015 | 0.035 | 0.069 | 0.124 | -0.027 | 0.074 | 0.041 |
| miR-197 | miR-146b | miR-10b | miR-181d | miR-34a | miR-122 | miR-99a | miR-29a | |
| AST | -0.257 | -0.331 | -0.225 | -0.252 | -0.084 | -0.225 | -0.138 | -0.322 |
| ALT | -0.131 | -0.147 | -0.080 | -0.215 | 0.122 | 0.103 | 0.077 | -0.143 |
| GGT | -0.327 | -0.225 | -0.303 | -0.4641 | -0.29 | -0.231 | -0.4791 | -0.221 |
| Triglyceride | 0.008 | -0.039 | 0.009 | -0.035 | -0.075 | 0.087 | -0.239 | -0.058 |
| HDL-C | 0.027 | 0.051 | -0.031 | 0.042 | 0.052 | 0.111 | 0.16 | 0.034 |
| LDL-C | 0.078 | 0.042 | 0.067 | 0.015 | 0.139 | 0.102 | 0.213 | 0.155 |
| HOMA-IR | -0.055 | 0.117 | 0.011 | -0.015 | 0.195 | 0.205 | 0.221 | 0.177 |
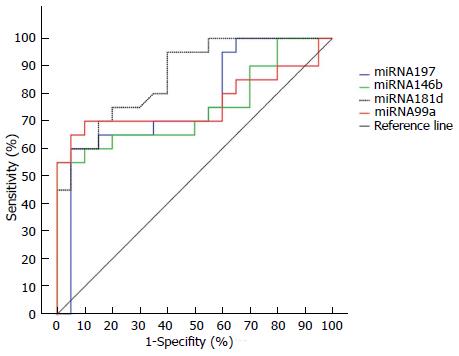
To determine the presence of NASH, we performed ROC curve analyses of miR-197, miR-146b, miR-181d and miR-99a (Figure 3). AUROC values were 0.77 (0.60-0.88), 0.75 (0.59-0.87), 0.86 (0.72-0.95), and 0.76 (0.60-0.88), respectively. mir-181d seemed to be the best marker for NASH, and there was no statistically significant difference between any two miRNA AUROC values (P > 0.05). Using the Youden index, best cut-off values and the related diagnostic measures are given in Table 5.
| miRNA | Cut-off value | SEN (95%CI) | SPE (95%CI) | PPV (95%CI) | NPV (95%CI) |
| miR-197 | ≤ -1.0144 | 60.0 (36.1-80.9) | 95.0 (75.1-99.9) | 92.3 (64.0-99.8) | 70.4 (49.8-86.2) |
| miR-146b | ≤ -0.7689 | 55.0 (31.6-76.9) | 100.0 (83.0-100.0) | 100.0 (71.3-100.0) | 69.0 (49.2-84.7) |
| miR-181d | ≤ -0.2861 | 70.0 (45.7-88.0) | 85.0 (62.1-96.6) | 82.4 (56.6-96.0) | 73.9 (51.6-89.7) |
| miR-99a | ≤ -0.1562 | 65.0 (40.8-84.5) | 95.0 (75.1-99.2) | 92.9 (66.1-98.8) | 73.1 (52.2-88.4) |
The microRNA expression pattern changes in NAFLD have been demonstrated in animal and human studies[4,5]. There is, however, only limited information regarding their detection in blood and their correlation with histological disease severity in patients with NAFLD. For this reason, we measured the serum levels of miR-181d, miR-10b, miR-122, miR-34a, miR-146b, miR-197, miR-99a and miR-29a in NAFLD patients. Of these microRNAs, miR-181d, miR-99a, miR-197 and miR-146b were expressed at lower levels in NASH patients than in controls.
miR-29a was significantly down-regulated in fibrotic/cirrhotic livers compared with nonfibrotic livers[7]. Roderburg et al[7] also showed that significantly lower serum levels of miR-29a were observed in fibrosis patients compared with healthy controls. In our study, miR-29a serum levels were not different from controls. miRNA-10b significantly increased the triglyceride level and lipid content in human hepatocyte cell line L02 cells. miRNA-10b was proven to regulate the steatosis level in human hepatocyte cell culture via the peroxisome proliferator-activated receptor-α pathway[8]. miR-122 was significantly underexpressed in liver samples of NASH subjects compared to controls[5]. Overexpression of miR-122 in cell culture resulted in a significant decrease in lipogenic genes[5]. Although miR-10b and miR-122 were found to regulate lipid content in hepatocyte cell cultures, serum expression patterns did not differ in patients with NASH in our study. The miR-34a expression pattern was not changed in our study, which was significantly overexpressed in NASH in human liver tissue[5].
There is insufficient data on miR-181d in NAFLD. miR-181d may play a role in regulating the lipid content of hepatocytes. miR-181d has been shown to decrease lipid droplets, and to reduce cellular triglycerides and cholesterol ester in cell culture[9]. In our study, serum levels of miR-181d were decreased in NAFLD by 2.49-fold compared to controls. Estep et al[10] studied miRNA expression in the visceral adipose tissue of patients with NAFLD. They found that significant changes in miRNA expression were characterized by overall down-regulation in the visceral adipose tissue. They also found that down-regulation of miR-197 and miR-99 in the visceral adipose tissue was significantly associated with pericellular fibrosis in NAFLD patients. We also found that miR-99 and miR-197 were down-regulated in the serum of NASH patients, but we did not find any relationship between fibrosis stages. A study conducted by Klöting et al[11] showed that the human adipose tissue level of miR-99a negatively correlated with FFA levels. NAFLD is significantly associated with obesity, type II diabetes mellitus, and the metabolic syndrome. Serum FFA levels are elevated in all of these disorders[16]. It was reported that the main source for TG in hepatocytes was derived from circulating FFAs in NAFLD[17]. Although we did not analyze serum fatty acid levels in our study, down-regulation of miR-99a in NASH was concordant with this evidence. Chartoumpekis et al[12] examined miRNA profiling after long-term high-fat diet-induced obesity in mice and found that miR-146a and miR-146b were up-regulated in adipose tissue. In one study, the miR-146a expression levels were significantly decreased in peripheral blood mononuclear cells from patients with Type 2 diabetes compared to control subjects. They also reported that reduced miR-146a levels are associated with insulin resistance and poor glycemic control[18]. Although miR-146a and miR-146b are encoded on different chromosomes, their mRNA targets are predicted to overlap significantly[19]. The data in this study showed decreased miR-146b levels which seems reasonable in light of these findings.
During our research, the study by Cermelli et al[20] showed that serum levels of miR-122, miR-34a and miR-16 were significantly higher in NAFLD patients. They also found that miR-122 and miR-34a levels were correlated with liver enzyme levels, fibrosis stage and inflammation activity in NAFLD. These findings regarding miR-122 and miR-34a were not confirmed by our study results. This may be attributed to patient selection. We did not include hypertensive patients due to their impaired miRNA profile[21,22].
The current study provides evidence that biopsy proven NAFLD is associated with altered serum miRNA expression. The potential targets of differentially expressed miRNAs are known to play a role in metabolism, immune function, cell proliferation, apoptosis, tissue development and differentiation; all these are key processes involved in the development and progression of NASH[5]. While our findings do not provide direct proof of the involvement of these differentially expressed miRNAs in the development, progression and systemic effect of NASH, they serve as a resource for future studies on the role of miRNAs in the non-invasive assessment of NASH. This was a principal aim of this study.
The pattern of miRNA expression can be affected by hypertension[21,22]. This possibility was minimized by excluding patients with hypertension. The miRNA expression pattern is also possibly affected by the development of cirrhosis. We excluded patients with cirrhosis from this study. We want to point out that the aim of this study was to determine whether biopsy proven NAFLD was associated with altered serum miRNA expression without hypertension. Therefore, only subjects without hypertension were studied. At the same time, this study does not provide any information on the serum miRNA expression pattern in those with isolated hepatic steatosis due to insufficient funding. Differential expression of microRNA with two distinct clinical entities, in simple steatosis and NASH, may exist. It will be very interesting to investigate the serum expression pattern of miRNA in simple steatosis and NASH. In addition, this study had a cross-sectional design. Due to the financial constraints we selected these eight miRNAs from earlier studies which were related to NAFLD progression and pathogenesis and were also related to serum FFA, insulin resistance and adipose tissue.
This study showed the following: (1) miR-181d, miR-99a, miR-197 and miR-146b expression was significantly lower in the serum of biopsy proven NAFLD patients than in healthy controls; (2) The serum levels of miR-197 and miR-10b were inversely correlated with degree of inflammation; and (3) miR-181d and miR-99a were inversely correlated with serum GGT levels in NASH patients. In conclusion, biopsy proven NAFLD is associated with an altered serum miRNA expression pattern. This study provides clues for defining the non-invasive diagnosis of NAFLD.
Due to the limitations of liver biopsy, the use of non-invasive markers has emerged in recent years. MicroRNAs (miRNAs) are a class of naturally occurring small noncoding RNAs that regulate gene expression. Altered miRNA expression has been reported in animal and human liver samples in non-alcoholic fatty liver disease (NAFLD). There is, however, only limited information regarding their detection in blood and their correlation with histological disease severity in patients with NAFLD.
miRNAs are a class of naturally occurring small noncoding RNAs of approximately 22 nucleotides in length that regulate gene expression either by promoting mRNA degradation or by attenuating protein translation. miRNAs can influence NAFLD through pathways involving inflammation, fibrosis, insulin resistance, lipid metabolism and the metabolic syndrome. A research hotspot is the evaluation of miRNAs to determine hepatic damage and fibrosis in patients with NAFLD.
miR-181d, miR-99a, miR-197 and miR-146b were expressed at lower levels in non-alcoholic steatohepatitis (NASH) patients than in controls. Serum levels of miR-197 and miR-10b were inversely correlated with degree of inflammation and miR-181d and miR-99a were inversely correlated with serum gamma glutamyl transferase levels in NASH patients.
Biopsy proven NAFLD is associated with an altered serum miRNA expression pattern. This study provides clues for defining the non-invasive diagnosis of NAFLD.
NAFLD is the most common cause of chronic liver disease in Western populations. It comprises a disease spectrum which includes variable degrees of simple steatosis (fatty liver), NASH and cirrhosis which are likely to be characterized by different pathogenesis and clinical history. The identification of novel non-invasive biomarkers for NAFLD is needed. miRNAs are a class of naturally occurring small noncoding RNAs of approximately 22 nucleotides in length that regulate gene expression either by promoting mRNA degradation or by attenuating protein translation. miRNAs can influence NAFLD through pathways involving inflammation, fibrosis, insulin resistance, lipid metabolism and the metabolic syndrome.
This is a good descriptive study in which authors evaluate the serum miRNA levels to determine hepatic damage and fibrosis in patients with NAFLD. The results are interesting and suggest that miRNAs may be useful to determine hepatic inflammation with a non-invasive tool in NAFLD patients. The data analysis and presentation were appropriate, and the manuscript was well prepared.
P- Reviewer: Ampuero J, Dirchwolf M, Ikura Y, Kobyliak NK, Lin CW S- Editor: Wen LL L- Editor: Webster JR E- Editor: Wu HL
| 1. | Yilmaz Y. Is nonalcoholic fatty liver disease the hepatic expression of the metabolic syndrome? World J Hepatol. 2012;4:332-334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 2. | Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3151] [Cited by in RCA: 3673] [Article Influence: 262.4] [Reference Citation Analysis (0)] |
| 3. | Shrivastava S, Mukherjee A, Ray RB. Hepatitis C virus infection, microRNA and liver disease progression. World J Hepatol. 2013;5:479-486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (1)] |
| 4. | Li S, Chen X, Zhang H, Liang X, Xiang Y, Yu C, Zen K, Li Y, Zhang CY. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J Lipid Res. 2009;50:1756-1765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 162] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 5. | Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810-1820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 583] [Cited by in RCA: 550] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 6. | Li YY. Genetic and epigenetic variants influencing the development of nonalcoholic fatty liver disease. World J Gastroenterol. 2012;18:6546-6551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 62] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 7. | Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 656] [Cited by in RCA: 661] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 8. | Zheng L, Lv GC, Sheng J, Yang YD. Effect of miRNA-10b in regulating cellular steatosis level by targeting PPAR-alpha expression, a novel mechanism for the pathogenesis of NAFLD. J Gastroenterol Hepatol. 2010;25:156-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 113] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 9. | Whittaker R, Loy PA, Sisman E, Suyama E, Aza-Blanc P, Ingermanson RS, Price JH, McDonough PM. Identification of MicroRNAs that control lipid droplet formation and growth in hepatocytes via high-content screening. J Biomol Screen. 2010;15:798-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (1)] |
| 10. | Estep M, Armistead D, Hossain N, Elarainy H, Goodman Z, Baranova A, Chandhoke V, Younossi ZM. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2010;32:487-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 112] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 11. | Klöting N, Berthold S, Kovacs P, Schön MR, Fasshauer M, Ruschke K, Stumvoll M, Blüher M. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS One. 2009;4:e4699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 267] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 12. | Chartoumpekis DV, Zaravinos A, Ziros PG, Iskrenova RP, Psyrogiannis AI, Kyriazopoulou VE, Habeos IG. Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS One. 2012;7:e34872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 188] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 13. | Sanyal AJ, Brunt EM, Kleiner DE, Kowdley KV, Chalasani N, Lavine JE, Ratziu V, McCullough A. Endpoints and clinical trial design for nonalcoholic steatohepatitis. Hepatology. 2011;54:344-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 573] [Cited by in RCA: 579] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 14. | Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6807] [Cited by in RCA: 8208] [Article Influence: 410.4] [Reference Citation Analysis (5)] |
| 15. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 133349] [Article Influence: 5556.2] [Reference Citation Analysis (1)] |
| 16. | Bradbury MW. Lipid metabolism and liver inflammation. I. Hepatic fatty acid uptake: possible role in steatosis. Am J Physiol Gastrointest Liver Physiol. 2006;290:G194-G198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 255] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 17. | Donnelly KL, Smith CI, Schwarzenberg SJ, Jessurun J, Boldt MD, Parks EJ. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest. 2005;115:1343-1351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2112] [Cited by in RCA: 2588] [Article Influence: 129.4] [Reference Citation Analysis (0)] |
| 18. | Balasubramanyam M, Aravind S, Gokulakrishnan K, Prabu P, Sathishkumar C, Ranjani H, Mohan V. Impaired miR-146a expression links subclinical inflammation and insulin resistance in Type 2 diabetes. Mol Cell Biochem. 2011;351:197-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 191] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 19. | Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495-7498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 244] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 20. | Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 466] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 21. | Yamakuchi M. MicroRNAs in Vascular Biology. Int J Vasc Med. 2012;2012:794898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 22. | Schroen B, Heymans S. MicroRNAs and beyond: the heart reveals its treasures. Hypertension. 2009;54:1189-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |