INTRODUCTION
The smallest virus known to infect humans, hepatitis delta virus (HDV), is increasingly again becoming a cause of fulminant hepatitis or a more rapid progression of liver disease in the setting of chronic hepatitis B virus (HBV) infection[1]. HDV is a defective satellite RNA virus which requires the helper function of HBV for its replication and assembly of new virions[2]. An estimated 15-20 million individuals with HBV worldwide are found infected with HDV[2], highlighting a need to exactly understand the pathogenesis and molecular biology of the virus.
GENOMIC STRUCTURE AND TAXONOMIC CLASSIFICATION
HDV is a small, spherical virus with a diameter of about 36 nm[1,3]. The viral genome is a circular, single-stranded, negative sense RNA molecule with an internal core delta antigen surrounded by an envelope derived from HBV surface proteins[4,5]. The genomic RNA of HDV is composed of approximately 1700 nucleotides, packaged with about 200 molecules of hepatitis delta antigen (HDAg)[6] to form a single viral particle. Due to extensive base-pairing within the RNA molecule, the genome appears as a double-stranded rod like structure and thus it remarkably resembles viroids (plant pathogens)[1,7]. However, unlike viroids, the HDV has more nucleotides in its genome compared to 200-400 nucleotides in a viroid RNA. Furthermore, viroids lack protein coding ability whereas HDV codes for its own HDAg and requires HBV for its propagation[7]. The envelope surrounds the genome and HDAg composed of all three HBV envelope proteins, namely small hepatitis B surface antigen (S-HBsAg), medium HBsAg and large HBsAg. The HDV genome also exhibits a self-cleavage activity by encoding a ribozyme domain about 80-100 nucleotides long. HDV seems to be a unique animal virus because of its high GC nucleotide content and circle rolling mechanism of replication[8-10]. The virus is currently assigned the separate genus deltavirus and yet awaits a complete taxonomic classification, including order and family.
Unlike most RNA viruses, HDV does not encode its own replicase or RNA-dependant RNA polymerase to replicate its genome. Rather, it makes use of cellular RNA polymerases which are DNA-dependant RNA polymerases. Since HDV genomic RNA has negative or anti-messenger polarity, during replication three different forms of RNA are being made: circular genomic RNA, circular complementary anti-genomic RNA and a linear anti-genomic RNA through a circle rolling mechanism. The circle rolling mechanism involves unidirectional replication of nucleic acids to form multiple copies of circular genome using cellular RNA polymerases[11]. Linear polyadenylated anti-genomic RNA serves as messenger RNA (mRNA) to produce HDAg.
Since HDV is presented with a limited protein coding capacity encoding only one HDAg with two isoforms, it thus makes use of host cellular machinery (cellular proteins) to accomplish the processes which are essential for its life cycle, such as transcription, replication, post-transcriptional and translational modifications[9,12].
HDV ANTIGEN
HDV genome has been found to contain several open reading frames (ORFs)[13]. Out of all the open reading frames, only one appears to be actively transcribed and encodes for antigen (HDAg)[5], the function of the rest of the ORFs is still unknown. There are two isoforms of HDAg, small HDAg 24 kDa (S-HDAg) composed of 195 amino acids and large HDAg 27 kDa comprising of 214 amino acids (L-HDAg). The open reading frame transcribes into a mRNA using host cell RNA polymerase IIwhich translates to produce S-HDAg. A post-transcriptional modification by the cellular enzyme adenosine deaminase-1 (ADAR 1) replaces the stop codon (UAG at position 196) on the mRNA by a tryptophan (codon UGG), extending the reading frame by an additional 19 amino acids leading to the production of L-HDAg[5,11,14]. The nineteen extra amino acids added at the carboxyl terminal of L-HDAg confer it functional properties that are different from S-HDAg. S-HDAg is required for the initiation of the viral genome replication, whereas L-HDAg which is synthesized in the late stage of viral replication serves as a principal inhibitor of replication and is essential for the assembly of new virion particles[8,9]. L-HDAg not only regulates HDV genome replication but also its own synthesis by inhibiting viral replication which prevents editing of amber/W site necessary for the expression of L-HDAg[15].
The HDAg contains different functional domains, such as RNA-binding domain, coiled-coil sequence and nuclear localization sequence. The L-HDAg in addition contains a few more domains which include virus assembly signal (VAS) and nuclear export signal[8,9]. VAS in the L-HDAg renders it obligate for virion assembly[16]. HDAg may directly activate transcription of the viral genome by binding to RNA. HDAg may also facilitate transcription elongation by replacing transcription repressor bound to RNA polymerase II[8,9,16]. In the absence of HBsAg, both the S-HDAg and L-HDAg tend to localize in the nucleus as they carry nuclear localization signals. Thus, HBsAg is essential for cytoplasmic translocation of L-HDAg as only L-HDAg bears nuclear export signal for the purpose of its established role in virion assembly[17].
POST-TRANSLATIONAL MODIFICATIONS OF HDAG
The post-translational modifications of HDAg have been reported[8,7,18] and are of immense importance as these may modulate HDAg function and may lead to progression of the viral cycle[17]. These post-translational modifications include serine and threonine phosphorylation, lysine acetylation, arginine methylation, lysine sumoylation and cysteine farnesylation[9,12]. Since HDV is deficient in enzymes responsible for post-translational modifications, HDV precisely depends upon cellular proteins to accomplish these processes.
Phosphorylation occurs at the serine and threonine residues of S-HDAg, whereas only serine residues are phosphorylated in L-HDAg[9,19]. Post-translational phosphorylation of S-HDAg at serine-177 is crucial for its interaction with the cellular RNA polymerase II[20], which is responsible for genomic HDV RNA synthesis (HDV antigenome RNA replication). Phosphorylation of S-HDAg at serine-177 and phosphorylation at serine-2 residue[21] therefore enhances HDV replication. Furthermore, RNA-binding activity of HDAg has found to be mediated by phosphorylational modification and thus seems essential for viral replication[21].
Acetylation of lysine residues of both the S-HDAg and L-HDAg have been reported and are being associated with modulation of HDV replication. Mu et al[22] found that a substitution of acetylated lysine-72 of S-HDAg by an alanine re-localized the mutant S-HDAg into the cytoplasm and was associated with the diminished viral RNA accumulation and earlier L-HDAg appearance. Methylation of the arginine residue of S-HDAg has been observed to influence HDV replication[17,18] and thus methylation inhibitors result in an inhibition of HDV replication.
Sumoylation is a newly known post-translational modification which involves the conjugation of S-HDAg with a small ubiquitin-related modifier isoform-1 (small ubiquitin-like protein), results in sumoylation of multiple lysine residues in S-HDAg and an enhanced genomic RNA and mRNA synthesis with no effect on anti-genomic transcription[9,23].
Isoprenylation is the most important modification which results in farnesylation of the cysteine-211 which resides in the isoprenylation signal sequence located at the carboxyl terminal of L-HDAg. Farnesylated L-HDAg inhibits viral RNA replication and facilitates virion assembly by mediating the direct binding between L-HDAg and HBV envelope proteins[9]. Farnesyl inhibitors, although in their early phase of development, may improve disease outcome.
CELL ATTACHMENT, ENTRY, UNCOATING AND REPLICATION
The mechanism of entry of the HDV into the hepatocytes is not clearly understood, however, it is thought to be similar to HBV. HDV enters hepatocytes by binding to the carbohydrate side chains of heparin sulphate proteoglycan present on the surface of hepatocytes[24]. The N-terminal aminoacids of the pre-S1 domain of L-HBsAg are thus obligatory to HDV entry into hepatocytes. Mutations/deletions in highly conserved pre-S1 sequence and acetylation or myristoylation of pre-S1 N-terminal amino acids have been found to inhibit HDV entry into hepatocytes[25]. Recently, Yan et al[26] have identified a putative receptor for the entry of HBV and HDV into the hepatocytes. The authors proposed that pre-S1 domain of L-HBsAg interacts with sodium-taurocholate cotransporting polypeptide, an integral transmembrane glycoprotein involved in enterohepatic circulation, to facilitate HDV infection.
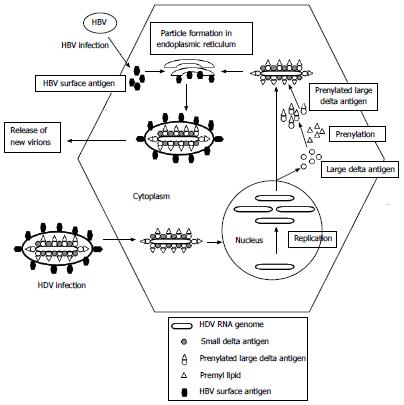
After HDV enters the cell, the uncoating of viral particle occurs and HDAg translocates the viral genome into the nucleus where RNA polymerases I and II are employed to replicate the genome (Figure 1). Polymerase I involves the transcription of antigenome from viral genome in the nucleolus, while polymerase II catalyzes genome replication from antigenome and transcription of mRNA in the nucleoplasm[27].
Figure 1 Hepatitis D virus replication cycle.
HBV: Hepatitis B virus; HDV: Hepatitis D virus.
The process of replication starts with the transcription of antigenome using the viral genome through a circle rolling mechanism which produces an antigenomic RNA of more than one unit length. The antigenomic RNA is then self-cleaved by intrinsic ribozyme activity and ligated to form a circular antigenome using cellular ligases. The antigenomic RNA is then used to produce genomic RNA in the nucleoplasm. The mRNA is transcribed using the same genomic transcript and is translated to produce HDAg. It is therefore evident that HBV plays no role in HDV replication and it can proceed even in the absence of the helper virus. It is required only for cell entry, virion assembly and export.
PATHOGENESIS
HDV replicates only in the hepatocytes. The cellular damage associated with HDV infection thus involves mainly the liver. Immune-mediated liver damage is thought to be implicated in HDV infection[28]. However, data from experimental chimpanzees has also suggested a direct cytopathic effect of HDV on hepatocytes, particularly in acute hepatitis setting[29-32]. It is postulated that in acute HDV infection, infected hepatocytes undergo degenerative changes characterized by shrunken eosinophilic cytoplasm and pyknotic nuclei as well as the presence of minimal inflammatory cells in the liver parenchyma, consistent with cytopathic hepatocellular damage. These findings are also evident from in vitro (cell culture system)[33] and human studies[34,35]. Small delta antigen expressed by infected hepatocytes is thought to be responsible for this direct cytopathic effect of HDV[33], while large delta antigen per se is non-cytotoxic, promotes persistence of HDV (chronicity) and makes hepatocytes susceptible to immune-mediated damage.
Experimental woodchuck models have proven very helpful in furthering our knowledge of HDV pathogenesis and the chronicity associated with HDV superinfection[36,37], owing to the marked resemblance between the course of disease in woodchuck models and the outcome of HDV superinfection in humans. In addition, these models are also invaluable for testing the efficacy and protective role of new treatments for HDV, including vaccine candidates. Studies on these experimental models have disclosed that both the protein immunization and DNA immunization for HDV are insignificant in protecting against HDV superinfection[38,39], highlighting the need of adopting different approaches to develop an HDV vaccine.
Variation in immune-mediated responses during acute and chronic HDV infection has been noticed[37,40], which may explain the persistence and chronicity of HDV superinfection. Cytotoxic T lymphocytes are mainly responsible for clearing the virus by destroying HDV-infected cells. Delayed and insufficient immune response with ability of recognizing only limited viral epitopes has been implicated in failure to clear the infection coupled with establishment of chronic infection. Fulminant hepatic failure has been observed in 1% of HBV/HDV co-infected patients while in 5% of those superinfected with HDV. An exaggerated immune response, particularly a cell-mediated one, is proposed to be involved in causing massive hepatocyte necrosis and liver damage in fulminant hepatic failure[41,42].
The pathogenesis of HDV is also thought to be influenced by the interaction of HDV with the HBV[28,43], which has not yet been clearly elucidated by investigating bodies. HDV infection is known to occur either as a coinfection or a superinfection. A coinfection with HBV/HDV usually eradicates both the organisms and often results in complete recovery, while a superinfection frequently progresses into chronic hepatitis D infection[44]. Patients with chronic hepatitis B who develop superinfection with hepatitis D may also go into acute on chronic liver failure, leading to ascites and hepatic encephalopathy.
HBV-HDV INTERACTION
HBV coinfection with other hepatitis viruses is associated with various patterns of reciprocal inhibition of viral replication. HDV has been frequently shown to suppress HBV replication[45]. L-HDAg up-regulates the myxovirus resistance-A transcription, an interferon-inducible antiviral response mediator which is involved in suppression of HBV replication. It is therefore suggested that the liver disease in HBV/HDV superinfection is mainly due to HDV[8]. Chronic HDV/HBV infection causes a more severe liver disease than HBV monoinfection alone; the disease runs a rapidly progressive course, leading to early cirrhosis, decompensation and hepatocellular carcinoma (HCC), and a shorter 5 year survival[46].
Suppression of HBV replication by HDV is not sustained for an overall period of the disease as the viral response changes over time[47]. One study revealed significant HBV replication in about half of the patients[48]. Taken together, three phases of chronic hepatitis D have been proposed: (1) early active phase with active HDV replication and suppression of HBV; (2) a second moderately active one with decreasing HDV and reactivating HBV; and (3) late phase with the development of cirrhosis and hepatocellular carcinoma caused by replication of either virus or with remission resulting from the marked reduction of both viruses[49].
INTERFERON-ALPHA SIGNALING INHIBITION
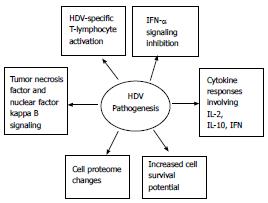
The pathogenesis of HDV mainly involves interferon-α (IFN-α) signaling inhibition[50], HDV-specific T-lymphocyte activation and cytokine responses[51,52,53], tumor necrosis factor-alpha (TNF-α) and nuclear factor kappa B signaling[54,55], together with modifications in cell proteome and an associated increased cell survival potential[56] (Figure 2).
Figure 2 Depicts hepatitis D virus pathogenesis.
HDV: Hepatitis D virus; IL: Interleukin; IFN: Involves interferon.
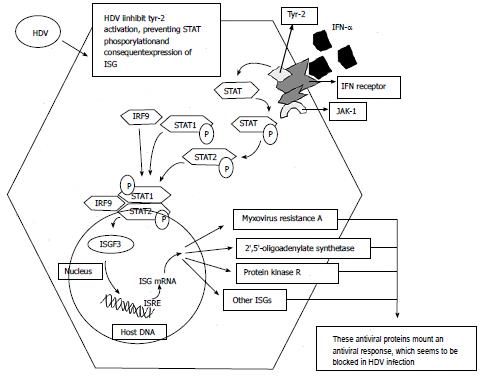
IFN-α signaling by the virus-infected cells to warn their neighboring cells of a viral presence is a first line of defense of the host to eradicate viruses. IFN-α is induced by the double-stranded RNA presented during viral replication[57]. The IFNs thus produced exert their effect by binding to α and β-IFN receptors on the cell surface, resulting in activation of the tyrosine kinases of the janus kinase (JAK) family which in turn phosphorylate tyrosine residues of the cytoplasmic transcription factors acting as signal transducer and activator of transcription (STAT). Activation of JAK/STAT signaling pathway stimulates the expression of IFN-induced genes. The IFN-α-stimulated genes then code for the antiviral proteins, namely myxovirus resistance A, double-stranded RNA (dsRNA)-activated protein kinase and 2’,5’-oligoadenylate synthetase which, in turn, mount an antiviral response[58]. The resultant activated non-specific innate and specific acquired immune responses help combat the viral infection, but this is not the case in the setting of HDV infection.
Since HDV is composed of a single-stranded RNA molecule, it is not expected that it would stimulate IFN-α release[59]. On the other hand, interferon-alpha signaling activation has shown to be inhibited in HDV infection[50]. HDV averts tyrosine kinase-2 (tyr-2) activation preventing phosphorylation of STAT-1 and STAT-2 and their intra-nuclear translocation for the expression of IFN-induced genes. HDV thus interferes with the activation of JAK/STAT signaling pathway by IFN-α signaling inhibition[50] which may be implicated in viral persistence and treatment failure (Figure 3).
Figure 3 Interferon-α signaling and inhibition by hepatitis D virus.
HDV: Hepatitis D virus; INF: Interferon; STAT: Signal transducer and activator of transcription; JAK: Janus kinase; IRF9: Interferon regulatory factor 9; ISGF3: INF-stimulated gene factor 3; ISG: INF stimulated genes; ISRE: INF-stimulated response elements; P: Phosphorylated residues.
IFN-α is also known to stimulate the cellular enzyme ADAR 1 and thus L-HDAg expression by increased editing of mRNA[60]. IFN-α signaling failure in HDV infection may prevent an early production of L-HDAg and cessation of viral replication. From the above description, it could be concluded that inhibition of IFN-α signaling in an HDV infected individual plays a pivotal role in failure to clear the virus and also confers resistance to IFN-α treatment. Babiker et al[61] tested an interferon-sparing antiviral treatment regime consisting of tenofovir disoproxil fumarate and lamivudine in a patient with severe acute HDV infection. Successful suppression of HDV RNA was achieved after 16 mo of treatment, along with significant reductions in HBV DNA and HBsAg levels. The interferon-sparing regimens should undergo more clinical trials to establish their efficacy for the treatment of hepatitis D.
T-CELL RESPONSES
It is already established that the pathogenesis of liver injury in HDV infection is not directly cytopathic but immune-mediated mechanisms are known to be involved. After the entry of virus into hepatocytes, its antigen is processed in endoplasmic reticulum of cell cytoplasm and presented on the cell surface in association with major histocompatibility complex-I (MHC-I) protein. CD8+ cytotoxic T lymphocytes recognize endogenously synthesized antigen presented in association with class-I MHC proteins and kill the virus infected cell by two mechanisms: release of perforins and granzymes and an interaction between FAS receptor and FAS ligand. Both of these mechanisms lead to DNA fragmentation and apoptosis of the target cells[62].
Secondly, exogenous antigens and noninfectious viral particles are endocytosed by the surrounding antigen-presenting cells (APCs) (which include macrophages, B lymphocytes and dendritic cells) which present the antigens on their surface in association with class II MHC proteins. CD4+ helper T-cells recognize antigens when they are presented in association with class II MHC proteins. A virus-specific clone of helper T-cells is thus activated and starts proliferating to bring about clonal expansion in order to clear the viral infection. Proliferating helper T-cells are categorized into three subtypes: type 0 (Th-0), Th-1 and Th-2 T-cells on the basis of their functions and cytokines they produce. Thus, an HDV-specific activated clone of helper T-cell is the key component around which the pathogenesis of HDV revolves[52].
CYTOKINES
The pathogenesis of HDV involves activation of a clone of HDV-specific helper T-cell which, in turn, expresses cytokines, mainly interleukin-2 (IL-2), IL-2 receptor, IL-10 and IFN-γ[51,52,63]. IL-2, being a T-cell growth and differentiation factor, stimulates both HDV-specific helper T-cell and CD8+ cytotoxic T-cell to proliferate and undergo clonal expansion. Cytotoxic T-cells destroy virus-infected cells while the activated helper T-cells under the influence of IL-12 develop into subtype Th-1 cells and specifically secrete IFN-γ.
In HDV infection, HDV-specific Th-1 and cytotoxic T-cells have been shown to produce a large amount of IFN-γ[52] which stimulates phagocytosis and killing by macrophages and up-regulates the expression of class-I and class-II MHC proteins on cell surfaces. IFN-γ may inhibit viral replication directly or from its immune-modulatory and immune-stimulatory effects[51]. IFN-γ also stimulates the secretion of IFN-γ induced protein-10 (CXCL-10), a chemoattractant which recruits natural killer cells (NK cells), T-cells, macrophages/monocytes and dendritic cells to destroy HDV-infected cells with the consequent viral clearance at the cost of inflammatory infiltrates causing liver damage[61].
It is also postulated that in chronic hepatitis, production of IFN-γ by the Th-1 helper T-cells induces hepatocytes to express class-II MHC protein in addition to class-I MHC protein in an attempt to increase their capacity to clear the infection[53]. This may be associated with severe liver necrosis and increased severity of hepatitis in HDV- infected patients as more uninfected hepatocytes expressing MHC-II protein will also be recognized and killed by helper T and cytotoxic T-cells.
Since HDV-specific CD4+ T-cell response has been identified in HDV infected patients in response to HDAg, there are specific sequences on HDAg identified which are being recognized and processed by APCs[52] and presented on the cell surface in association with MHC-II protein to mount an HDV-specific antiviral response by CD4+ helper T-cells. Nisini et al[52] thus employed synthetic peptides spanning the entire HDAg sequence in an attempt to determine HDAg-specific helper T-cell recognition of specific antigenic determinants. They studied their fine specificity to identify immunogenic epitopes of HDAg that could be used to generate a vaccine for the prophylaxis and treatment of HDV infection[52]. Furthermore, two studies have identified a low degree of heterogeneity in HDAg which encourage the development of a vaccine using these immunogenic sequences of HDAg[64,65].
The study results of Nisini et al[52] also suggested that HDAg-specific T-cell response in peripheral blood of HDV-infected individuals is associated with reduced HDV replication and anti-HD IgM disappearance with the consequent reduced activity of HDV-induced liver disease[52].
A vigorous immune response involving HDAg-specific T-cell response and cytotoxic killing of HDV-infected cells following HDV infection thus results in both viral clearance and augmented liver damage.
CELL PROTEOME MODIFICATIONS AND ROLE OF CELLULAR PROTEINS
Since HDV lacks essential enzymes to carry out its own replication and cell cycle, host cell proteins and components are thus intimately involved in the HDV pathogenesis [9,12].
There are a number of cellular proteins identified to date which interact with HDV RNA and HDAg to accomplish HDV life cycle processes. The interaction of cellular proteins with HDV RNA and HDAg significantly alters cell proteome. Cellular RNA polymerase subunits, helicases, RNA-binding proteins, heterogeneous ribonucleoproteins (hnRNPs) and transcriptional and splicing factors have been known to interact with S-HDAg to facilitate HDV RNA transcription and translation. Host transcription factors interacting with HDAg may remarkably alter cellular gene expression resulting in enhanced cytokines, inflammatory enzymes, growth factors and anti-apoptotic proteins[9], which may suggest severe necroinflammation, amplified liver damage and a concomitent increased cell survival in HDV infected patients.
Cellular proteins involved in pathways, such as regulation of cell metabolism and energy pathways, nucleic acid and protein metabolism, apoptosis and cell growth and maintenance, demonstrate a significantly altered expression profile in the presence of HDV components.
HDV AND CIRRHOSIS
Progression of liver disease in HDV infection has been demonstrated to be influenced by many factors, including mode of infection (i.e., coinfection or superinfection), specific HDAg variants[66] and HDV[67] and HBV[68] genotypes. Superinfection with HDV in chronic HBV is associated with a more severe form of liver disease owing to its exacerbation of pre-existing liver damage due to HBV[69]. HBV genotypes have much less convincing evidence of directly influencing liver pathology, rather it regulates HDV viral loads which adversely affect disease outcome[70].
Since HDV infection causes fulminant hepatitis and liver cirrhosis, L-HDAg has been shown to stimulate transforming growth factor-β (TGF-β) and c-Jun-induced signaling cascades which in turn may induce epithelial-mesenchymal transition and fibrogenesis[70,71]. Chronic HDV may thus induce liver cirrhosis by up-regulating the expression of TGF-β. This process is specifically accomplished by the isoprenylation ( farnesylationn) of L-HDAg[72]. Isoprenylation inhibitors, which are still in their early phase of development, may play a key role in preventing these undesirable outcomes following HDV infection.
HDV AND HEPATOCELLULAR CARCINOMA
It has already been mentioned that cells infected with HDV appear to have altered gene expression and cellular responses, which is also evident from augmented expression of pro-inflammatory, growth and anti-apoptotic factors. It is thus explanatory that severe liver damage and a concomitant increased hepatic cell survival in HDV-infected patients may lead to HCC.
It is well known that nuclear factor kappa B (NF-κB) dysregulation is associated with inflammation and cancer[73]. L-HDAg has been shown to activate nicotinamide adenosine denucleotide hydro-phosphoric acid oxidase-4 which in turn induces oxidative stress. L-HDAg is therefore able to activate the signal transducer and activator of transcription-3 and the NF-κB through the oxidative stress pathway[54]. L-HDAg may also stimulate TNF-α induced NF-κB, probably via TNF receptor-associated factor 2 (TRAF2), a protein involved in the early signal transduction events[56]. This may underscore a possible underlying cause of severe necroinflammation in HDV infection and its progression to HCC. A clinical study has also suggested that HCC in HDV infection may be a secondary effect of the necroinflammation and cirrhosis[74]. In this study, decreased liver size was noticed more in cases of HDV HCC compared to HBV monoinfection group where the liver size was normal or increased. HDV patients had lower platelets and larger varices on endoscopy.
Both L-HDAg and S-HDAg have been shown to enhance clusterin gene expression[56] which is found up-regulated in tumor cells and evidently implicated in tumorigenesis[75]. HDAg amplifies the expression of clusterin gene by enhanced acetylation of histone H3 in clusterin gene promoter region. The similar modification has also been observed in several oncogenic viruses associated with the expression of specific proteins, such as the adenovirus protein E1A[76], the simian virus 40T antigen[77] and the E7 protein of the human papilloma virus[72]. An increased histone acetylation and clusterin protein production are associated with increased survival of HDV-infected cells[56]. This may reliably be implicated in the development of HCC in HDV-infected patients.
RELEVANCE OF GENOTYPES
To date, there are eight genotypes of HDV which have been reported with unexplained variations in their pathogenecity. Furthermore, HDV genotypes have a distinct geographical distribution, apart from HDV genotype 1 which has been observed universally[47].
The available literature is apparently unable to outline a specific HDV genotype associated with the disease severity in HDV infection as different genotypes are prevalent in different parts of the world and none could be correlated with more severe disease outcome with certainty. Any geographical area which reports HDV associated disease severity primarily belongs to the genotypes prevalent in that area. This entity thus requires further research to establish an association between disease severity and different HDV genotype infections. However, Su et al[78] have reported genotype I HDV and older age associated with adverse outcomes in HDV/HBV infection. There are patterns of disease suggesting cytopathic viral illness, as was indicated by outbreaks of severe hepatitis in the northern part of South America[79]. These cases were mostly caused by HDV genotype 3.
CONCLUSION
Although a global decline in HDV prevalence has now been observed due to better prevention practices, mass awareness and expanded HBV vaccination in industrialized countries, it is still a major health concern in the Asia-Pacific region where circumstances favor the spread of hepatitis B and related infections. A scarcity of literature about the HDV life cycle and pathogenesis is clearly evident. Furthermore, severity of HDV-associated liver disease with its adverse outcomes and lack of an efficient treatment regime encourage research into HDV biology. A better understanding of the viral pathogenesis will certainly help the investigating bodies to develop new treatment approaches which would be able to cope with both disease severity and outcome. An innovative approach towards generating a vaccine against HDV is also needed.