Published online Oct 27, 2014. doi: 10.4254/wjh.v6.i10.752
Revised: August 13, 2014
Accepted: September 6, 2014
Published online: October 27, 2014
Processing time: 81 Days and 11.2 Hours
AIM: To investigate the role of sorafenib (SFN) in autophagy of hepatocellular carcinoma (HCC). We evaluated how SFN affects autophagy signaling pathway in human HCC cell lines.
METHODS: Two different human HCC cell lines, Hep3B and Huh7, were subjected to different concentrations of SFN. Cell viability and onset of apoptosis were determined with colorimetric assay and immunoblotting analysis, respectively. The changes in autophagy-related proteins, including LC3, ULK1, AMPK, and LKB, were determined with immunoblotting analysis in the presence or absence of SFN. To assess autophagic dynamics, autophagic flux was measured with chloroquine, a lysosomal inhibitor. The autophagic responsiveness between different HCC cell lines was compared under the autophagy enhancing conditions.
RESULTS: Hep3B cells were significantly more resistant to SFN than Huh7 cells. Immunoblotting analysis revealed a marked increase in SFN-mediated autophagy flux in Huh7 cells, which was, however, absent in Hep3B cells. While both starvation and rapamycin enhanced autophagy in Huh7 cells, only rapamycin increased autophagy in Hep3B cells. Immunoblotting analysis of autophagy initiation proteins showed that SFN substantially increased phosphorylation of AMPK and consequently autophagy in Huh7, but not in Hep3B cells.
CONCLUSION: The autophagic responsiveness to SFN is distinct between Hep3B and Huh7 cells. Resistance of Hep3B cells to SFN may be associated with altered autophagy signaling pathways.
Core tip: Hepatocellular carcinoma (HCC) is difficult to treat. Sorafenib (SFN) is one treatment option. Autophagy has been proposed to play a pivotal role in HCC. In the present study we investigated the role of autophagy in SFN-treated HCC cells. We found that the autophagic responsiveness to SFN is markedly distinct between Hep3B and Huh7 cells.
- Citation: Fischer TD, Wang JH, Vlada A, Kim JS, Behrns KE. Role of autophagy in differential sensitivity of hepatocarcinoma cells to sorafenib. World J Hepatol 2014; 6(10): 752-758
- URL: https://www.wjgnet.com/1948-5182/full/v6/i10/752.htm
- DOI: https://dx.doi.org/10.4254/wjh.v6.i10.752
Hepatocellular carcinoma (HCC), typically occurring in the setting of cirrhosis and chronic hepatitis, is fifth most common cancer diagnosed worldwide and more than 600000 patients are dying of this disease each year[1]. The incidence of HCC is also rising in the United States. Despite recent advances in the screening and management of HCC, treatment of this pernicious disease is still far from complete, mostly due to its complex mechanisms underlying proliferation, tissue invasion and metastasis of HCC.
Therapies for HCC include chemoembolization, ablation, surgical resection and transplantation[2], but these interventions are highly invasive and often require prolonged hospitalization of the patients. Recently, sorafenib (SFN), an oral multi-kinase inhibitor, has been shown to inhibit tumor-cell proliferation and tumor angiogenesis through its inhibition of vascular endothelial growth factor receptor 2 and other receptor tyrosine kinases[3,4]. In a placebo-controlled phase III study, SFN displayed about 3-mo extension of survival in advanced and inoperable HCC cases, leading to Food and Drug Administration (FDA) approval[5]. It is, however, noteworthy that some HCC patients show unresponsiveness or acquired resistance to SFN[6]. It is unclear why the efficacy of SFN is limited, although the activation of survival pathways like PI3K/AKT has been proposed to cause the development of SFN resistance[7].
Autophagy is an evolutionary conserved cellular process that degrades both long-lived cytoplasmic proteins and surplus or dysfunctional organelles by lysosome-dependent machinery[8]. Impaired and insufficient autophagy is causatively linked to pathogenesis of ischemia/reperfusion injury and drug-induced toxicity in the liver[9-11]. Growing evidence is accumulating that autophagy also plays a pivotal role in carcinogenesis, tumor proliferation, and resistance to chemotherapy[12]. In addition, recent studies on an anti-cancerous role of autophagy raise a possibility that the modulation of autophagy could be a new therapy against cancer[13,14]. However, a pro-cancerous role of autophagy has also been suggested[15]. Thus, the precise role of autophagy in HCC is largely yet to be elucidated.
In the present study, we investigated the role of autophagy in HCC using two human HCC cell lines, Hep3B and Huh7 cells. Our results demonstrate that autophagic response to SFN and autophagy signaling pathways are markedly distinct between these two HCC cells.
SFN and rapamycin were purchased from LC Laboratories (Woburn, MA) and dissolved in DMSO. Chloroquine was purchased from Sigma Chemical Co (St. Louis, MO) and dissolved in phosphate-buffered saline (PBS; 2.7 mmol/L KCl, 137 mmol/L NaCl 10.1 mmol/L Na2HPO4, and 1.8 mmol/L KH2PO4, pH7.4). Antibodies against ULK1 were purchased from Sigma Chemical Co (St. Louis, MO). All other primary antibodies were purchased from Cell Signaling Technology (Danvers, MA).
The human HCC cell lines, Hep3B and Huh7 cells, were purchased from American Type Culture Collection (Manassass, VA) and were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Mediatech, Manassass, VA) supplemented with 10% fetal bovine serum (FBS; Sigma) and 1% penicillin/streptomycin (Mediatech) in 5% CO2 at 37 °C. Cells were used for experiments at approximately 80% confluence. For immunoblotting experiments, cells were plated in 60 mm culture dishes at 6 × 105 cells. DMSO was used for a vehicle control. To induce nutrient depletion and starvation, cells were incubated in Krebs-Ringer-hydroxyethylpiperazine-N-2 ethanesulfonic acid (HEPES) (KRH) medium containing 115 mmol/L NaCl, 5 mmol/L KCl, 2 mmol/L CaCl2, 1 mmol/L KH2PO4, 1.2 mmol/L MgSO4, and 25 mmol/L HEPES at a pH of 7.4.
To determine cell viability, 3-(4,5-dimethyl-thiazole-2-yl)-2,5-biphenyl tetrazolium (MTT) assay was used with the different concentrations of SFN and rapamycin. HCC cells were plated on a 96-well microplate at 5 × 103 cells per well for up to 72 h in DMEM medium. DMSO was used as a vehicle control. The MTT salt dissolved in PBS (5 mg/mL) was added to the medium and incubated for 2 h[16]. The medium was subsequently removed and replaced with isopropyl alcohol. The optical density was read at 562 nm in SpectraMax M2e Microplate Reader (Molecular Devices Corporation, Sunnyvale, CA). The results were shown as a ratio of viability of treated to vehicle groups.
Whole cell lysates were prepared by extracting proteins with radioimmunoprecipitation (RIPA) buffer (150 mmol/L NaCl, 25 mmol/L Tris-HCl (pH 8, 0.1% sodium dodecylsulfate, 1% sodium deoxycholic acid, 1% TritonX-100 and 5 mmol/L EDTA) with 1% protease and 1% phosphatase inhibitors. Protein concentrations were determined by BCA protein assay kit (Pierce, Rockford, IL). Proteins (10 or 15 μg) were separated by the electrophoresis through 4%-12% polyacrylamide gels (InVitrogen, Carlsbad, CA) or by sodium dodecyl sulfate polyacrylamide gel and transferred to polyvinylidene difluoride or nitrocellulose membranes. The expression of LC3, LKB1, phospho-AMPKα (Thr172), AMPKα, PARP and GAPDH were detected using primary polyclonal antibodies. After overnight incubation with primary antibodies at 4 °C, the membranes were incubated with donkey anti-rabbit IgG-HRP (Santa Cruz Biotechnology, Santa Cruz, CA) and subsequently visualized by enhanced chemiluminescence. Changes in protein expression were determined using the ImageJ software (National Institutes of Health, Bethesda, MD)
Small interfering RNA (siRNA) for AMPKα1/α2 (sc-45312) and the siRNA Reagent System (sc-45064) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). In a six well tissue culture plate, Huh7 cells were cultured until approximately 70% confluent. For each transfection, 80 pmol of siRNA was diluted into 100 μL of siRNA transfection medium, as suggested by the manufacturer. Huh7 cells were incubated with siRNA transfection reagents. Scrambled siRNA (sc-37007) was used for the control experiments.
Results were evaluated using unpaired two-tailed Student’s t test. Data are expressed as mean ± SE and P < 0.05 denotes statistical significance. All values are representative of at least three different experiments per group.
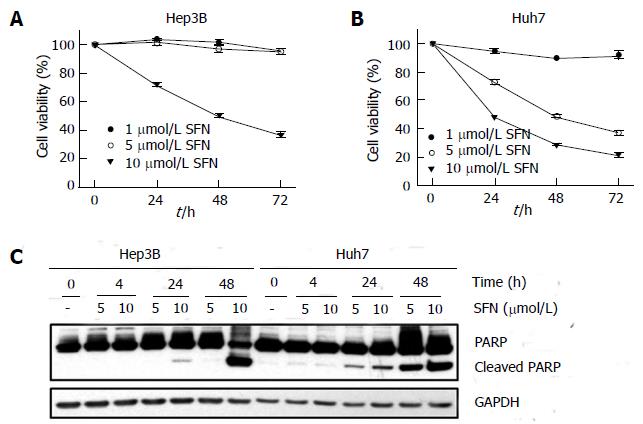
To determine the chemosensitivity of HCC cells to SFN, Hep3B and Huh7 cells were treated with different concentrations of SFN for up to 72 h and cell death was evaluated with the MTT assay (Figure 1). There was a marked difference of cell death between two cells. While 5 μmol/L SFN induced virtually no cell death in Hep3B, this concentration caused a significant cell death in Huh7 in a time dependent manner (Figure 1A). Although 10 μmol/L SFN induced cell death in both cell types, the extent of cell killing was substantially greater in Huh7 cells than in Hep3B cells. To confirm the differential sensitivity to SFN between two cell types, the onset of apoptosis was determined with immunoblotting of poly (ADP ribose) polymerase (PARP) cleavage (Figure 1B). Similar to the results from the MTT assay, SFN substantially induced apoptotic cell death in Huh7 cells. Taken together, these results suggest that Hep3B cells are intrinsically more resistant to SFN than Huh7 cells.
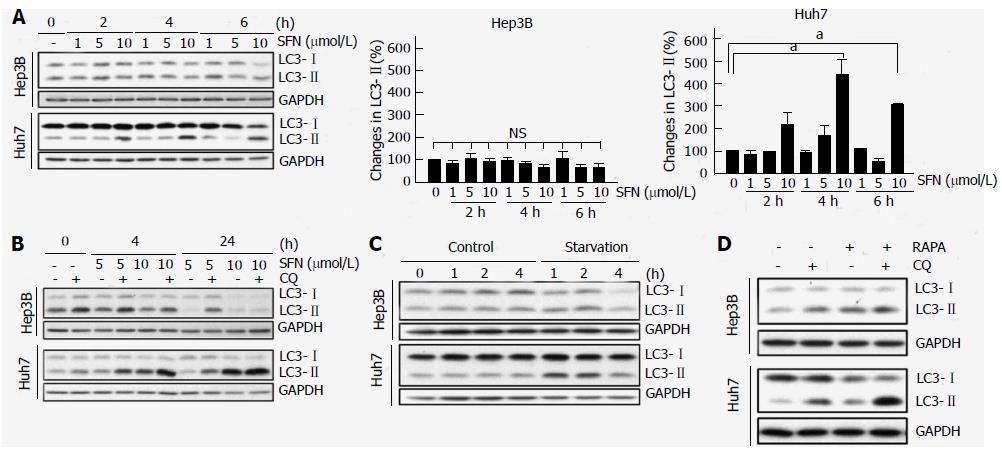
Autophagy is a pro-survival mechanism in normal cells and has also been associated with chemoresistance in cancer cells[17-19]. Accordingly, we investigated if the autophagic responsiveness to SFN is different between two cells. Microtubule-associated protein 1 light chain 3-II (LC3-II), a mammalian orthologue of Atg8, is a specific autophagy marker[20]. Immunoblotting of LC3 showed that SFN at three different concentrations barely changed the expression of LC3-II in Hep3B cells (Figure 2A). However, in Huh7 cells, 10 μmol/L SFN significantly increased LC3-II expression, suggesting that this concentration of SFN alters autophagy in Huh7 cells, but not in Hep3B. Autophagy is a dynamic process between autophagosomal entrapment and autolysosomal clearance[8]. Thus, the increase in LC3-II by SFN in Huh7 cells could be due to either an increase in autophagosome formation or a decrease in autolysosome formation. To distinguish these two possibilities, we measured autophagic flux with chloroquine (CQ), a lysosomotropic agent that inhibits autolysosomal clearance[10]. Immunoblotting analysis of LC3 in the presence and absence of CQ revealed that SFN substantially increased autophagic flux only in Huh7 cells (Figure 2B). Therefore, these data demonstrate that Huh7 cells have higher autophagic responsiveness to SFN than Hep3B cells.
Starvation or nutrient depletion is a powerful stimulus for autophagy[9]. Next, we examined autophagic response of two cells to starvation. To induce starvation, cells were incubated in amino acid- and serum-free KRH for up to 4 h and changes in LC3 were evaluated with immunoblotting (Figure 2C). Autophagy was rapidly increased in Huh7 cells during 1 h of starvation, as judged by increased LC3-II expression. However, this starvation-induced increase in LC3-II was not evident in Hep3B cells, implying that starvation fails to induce autophagy in Hep3B cells.
Rapamycin enhances autophagy through mTOR inhibition[21]. To investigate if rapamycin can induce autophagy in HCC cells, either Hep3B or Huh7 cells were treated with 5 nmol/L rapamycin and changes in LC3 were determined in the presence and absence of CQ (Figure 2D). In a striking contrast to both SFN and starvation, rapamycin increased the expression of LC3-IIand autophagic flux in Hep3B cells as well as Huh7 cells. These findings suggest that mTOR-mediated autophagy is functional and operative in both Hep3B and Huh7 cells. Collectively, our data demonstrate that the autophagic responsiveness of Hep3B cells to SFN and starvation is markedly distinct from that of Huh7 cells.
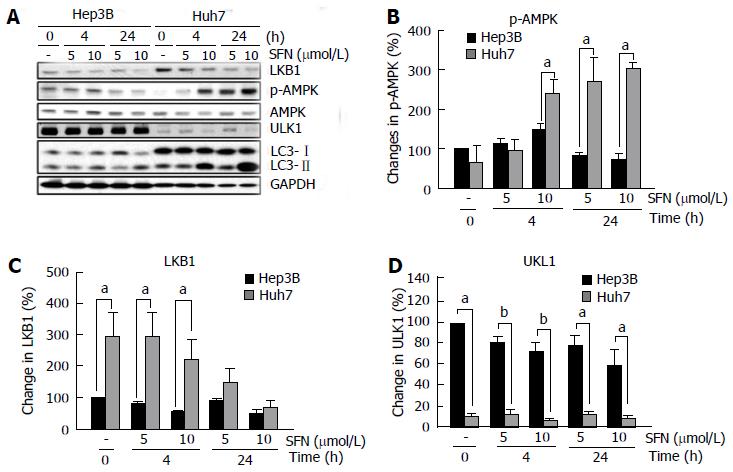
Autophagy is a multi-step process consisting of initiation, elongation and completion[8]. At the initiation stage of autophagy, two proteins play an integral role in cargo selection and phagophore formation: 5’-adenosine monophosphate-activated protein kinase (AMPK), a molecular energy sensor, and Unc-51 Like Autophagy Activating Kinase 1 (ULK1)[22], a mammalian homolog of yeast Atg1. To investigate whether SFN affects these proteins, we analyzed changes in AMPK and ULK1 expression with immunoblots (Figure 3). Densitometric analysis revealed a significant difference in the status of phospho-AMPK (p-AMPK) between two cells when SFN was added (Figure 3A and B). While the basal levels of p-AMPK were comparable between two cells, administration of SFN to Huh7 cells significantly increased p-AMPK expression. Changes in p-AMPK in Hep3B in the presence of SFN were, however, minimal. Total AMPK levels remained unchanged in both cells. Interestingly, the basal levels of liver kinase B1 (LKB1), a Ser/Thr kinase phosphorylating AMPK[23], was also significantly higher in Huh7 cells than in Hep3B cells (Figure 3A and C). When Huh7 cells were treated with SFN, the expression of LKB1 gradually decreased but remained higher than Hep3B cells especially during the early period of SFN treatment. On the contrary, the basal levels of ULK1 were significantly higher in Hep3B cells than in Huh7 cells (Figure 3A and D). Administration of SFN did not change ULK1 expression in both cell types throughout 24 h of treatment. Therefore, these results demonstrate that the initiation signaling pathways of autophagy are noticeably different between two HCC cells.
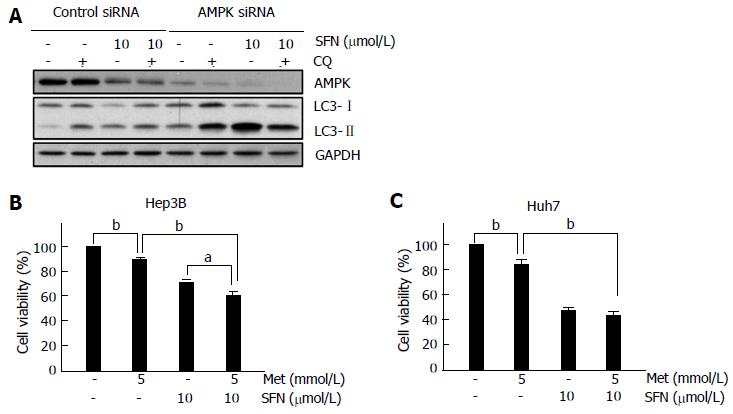
To further investigate how AMPK affects autophagy in Huh7 cells, AMPK was silenced with siRNA-mediated approaches (Figure 4A). Knocking down of AMPK substantially increased the levels of LC3-II, suggesting an integral role of AMPK in the basal autophagy of Huh7 cells. Co-addition of SFN further enhanced LC3-II expression more than 300%, as compared to the treatment with siRNA alone, implying that SFN may regulate multiple targets of autophagy process other than AMPK. Notably, CQ did not increase LC3-II under this condition.
Next, we explored the effects of AMPK activation on HCC cell death. HCC cells were treated either with 5 mmol/L metformin, an AMPK activator, or with 10 μmol/L SFN for 24 h. Some cells were treated with both. In Hep3B cells, metformin itself significantly increased cell death, which was further enhanced by SFN (Figure 4B). On the contrary, the addition of metformin failed to increase SFN-dependent cell death in Huh7 cells, suggesting that the responsiveness to metformin in the presence of SFN is distinct between two cells.
HCC is prevailing worldwide and more than 20000 new cases are reported in the United States each year[1]. The onset of HCC from hepatitis C virus infection is the one of the fastest-rising cause of cancer-mediated death in the United States. During the past two decades, the incidence of HCC in the United States has tripled while the survival for 5 years has remained below 12%[24]. Thus, HCC remains a difficult and challenging cancer to treat. SFN is an oral multi-kinase inhibitor and inhibits tumor-cell proliferation and tumor angiogenesis[3,4]. Although this FDA-approved dug can prolong the survival of advanced and inoperable HCC patients, the average extension of survival is limited to about 3-mo. Furthermore, some patients do not respond to SFN[6]. The mechanisms behind such a limited efficacy of SFN remain unknown. Autophagy is an evolutionary conserved cellular process and has been proposed to play a pivotal role in HCC. In the present study, using human HCC cell lines, we show that Hep3B cells are resistant to SFN-mediated apoptosis, while Huh7 cells are prone to apoptosis in the presence of SFN. Furthermore, we demonstrate that SFN induces a substantial enhancement of autophagy in Huh7 cells, but not in Hep3B cells, and that autophagy signaling pathways are noticeably distinct between two HCC cells.
Autophagy mainly plays a pro-survival role in normal cells by providing cells and tissues with nutrients. However, autophagy can cause cell death through a selective degradation of essential proteins and constituents in the cells[25]. The SFN-sensitive HCC cell line, Huh7 cells, exhibited a substantial difference in both cell death and autophagic responsiveness, compared to the SFN-resistance cell line, Hep3B cells (Figures 1 and 2). Immunoblotting analysis of LC3 and autophagic flux showed that SFN induced a marked increase in autophagy in Huh7 cells, which was, however, absent in Hep3B cells (Figure 2). Lack of the autophagic responsiveness in Hep3B cells was also observed under the condition of starvation, a powerful stimulus of autophagy[9,12], suggesting that these two HCC cells have an intrinsically distinct autophagy. Furthermore, our results suggest that the enhancement of autophagy by SFN in Huh7 cells may be associated with the activation of p-AMPK, a key protein involved in autophagy initiation[22] (Figure 3). The importance of p-AMPK in SFN-dependent activation of autophagy is further supported by our findings that Hep3B cells fail to increase p-AMPK expression upon treatment of SFN. The necessity of AMPK activation for autophagy induction in HCC has been recently reported[26,27].
The contradictory effects of SFN on Hep3B and Huh7 cells have been reported[28] and may be linked to different autophagic responsiveness to this agent. Our results show that starvation or nutrient depletion fails to induce autophagy in Hep3B cells (Figure 2C). The absence of autophagy enhancement by either SFN or starvation could stem from defective or impaired autophagy in Hep3B cells. However, autophagic flux analysis revealed that Hep3B cells, indeed, have a considerable basal and mTOR-dependent autophagic capacity, as judged by the increase in LC3-II either with CQ (Figure 2B) or with rapamycin (Figure 2D). These results imply that the signaling pathways of starvation-mediated autophagy may be altered in Hep3B cells. Although the mechanisms underlying SFN-induced autophagy remain to be elucidated, lack of autophagic response to both starvation and SFN in Hep3B cells led us to speculate that SFN-dependent autophagy requires a similar signaling pathway of the starvation-mediated autophagy. Since the only known difference in signaling mechanism between starvation- and rapamycin-induced autophagy exists in the initiation stage of autophagy process[8], we reasoned that events upstream to autophagy signaling pathways might be altered in Hep3B cells. In agreement with this view, we observed that the activation of AMPK, a critical event in autophagy induction under the condition of starvation, was evident only in Huh7 cells, but not in Hep3B cells, upon SFN administration. The precise mechanisms behind SNF-induced autophagy warrant future studies.
When normal, non-tumorigenic cells are subjected to stresses such as ischemia/reperfusion, alcohol and drug, autophagy becomes activated as an adaptive response to these stresses[12]. In contrast, autophagy can prevent and promote tumor development. These seemingly contradictory roles of autophagy in tumor stem from the complexity of tumorgenesis[12]. Prior to tumor establishment, autophagy clears damaged organelles and proteins, leading to preventing neoplastic transformation. However, when tumor develops, the demand for metabolic supplies is progressively increasing. As a consequence, autophagy becomes fully activated to provide the tumor cells with nutrients and amino acids. Thus, autophagy is a “double-edged sword” in cancer where it initially acts as a tumor suppressor, but later acts as a tumor promoter when tumor is established[15].
In conclusion, we have shown that Hep3B cells responds differently to various autophagy stimuli, compared to Huh7 cells. Although the modulation of autophagy could have a new therapeutic potential against cancer, our study demonstrates that caution should be taken before considering autophagy as anticancer regimes in HCC patients.
The incidence of hepatocellular carcinoma (HCC) is prevailing worldwide but its treatment is still disappointing. Sorafenib (SFN), an oral multi-kinase inhibitor, is one treatment option for HCC patients but its efficacy is limited. Autophagy is a cellular catabolic process that degrades both long-lived cytoplasmic proteins and surplus or dysfunctional organelles by lysosome-dependent machinery. Autophagy also plays a pivotal role in carcinogenesis, tumor proliferation, and resistance to chemotherapy. However, the role of autophagy in HCC remains unclear.
HCC is fifth most common cancer diagnosed worldwide and the incidence of this pernicious disease is also rising in the United States. However, the treatment of HCC is still far from complete.
In this study authors evaluated the effects of SFN on autophagy in HCC cell lines. Authors found that individual HCC cells respond quite differently to various autophagy stimuli due to distinct autophagy signaling pathways between HCC cells.
Although the modulation of autophagy could have a new therapeutic potential against cancer, authors demonstrate here that caution should be taken before considering autophagy as anticancer regimes in HCC patients.
The most important terms in this article are: HCC, SFN and autophagy.
The authors have an idea to find SFN effects in HCC cell lines. To this end, they analyzed the changes of autophagy-related proteins in the cells, and found the different responsiveness to SFN involves autophagy signaling pathway. The paper is interesting.
P- Reviewer: Kovacs SJ, Lu WY, Wozniak M, Yang J S- Editor: Song XX L- Editor: A E- Editor: Wu HL
| 1. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11128] [Cited by in RCA: 11836] [Article Influence: 845.4] [Reference Citation Analysis (4)] |
| 2. | Davis GL, Dempster J, Meler JD, Orr DW, Walberg MW, Brown B, Berger BD, O’Connor JK, Goldstein RM. Hepatocellular carcinoma: management of an increasingly common problem. Proc (Bayl Univ Med Cent). 2008;21:266-280. [PubMed] |
| 3. | Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64:7099-7109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2966] [Cited by in RCA: 3147] [Article Influence: 149.9] [Reference Citation Analysis (0)] |
| 4. | Liu L, Cao Y, Chen C, Zhang X, McNabola A, Wilkie D, Wilhelm S, Lynch M, Carter C. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66:11851-11858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1042] [Cited by in RCA: 1205] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 5. | Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9016] [Cited by in RCA: 10270] [Article Influence: 604.1] [Reference Citation Analysis (2)] |
| 6. | Gauthier A, Ho M. Role of sorafenib in the treatment of advanced hepatocellular carcinoma: An update. Hepatol Res. 2013;43:147-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 7. | Chen KF, Chen HL, Tai WT, Feng WC, Hsu CH, Chen PJ, Cheng AL. Activation of phosphatidylinositol 3-kinase/Akt signaling pathway mediates acquired resistance to sorafenib in hepatocellular carcinoma cells. J Pharmacol Exp Ther. 2011;337:155-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 256] [Article Influence: 18.3] [Reference Citation Analysis (1)] |
| 8. | Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2780] [Cited by in RCA: 2972] [Article Influence: 141.5] [Reference Citation Analysis (0)] |
| 9. | Kim JS, Nitta T, Mohuczy D, O’Malley KA, Moldawer LL, Dunn WA, Behrns KE. Impaired autophagy: A mechanism of mitochondrial dysfunction in anoxic rat hepatocytes. Hepatology. 2008;47:1725-1736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 159] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 10. | Wang JH, Ahn IS, Fischer TD, Byeon JI, Dunn WA, Behrns KE, Leeuwenburgh C, Kim JS. Autophagy suppresses age-dependent ischemia and reperfusion injury in livers of mice. Gastroenterology. 2011;141:2188-2199.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 115] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 11. | Ni HM, Bockus A, Boggess N, Jaeschke H, Ding WX. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology. 2012;55:222-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 355] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 12. | Czaja MJ, Ding WX, Donohue TM, Friedman SL, Kim JS, Komatsu M, Lemasters JJ, Lemoine A, Lin JD, Ou JH. Functions of autophagy in normal and diseased liver. Autophagy. 2013;9:1131-1158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 369] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 13. | Wang Y, Qiu Q, Shen JJ, Li DD, Jiang XJ, Si SY, Shao RG, Wang Z. Cardiac glycosides induce autophagy in human non-small cell lung cancer cells through regulation of dual signaling pathways. Int J Biochem Cell Biol. 2012;44:1813-1824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 14. | Yang WL, Perillo W, Liou D, Marambaud P, Wang P. AMPK inhibitor compound C suppresses cell proliferation by induction of apoptosis and autophagy in human colorectal cancer cells. J Surg Oncol. 2012;106:680-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 15. | Giuliani CM, Dass CR. Autophagy and cancer: taking the ‘toxic’ out of cytotoxics. J Pharm Pharmacol. 2013;65:777-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Denizot F, Lang R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J Immunol Methods. 1986;89:271-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3523] [Cited by in RCA: 3668] [Article Influence: 94.1] [Reference Citation Analysis (0)] |
| 17. | Song J, Qu Z, Guo X, Zhao Q, Zhao X, Gao L, Sun K, Shen F, Wu M, Wei L. Hypoxia-induced autophagy contributes to the chemoresistance of hepatocellular carcinoma cells. Autophagy. 2009;5:1131-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 147] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 18. | Ding ZB, Hui B, Shi YH, Zhou J, Peng YF, Gu CY, Yang H, Shi GM, Ke AW, Wang XY. Autophagy activation in hepatocellular carcinoma contributes to the tolerance of oxaliplatin via reactive oxygen species modulation. Clin Cancer Res. 2011;17:6229-6238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 148] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 19. | Shimizu S, Takehara T, Hikita H, Kodama T, Tsunematsu H, Miyagi T, Hosui A, Ishida H, Tatsumi T, Kanto T. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int J Cancer. 2012;131:548-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 215] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 20. | Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313-326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3876] [Cited by in RCA: 3803] [Article Influence: 253.5] [Reference Citation Analysis (0)] |
| 21. | Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 363] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 22. | Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456-461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2125] [Cited by in RCA: 2047] [Article Influence: 146.2] [Reference Citation Analysis (0)] |
| 23. | Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 735] [Cited by in RCA: 835] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 24. | El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2881] [Cited by in RCA: 3088] [Article Influence: 220.6] [Reference Citation Analysis (0)] |
| 25. | Gozuacik D, Kimchi A. Autophagy and cell death. Curr Top Dev Biol. 2007;78:217-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 330] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 26. | Zhang H, Chen GG, Zhang Z, Chun S, Leung BC, Lai PB. Induction of autophagy in hepatocellular carcinoma cells by SB203580 requires activation of AMPK and DAPK but not p38 MAPK. Apoptosis. 2012;17:325-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Yu HC, Lin CS, Tai WT, Liu CY, Shiau CW, Chen KF. Nilotinib induces autophagy in hepatocellular carcinoma through AMPK activation. J Biol Chem. 2013;288:18249-18259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 28. | Martin AP, Park MA, Mitchell C, Walker T, Rahmani M, Thorburn A, Häussinger D, Reinehr R, Grant S, Dent P. BCL-2 family inhibitors enhance histone deacetylase inhibitor and sorafenib lethality via autophagy and overcome blockade of the extrinsic pathway to facilitate killing. Mol Pharmacol. 2009;76:327-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |