Published online Apr 27, 2012. doi: 10.4254/wjh.v4.i4.139
Revised: September 6, 2011
Accepted: April 24, 2012
Published online: April 27, 2012
AIM: To investigate the impact of hepatitis B virus (HBV) infection on cellular gene expression, by conducting both in vitro and in vivo studies.
METHODS: Knockdown of HBV was targeted by stable expression of short hairpin RNA (shRNA) in huH-1 cells. Cellular gene expression was compared using a human 30K cDNA microarray in the cells and quantified by real-time reverse transcription-polymerase chain reaction (RT-PCR) (qRT-PCR) in the cells, hepatocellular carcinoma (HCC) and surrounding non-cancerous liver tissues (SL).
RESULTS: The expressions of HBsAg and HBx protein were markedly suppressed in the cells and in HBx transgenic mouse liver, respectively, after introduction of shRNA. Of the 30K genes studied, 135 and 103 genes were identified as being down- and up-regulated, respectively, by at least twofold in the knockdown cells. Functional annotation revealed that 85 and 62 genes were classified into four up-regulated and five down-regulated functional categories, respectively. When gene expression levels were compared between HCC and SL, eight candidate genes that were confirmed to be up- or down-regulated in the knockdown cells by both microarray and qRT-PCR analyses were not expressed as expected from HBV reduction in HCC, but had similar expression patterns in HBV- and hepatitis C virus-associated cases. In contrast, among the eight genes, only APM2 was constantly repressed in HBV non-associated tissues irrespective of HCC or SL.
CONCLUSION: The signature of cellular gene expression should provide new information regarding the pathophysiological mechanisms of persistent hepatitis and hepatocarcinogenesis that are associated with HBV infection.
- Citation: Fukuhara Y, Suda T, Kobayashi M, Tamura Y, Igarashi M, Waguri N, Kawai H, Aoyagi Y. Identification of cellular genes showing differential expression associated with hepatitis B virus infection. World J Hepatol 2012; 4(4): 139-148
- URL: https://www.wjgnet.com/1948-5182/full/v4/i4/139.htm
- DOI: https://dx.doi.org/10.4254/wjh.v4.i4.139
Hepatitis B virus (HBV) is a major causative agent of chronic liver diseases that lead to the development of hepatocellular carcinoma (HCC) worldwide[1]. Vaccination against HBV has been proven efficacious for the prevention of virus transmission and has markedly reduced the carrier rate[2]. Several anti-viral agents, such as entecavir and interferon-α, also exert therapeutic effects by reducing virus titer[3]. Unfortunately, these preventative and therapeutic treatments are not widely administered, especially in areas with HBV epidemics[4]. Small molecule therapies and interferon treatments suffer from a number of drawbacks, including the selection of drug-resistant mutants, toxicity and limited efficacy[5,6]. Furthermore, the increase in international travel has introduced different genotypes of HBV to the world’s populations, which may not be efficiently protected by the vaccine developed against the endemic genotype[7]. The World Health Organization reported that an estimated 350 million people worldwide are chronically infected with HBV and a significant proportion of chronic infections terminate in HCC, leading to more than half a million deaths annually (http://www.who.int/mediacentre/factsheets/fs204/en/). Thus, the need to elucidate the detailed pathophysiology of HBV infection is great.
Recent advancements in technology allow us to evaluate the proteome or transcriptome during pathogenic processes, providing new insight in terms of host-pathogen interactions. So far, in vivo cellular reactions associated with HBV infection have mainly been evaluated by comparing HBV-associated HCC [HCC(B)] with other liver tissues. Kim et al[8] reported a characteristic protein profile of HCC(B) in comparison with hepatitis C virus (HCV)-associated HCC [HCC(C)]. Differential gene expression profiles have also been reported in HCC(B) in comparison with corresponding surrounding liver tissues (SL)[9] or HCC(C)[10]. Although reduced tumorigenicity after knockdown of HBx protein has been reported in PLC/PRF/5 HCC cells[11], it is unclear whether HBV still has significant effects on cellular gene expression once the cells have been transformed because, at the time of HCC development, tumor cells no longer allow efficient viral expression[12,13]. In addition, it is reasonable to assume that malignant transformation causes a significant alteration of the gene expression signature and may overcome the impact of HBV on the profile. Thus, a simple HCC-oriented observation may not accurately reflect the cellular events induced by HBV infection.
Artificial control of HBV expression is another approach to studying differential cellular gene expression. Otsuka et al[14] reported that, in comparison with parental cells, several cellular genes were specifically up- or down-regulated in HepG2.2.15 cells, which are derived from HepG2 cells by transfecting them with plasmids containing HBV DNA, leading to the production of HBV proteins. Alteration of cellular gene expression has also been reported in HepG2.2.15 cells after knockdown of HBV through RNA interference (RNAi)[15]. Furthermore, microarray analysis has revealed differential cellular gene expression between wild-type and HBV transgenic mouse livers[16,17]. There are concerns, however, that the methodologies employed may have direct effects on cellular gene expression. There are inconsistencies in the genes that have been reported to be altered as a result of HBV infection, not only among studies using different models of HBV infection, but also using the same methodologies[18].
In this report, we elucidate the differentially expressed cellular genes associated with HBV infection by sequentially applying two processes: (1) selection of candidate genes by knockdown of HBV expression using RNAi in cells derived from a HBV-associated case; and (2) quantification of the selected gene expression in various liver tissues from both HBV-infected and non-infected patients. The advantage of our approach and the pathophysiological implications of our results are discussed.
Seventeen HBV genome sequences from GenBank were aligned and analyzed to identify the conserved regions containing at least nineteen contiguous nucleotides spanning within the region that was shared by all four open reading frames. Nineteen nucleotides following AA were common for all genotypes except for F and H, which are quite rare in Japan, and were further analyzed by BLAST to ensure that the sequence does not have significant homology with known human genes. Finally, the selected sequence, 5’-TGTCAACGACCGACCTTGA-3’, was designed to form a hairpin structure when transcribed and cloned into pSUPER.retro (OligoEngine, WA, United States), which generates 3’-UU overhanging transcripts without a poly-A tail under the control of the polymerase-III H1-RNA gene promoter. Plasmids containing the target sequence or the same sequence with an A to G transition at the ninth nucleotide were designated pSUPER.HB4 or pSUPER.HB4G, respectively.
huH-1 cells; JCRB0199, were obtained from the JCRB cell repository and transfected with 2 μg of plasmids using Effectene transfection reagent (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. In brief, 5.0 × 105 huH-1 cells were seeded into a 60 mm dish a day before transfection and the plasmids were mixed with 16 μL of enhancer followed by the addition of 50 μL Effectene reagents. After mixing with medium, the mixture was applied onto the culture plate. The stable transformants with pSUPER.retro, pSUPER.HB4 and pSUPER.HB4G were named huHpS, huHB4 and huHB4G, respectively.
The cells of 1 × 106/mL were subjected to culture and supernatants were collected on days 2 and 4 and stored at -20 °C until use. HBsAg was quantified in the medium by a chemiluminescence immunoassay using ARCHITECT HBsAg QT (Abbott Japan Co. Ltd., Chiba, Japan).
Hydrodynamic gene delivery was employed[19] to target HBx in HBx transgenic mice[20] by injecting 100 μg of pSUPER.retro, pSUPER.HB4 or pSUPER.HB4G. Immunohistochemistry was performed on the liver specimens, which were obtained 48 hours after delivery, by a standard avidin-biotin complex method[21] that involved incubating the sections with primary antibodies of rabbit polyclonal anti-HBx[22].
Total RNA was isolated from huHpS and huHB4 cells using IsoGen (Nippon Gene Co. Ltd., Tokyo, Japan) and stored at -80 °C. After amplification by T7 polymerase, cDNAs were labeled with the fluorescent dyes cy3 or cy5, and hybridized with AceGene human 30K cDNA microarrays (DNA Chip Research Inc. Yokohama, Japan), which contain 32 000 sequences verified by the mouse IMAGE consortium (http://image.hudsonalpha.org/). After washing, the arrays were scanned and the signal intensity of a spot was considered significant if the intensity was 200 times greater than the background; otherwise, the spot was flagged as “not found”. Using TIGR MIDAS software (http://www.tm4.org/), the data obtained from qualified spots were normalized by applying the LOWESS algorithm. After normalization, the spots were judged to be inconsistent between a pair of flip-dye replicates and filtered out, if log2(cy3/cy5)/log2(cy5/cy3) was outside the range from -1 to 1. We took a difference of spot intensities between huHB4 and huHpS of more than double or less than half as significant. The selected genes were fed to the DAVID functional annotation tool (http://david.abcc.ncifcrf.gov/home.jsp).
Total RNAs of huH-1 cells and liver tissues were purified with RNeasy Mini kits (QIAGEN KK, Tokyo, Japan) after digestion with RNase-free DNase I (Invitrogen Corporation, Carlsbad, United States) and were reverse transcribed for use in TaqMan Gene Expression Assays using a LightCycler (Roche Diagnostics, Mannheim, Germany). To quantify the expression of the candidate genes selected in microarray analyses and of the internal controls, hypoxanthine phosphoribosyltransferase 1 (HPRT1) and beta 2-microglobulin (β2M), commercially available primer and probe sets were employed, whereas custom primers, 5’-CCCGTCTGTGCCTTCTCA-3’ and 5’-GGTCGGTCGTTGACATTGCT-3’, and a probe, 5’-CCGTGTGCACTTCGCT-3’, sequences that are common to all four open reading frames, were designed by Custom TaqMan® Gene Expression Assay (Applied Biosystems Inc., Foster City, United States) for HBV expression. The results were analyzed using LightCycler software (Roche Diagnostics, Mannheim, Germany). A relative fluorescent intensity was calculated from a standard curve obtained by quantitative analyses using a serial dilution of total RNA from HepG2. Finally, a relative amount of each message was calculated as a ratio of threshold cycle after normalization against HPRT1 or β2M.
Liver tissue samples were obtained from surgical resections of HCC or other cancers from eighteen patients consisting of five HBV-positive cases, five HCV-positive cases, three neither HBV nor HCV positive cases, and five cases without chronic liver disease, which included one colon cancer, one common bile duct cancer and three rectal cancer cases. The three patients without viral hepatitis had autoimmune hepatitis, primary biliary cirrhosis and alcoholic liver cirrhosis. No cases showed positive reactions to HBsAg or anti-HBc, except for the HBV-positive cases. Two expert pathologists independently evaluated liver specimens. Each HBV-positive or HCV-positive group consisted of two chronic hepatitis cases and three cirrhotic cases. Written informed consent was obtained from each patient and the study protocol conformed to the ethical guidelines of the 2008 Declaration of Helsinki, as reflected in prior approval by the Niigata University Graduate School of Medical and Dental Sciences Human Research Committee.
Doubling times of the cells and quantity of HBsAg in cultured medium were compared using ANOVA analysis followed by post hoc Bonferroni’s multiple comparison tests, whereas the Mann-Whitney test was employed to compare HBV expression between HCC(B) and SL(B). In the DAVID annotation system, Fisher’s exact test was adopted to measure the gene enrichment in annotation terms by referencing the frequencies of 30 000 genes of the human genome. All analyses except for the functional annotation were performed using GraphPad Prism 5 (GraphPad Software, Inc. La Jolla, United States) and a two-sided P value less than 0.05 was considered statistically significant.
We cloned huHB4G and huHB4, in which shRNA for the HBx coding region is continuously expressed in huH-1 cells with and without nucleotide replacement at the center from A to G, respectively. The cells transfected with plasmids with no insert were referred to as huHpS.
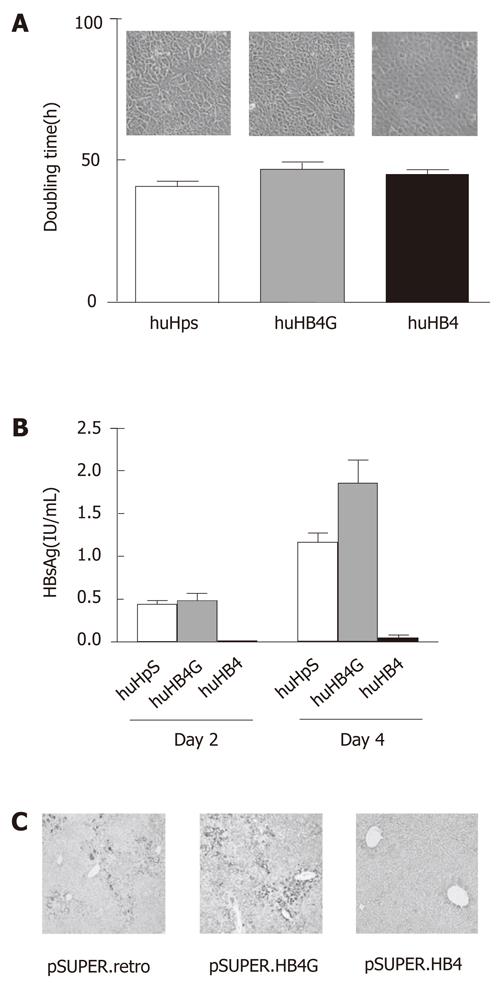
Morphologically, no remarkable differences were observed among the clones in culture, as shown in Figure 1A. The average doubling times in three independent cultures of huHpS, huHB4G and huHB4 were 40.9 ± 1.8 h, 46.8 ± 2.6 h and 45.1 ± 1.2 h, respectively, and were not significantly different between huHpS and huHB4, but were different between huHpS and huHB4G (P = 0.022). To evaluate the efficacy of the shRNA, HBsAg was quantified in the culture medium. Its concentrations were 0.44 ± 0.046 IU/mL, 0.48 ± 0.091 IU/mL and 0.010 ± 0.0016 IU/mL on the second day and 1.16 ± 0.11 IU/mL 1.86 ± 0.26 IU/mL and 0.050 ± 0.036 IU/mL on the fourth day, respectively (Figure 1B). On both days 2 and 4, HBsAg was significantly reduced in huHB4 compared with the other two clones, but was increased in huHB4G compared to huHpS (P = 0.0001 and P < 0.0001, respectively).
Next, the plasmids were delivered to hepatocytes in HBx transgenic mice using hydrodynamic gene delivery. After 48 h, the mouse livers were harvested and an immunohistochemical analysis was performed for HBx protein. As shown in Figure 1C, the positive signals were remarkably reduced in mice that received pSUPER.HB4 compared with other mice and were rather overrepresented in pSUPER.HB4G compared to pSUPER.retro.
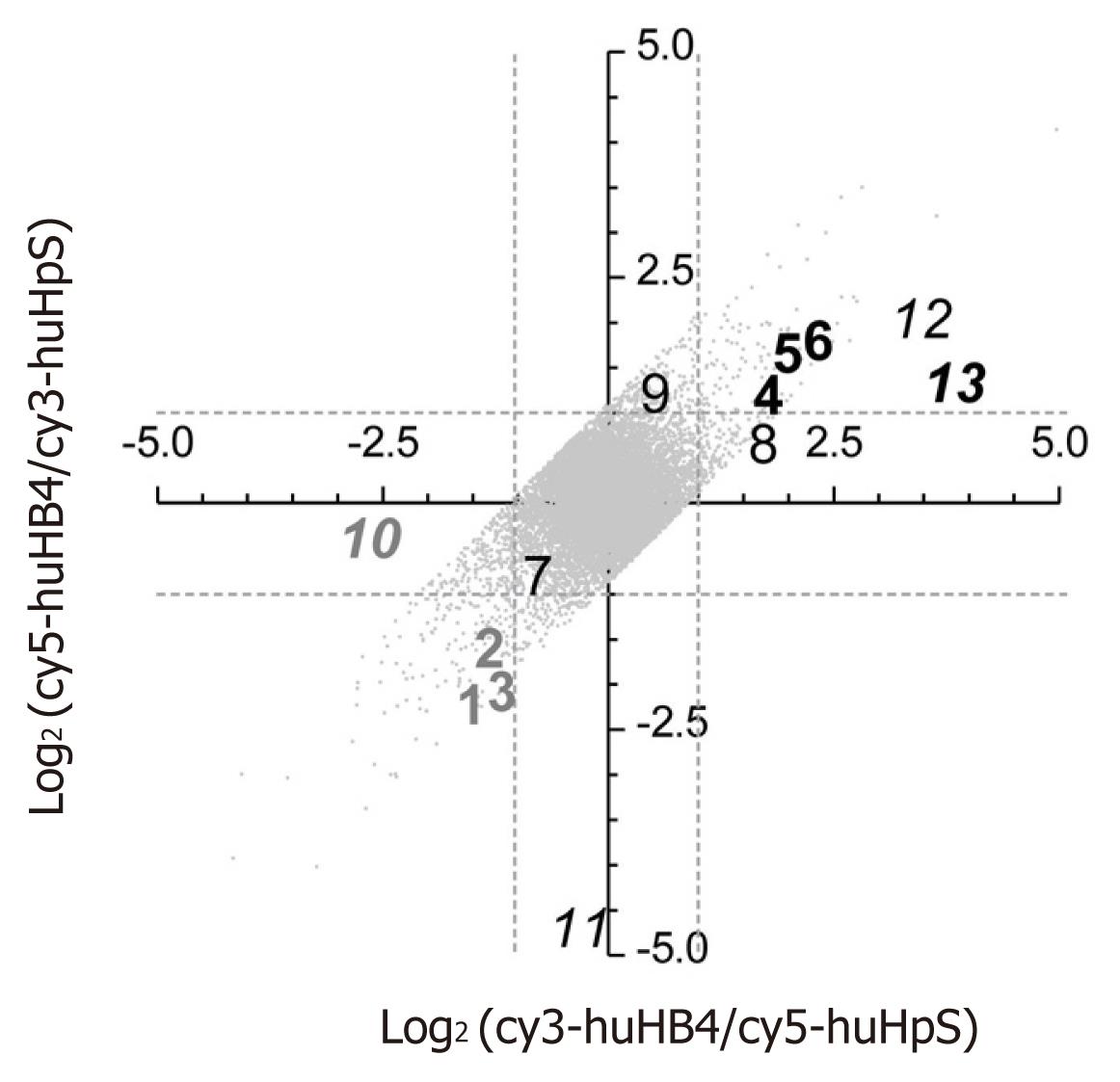
Because pSUPER.HB4G is suggested to have the potential enhancing HBV expression, gene expression signatures were compared between huHB4 and huHpS using a human cDNA microarray. Of 29 953 effective spots, LOWESS normalization validated 19 923 and 20 926 signals in each dye-sample combination, leading to final validation of 18 288 spots for both dye combinations. Additionally, 5276 signals were eliminated based on a flip-dye inconsistency greater than twofold, resulting in 13 012 genes available for further evaluation (Figure 2). Among those, 145 and 103 genes were down- and up-regulated, respectively, by at least twofold in huHB4 cells compared to huHpS cells (Supplementary Table 1, Supplementary material online). In order to exclude genes that might be repressed by off-targeting effects, the 13 012 genes were evaluated using a siRNA seed locator (http://www.dharmacon.com/seedlocator/default.aspx). A total of 1063 genes were found to have at least one match with our shRNA target sequence and ten genes were included in the 145 repressed genes. Thus, 135 repressed and 103 enhanced genes were selected as final candidates.
| No. | Genes | Microarray3 | qRT-PCR4 | Accession number5 | ||
| cy3/cy5 | cy5/cy3 | /β2M | /HPRT1 | |||
| 1 | CSTA1 | 0.35 | 0.26 | 0.23 | 0.23 | NM_005213 |
| 2 | APM21 | 0.46 | 0.31 | 0.22 | 0.23 | NM_006829 |
| 3 | SLPI1 | 0.47 | 0.31 | 0.08 | 0.09 | NM_003064 |
| 4 | CTGF1 | 3.36 | 2.026 | 3.49 | 3.93 | NM_001901 |
| 5 | NADE1 | 3.89 | 2.85 | 3.99 | 4.48 | AF187064 |
| 6 | TTR1 | 5.13 | 3.07 | 4.6 | 5.3 | NM_000371 |
| 7 | ARL3 | 0.59 | 0.52 | 0.54 | 0.62 | NM_004311 |
| 8 | GCNT3 | 3.2 | 1.97 | 1.74 | 1.95 | NM_004751 |
| 9 | NRF-1 | 1.42 | 2.16 | 0.98 | 1.12 | L22454 |
| 10 | KIAA180812 | 0.16 | 0.85 | 0.34 | 0.4 | AB058711 |
| 11 | HSPC1592 | 0.9 | 0.04 | 0.66 | 0.77 | NM_014181 |
| 12 | MAP2K62 | 11.08 | 3.76 | 1.68 | 1.89 | NM_002758 |
| 13 | SKAP212 | 13.45 | 2.33 | 2.03 | 2.34 | NM_003930 |
To confirm the microarray results, thirteen genes were randomly selected from the first and third quadrants of the log2(cy3-huHB4/cy5-huHpS)-log2(cy5-huHB4/cy3-huHpS) plot for the 18 288 LOWESS-validated genes and subjected to quantified by real-time reverse transcription-polymerase chain reaction (qRT-PCR) (Table 1). Of the thirteen genes, nine genes were chosen from the 13 012 flip-consistent genes, whereas the other four genes were selected from the 5276 flip-inconsistent genes. The flip-consistent genes were further divided into three groups of three genes each according to their intensity ratio of huHB4/huHpS: (1) less than 0.5 in both dye combinations; (2) more than 2 in both dye combinations; or (3) between 0.5 and 2 in at least one combination. In all flip-consistent genes, the relative quantities were not remarkably different between the two references of HPRT1 and β2M, and gene expression patterns were quite similar between microarray and qRT-PCR analyses. On the other hand, the four flip-inconsistent genes showed various expression patterns in qRT-PCR analysis. Two genes were found to express less or more than twofold in huHB4, whereas the expression differences were approximately twofold for the other two genes. These results suggest that the final 238 candidate genes are highly likely to have altered gene expression of a more than twofold magnitude in both microarray and qRT-PCR analyses.
To mine functional annotation, a list of the candidate genes was uploaded to DAVID program. Of 238 genes, 62 up- and 85 down-regulated genes were annotated and identified as being enriched into four and five representative functional categories, respectively, based on the controlled vocabulary of the Gene Ontology Consortium, as shown in Table 2. The up-regulated genes were classified into groups of lipid synthesis, glycoprotein, biopolymer metabolism and hydrolase activity. Genes that function as signal peptides, protease inhibitors and cytokines and in sensory perception and transport were significantly enriched in the down-regulated genes.
| Category | No. of genes | P value1 | Genes | Accession number2 |
| Up regulated | ||||
| Lipid synthesis | 3 | 0.01 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | NM_000859 |
| 7-dehydrocholesterol reductase | NM_001360 | |||
| Growth differentiation factor 1 | M62302 | |||
| Glycoprotein | 15 | 0.019 | Integrin beta 3 | NM_000212 |
| Interleukin 1 alpha | NM_000575 | |||
| Vitronectin | NM_000638 | |||
| Biopolymer metabolism | 16 | 0.02 | Cell division cycle 7 | NM_003503 |
| Colony stimulating factor 1 receptor | NM_005211 | |||
| Connective tissue growth factor | NM_001901 | |||
| Hydrolase activity | 6 | 0.046 | Bile acid coenzyme A: amino acid n-acyltransferase | NM_001701 |
| Inositol polyphoshpate-5-phosphatase | AF184215 | |||
| Poly(A)-specific ribonuclease | AJ005698 | |||
| Down regulated | ||||
| Signal | 17 | 0.001 | Cytotoxic T-lymphocyte-associated protein 4 | NM_005214 |
| Epithelial cell adhesion molecule | NM_002354 | |||
| Gamma-aminobutyric acid A receptor alpha 4 | NM_000809 | |||
| Protease Inhibitor | 3 | 0.024 | Cystatin A | BC010379 |
| Secretory leukocyte peptidase inhibitor | NM_003064 | |||
| Tissue factor pathway inhibitor | AF021834 | |||
| Sensory perception | 3 | 0.024 | Collagen type I alpha 2 | NM_000089 |
| GATA binding protein 3 | BC006839 | |||
| USHER syndrome 1C | NM_005709 | |||
| Transport | 7 | 0.031 | Amyloid beta precursor protein binding family A member 2 | BC007794 |
| ATP-binding cassette subfamily D member 4 | NM_020323 | |||
| Solute carrier family 17 member 2 | AL138726 | |||
| Cytokine activity | 4 | 0.046 | Chemokine ligand 25 | AB046579 |
| Kit ligand | NM_000899 | |||
| Prolactin | NM_000948 |
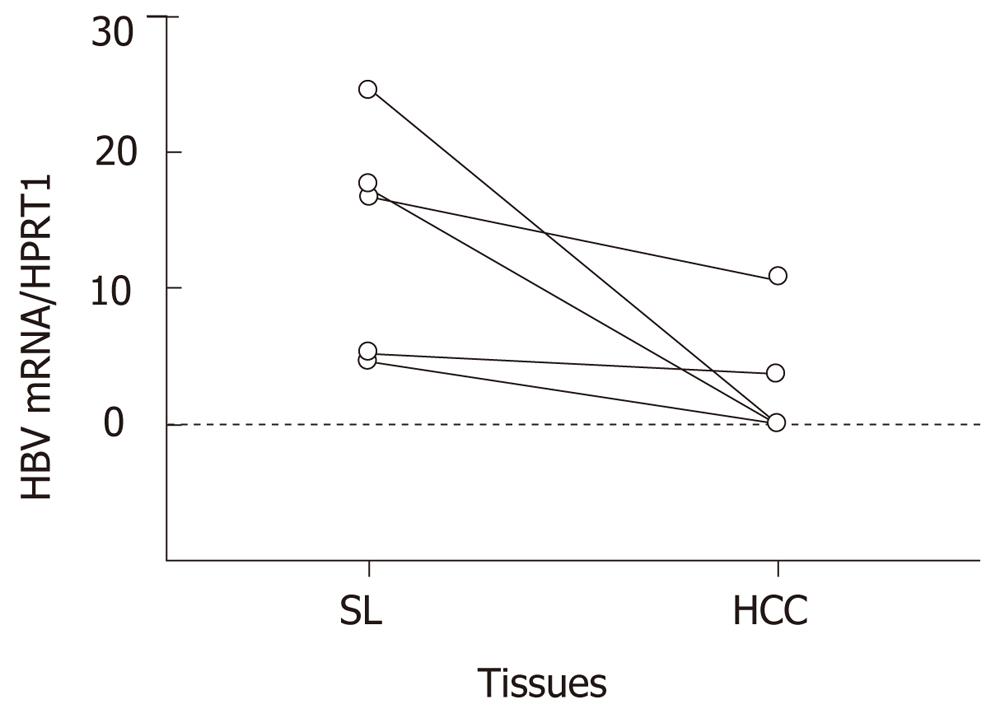
The differential expression in microarray analysis was validated by qRT-PCR using HCC and SL tissues from eighteen livers. Expression levels of the eight genes, which are listed as genes 1 to 6, 10 and 13 in Table 1, were quantified. Four genes were suppressed and the other four genes were enhanced more than twofold in huHB4. In HBV-associated tissues, HBV expression was also quantified. As shown in Figure 3, the average relative HBV expressions were 2.9 ± 4.7 and 13.7 ± 8.6 in HCC(B) and SL(B), respectively, and were a significantly decreased in HCC(B) (P = 0.032).
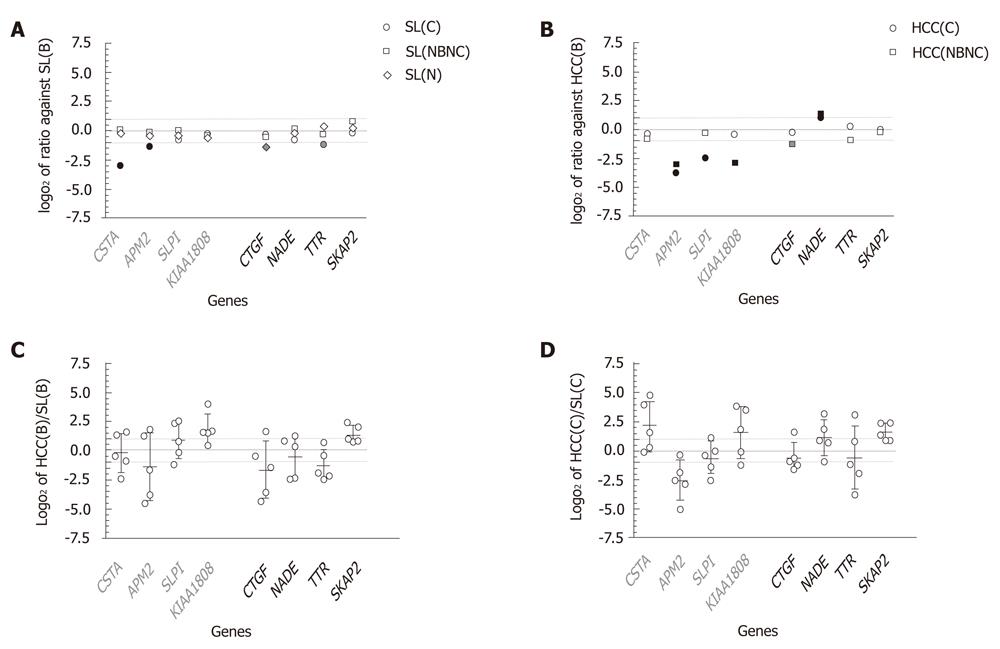
As shown in Figure 4A, all log2 ratios of SL(C), SL(NBNC) and SL(N) to SL(B) were distributed between -1 and 1, except for CSTA, APM2, CTGF and TTR, which showed expressions in SL(B) at magnitudes of 7.7-, 2.5-, 2.6- and 2.3-fold in comparison with SL(C), SL(C), SL(N) and SL(C), respectively. The reduced expression patterns in liver tissues without HBV infection are consistent with the results in huHB4 only for CSTA and APM2. In comparisons of HCC(C) and HCC(NBNC) with HCC(B), several genes showed expression differences of more than twofold that were consistent with differential expression in huHB4 (Figure 4B). Of the four genes that had low expression in huHB4, APM2 also had low expression in both HCC(C) and HCC(NBNC), with magnitudes of 0.074 and 0.11, respectively. SLPI and KIAA1808 expression were also decreased in HCC(C) and HCC(NBNC) to 0.17- and 0.13-fold, respectively. On the other hand, only NADE exhibited differential expression larger than twofold, a pattern consistent with that seen in huHB4, and it was expressed 2.0- and 2.4-fold higher in HCC(C) and HCC(NBNC), respectively.
On the other hand, the comparison of gene expression between HCC and SL from the same individual suffering from HBV infection showed wide variation among cases (Figure 4C). Only SKAP2 exhibited more than a twofold difference with relatively smaller variation and was expressed at 2.60 ± 1.07-fold higher in HCC(B). None of the eight candidates showed expression differences of more than twofold in all the five cases examined. Wide variation was also observed in HCV-associated cases, with a pattern similar to that seen in HBV-associated cases, as represented by SKAP2 (Figure 4D).
Consistent with previous reports[12,13], we found that HBV expression was significantly reduced in HCC(B) compared to the corresponding SL(B) in all of our cases. In order to mimic physiological repression of HBV expression through hepatocarcinogenesis, a sustained induction of RNAi against HBV was employed in this study. A single target of shRNA for a shared sequence in all four transcripts from HBV successfully achieved substantial reduction of HBsAg and HBx protein expressions in vitro and in vivo, respectively, without affecting cell morphology or replication rate.
RNAi is an evolutionarily conserved mechanism of post-transcriptional gene silencing induced by dsRNA and has been widely studied to prove its efficacy in reverse functional genomics and for therapeutic use[23,24]. Unfortunately, it is important to remember that the modulation of gene expression through RNAi can be extended to unintended genes due to various processes including off-target effects[25]. Off-target effects, which are RNAi-mediated events affecting the expression levels of dozens to hundreds of genes, are quite difficult to completely eliminate because off-targeting can be mediated by complementarity even between a hexamer seed region of dsRNA and the 3’ untranslated region of a target[26]. It has been reported that using different siRNA sequences that target HBV in HepG2.2.15 cells leads to different cellular gene expression profiles, suggesting a complicated influence of siRNA on cellular gene expression[18]. In this study, a seed locator program found at least one match in approximately 8% of 13 012 genes, even after the target sequence was carefully selected to make sure that no known cellular gene would show homology in its coding sequence with the target. Furthermore, pSUPER.HB4G unexpectedly up-regulated the expression of both HBsAg and HBx. It seems prudent to assume that cellular genes selected by knockdown experiments using RNAi against HBV involve genes directly affected by the RNAi process. In this regard, it is quite important to validate the results from in vitro experiments in liver tissues.
When human liver tissues were evaluated to find genes that are involved in hepatocarcinogenesis under HBV infection, gene expression profiles were generally compared between HCC(B) and SL(B). It is thought, however, that the transformation process itself requires a tremendous alteration of cellular gene expression, irrespective of etiology, that may outstrip the impact of HBV infection on the cellular gene expression. This concern seems to be relevant because qRT-PCR using liver tissues revealed that the differential expression patterns between HCC(B) and SL(B) were inconsistent with those between huHB4 and huHpS, but quite resembled the patterns between HCC(C) and SL(C). It would be ideal to evaluate the effect of HBV on cellular gene expression using non-tumorous liver tissues from the same individual at various time points with different levels of HBV expression, such as before and after natural seroconversion. In reality, however, it is quite difficult to collect liver specimens repeatedly in those settings.
To address this issue, HBV-associated liver tissues were compared with various liver tissues from the same individual and others. The major factors considered in comparisons among patients should be (1) with or without chronic liver diseases; (2) viral or non-viral liver diseases; and (3) cancerous or non-cancerous liver tissues. Thus, in the present study, gene expression levels in HCC(B) and SL(B) were compared with those in HCC(C), HCC(NBNC), SL(C), SL(NBNC) or SL(N). Because the presence or absence of viral infection presumably has a substantial impact on the cellular gene expression profile irrespective of virus species, the comparison between SL(B) and SL(NBNC) or between SL(B) and SL(N) does not reflect the specific influence of HBV infection, but of viral hepatitis in general. The cellular genes that are strictly regulated in connection with HBV infection should be differentially expressed, not only between HCC(B) and HCC(C), but also between SL(B) and SL(C). Among eight candidates, only APM2 fulfilled those criteria in this study. Thus, APM2, which is confirmed to be repressed after reduction of HBV expression in both RNAi knockdown experiments and liver tissues, is highly likely to be differentially expressed in association with HBV infection.
APM2 is a gene located on chromosome 10 at q23.2 and was originally identified as the second most abundant transcript in adipose tissue following adiponectin, APM1[27]. It is reported, however, that APM2 is expressed in a wide variety of tissues, including the liver[28], and is dysregulated in various cancers. Up-regulation of APM2 has been reported in pancreatic intraepithelial neoplasms[29], breast cancer tissues from patients with poor prognoses[30], and cisplatin-resistant gastric cancer cell lines[31]. Furthermore, overexpression of APM2 is reported to promote cisplatin resistance in a variety of cancer cell types[32]. Although the exact function of APM2 is currently unknown, these observations suggest that APM2 plays a role as an anti-apoptotic factor. HBV infection may induce sustained expression of APM2, leading to persistent viral infection and hepatocarcinogenesis. It would be interesting to determine whether HCC(B) is more resistant to cisplatin than HCC(C) and/or HCC(NBNC) because APM2 expression is substantially higher in HCC(B).
In the present study, 32 000 initial genes were evaluated, and of these genes, only one gene, APM2, was selected as a highly possible candidate gene that is differentially expressed due to HBV infection. Although we have so far evaluated only eight genes of 238 microarray candidates using human tissues, the concordance between the results from the cell lines and liver tissues is insufficient. It is true that the results obtained from the cell lines under shRNA overexpression technically involve many artificial limitations. However, another possible explanation for the low concordance is that cellular gene expression is regulated as a functional unit rather than by each gene. Functional annotations of genes selected in this study through microarray analysis indicated that the reduction of HBV expression led to disproportionately higher rates of increases or decreases of cellular gene expression in certain functional categories. The categories of genes with significantly enhanced or repressed expression after knockdown of HBV included lipid synthesis and protease inhibitors, respectively. In terms of lipid synthesis, for example, it has been reported that there is an inverse correlation between HBV and apolipoprotein expressions[33], and it is the largest functional category of lipid biosynthetic pathways to show differential expression between HBV transgenic and wild-type control mice[17]. Consistently, differential expression of cellular proteins were investigated in association with HBV infection and indicated that lots of proteins were up or down regulated as groups of several functional categories, including metabolisms[34-37]. Unfortunately, however, few reports applied the technologies of proteome or transcriptome analyses to human liver tissues instead of HBV-associated cell lines. To clarify the clinical significance of functional annotation, it is necessary to conduct proteome or transcriptome analyses on a large scale using liver tissues in various conditions.
In conclusion, it is suspected that the comparison between HCC(B) and SL(B) is not ideal for determining which genes are differentially expressed as a result of HBV infection. Because chronic viral hepatitis should have significant impacts on cellular gene expression, just as cellular transformation does, differential expression should be confirmed by comparing HBV-associated cases with HCV- or other viral-associated cases in terms of both HCC and SL. In the present study, the APM2 gene was selected from 32 000 human genes as the gene that was differentially expressed in those settings. In the future, differential cellular gene expression should be evaluated with respect to functional categories based on proteome and/or transcriptome analyses. The knowledge of cellular gene expression will help to elucidate the detailed mechanisms involved in chronic hepatitis B and HBV-associated hepatocarcinogenesis.
Supplementary Table 1 is available at Supplementary material online (http://www.wjgnet.com/1948-5182/g_info_20120428090827.htm).
Powerful preventive and therapeutic means have been developed for hepatitis B virus (HBV) infection, such as vaccination and nucleotide analogue, on the basis of our knowledge for HBV life cycle. Unfortunately, however, the World Health Organization reported that an estimated 350 million people worldwide are chronically infected with HBV and a significant proportion of chronic infections terminate in hepatocellular carcinoma (HCC), leading to more than half a million deaths annually. These dreadful situations request thorough understanding of the mechanisms for HBV to persuade sustained infection and hepatocarcinogenesis.
A recent technological advancement makes it possible to analyze an impact of a specific molecule such as HBV on the proteome and/or transcriptome in host cells. It is not difficult to assume that a deviation of gene expression profile plays a crucial role for immunological escape and cancer progression in HBV infection. The knowledge of HBV-host cell interaction must provide a powerful tool to fight HBV infection.
So far, an alteration of gene expression profile due to HBV infection has mainly been evaluated between non-infected parental cells and artificially-infected cells or between cells that were established from HBV positive case and HBV-knockdown cells. In terms of clinical materials, the comparisons between HBV-associated HCC and corresponding noncancerous liver tissues or between HBV-positive and HBV-negative cancer cells have been reported.
In the knockdown or artificial infection of HBV, it is quite difficult to completely eliminate the impact of knockdown/infection treatment itself on cellular gene expression. Furthermore, it is not difficult to assume that a malignant transformation process induces a tremendous change in cellular gene expression profile, which may overtake the effects of host-pathogen interactions. In order to avoid these confounding factors, the research team led by Takeshi Suda from Division of Gastroenterology and Hepatology, Niigata University took a two-step approach. Firstly, pick up candidate genes in a HBV-positive cell line by knocking HBV expression down, using small hairpin RNA, then confirm differential expression of the candidates in various liver tissues, including both cancer and noncancer associated with HBV, hepatitis C virus (HCV) or no HBV/HCV. Through this approach, the authors finally selected one gene, which differentially expressed in association with HBV infection, regardless of cancerous or noncancerous tissues.
The new insight of cellular reaction evoked by HBV infection must provide a clue to better understanding the mechanisms of sustained viral infection and cancer development, and will be implicated in the development of novel therapeutic options.
RNA interference is a process within living cells, in which double-stranded RNA directs a gene activity control with certain sequence specificity. shRNA is a small hairpin RNA, and short hairpin RNA is a sequence of RNA that makes a tight hairpin turn that can induce RNA interference. Off-targeting is a phenomenon where genes with incomplete complementarities with a target sequence are regulated in RNA interference. Dye flip is a strategy to account for dye bias in microarray experiments by labeling of DNA targets with the two dyes in reciprocal fashion. APM2 was discovered as the second most abundant transcript in adipose tissue following adiponectin and has been reported to be expressed in a variety of other tissues, including liver. APM2 gene overexpression is associated with malignant transformation and resistant to a chemotherapeutic agent have been reported.
The authors provided evidence indicating that silencing HBV mRNA altered cellular gene expressions in several functional categories including lipid synthesis. By stably targeting the common ORF region of the key HBV protein genes with shRNA, they observed a substantial decrease of HBsAg expression and up- or down-regulation of cellular genes, as revealed by DNA microarray. To verify in vitro assay results, they extended the observations to human liver tissues. The conclusions provide new insights into the mechanisms of HBV- or HCV-induced chronic liver diseases and hepatocarcinogenesis, and would be very useful for identification of novel drug targets against HBV infection.
Peer reviewers: Fei Chen, Associate Professor, Wayne State University, 259 Mack Avenue, Detroit, MI 48201, United States; George G Chen, Professor, Department of Surgery, The Chinese University of Hong Kong, Prince of Wales Hospital, CUHK, Shatin, NT, Hong Kong, China
S- Editor Wu X L- Editor Roemmele A E- Editor Wu X
| 1. | Shariff MI, Cox IJ, Gomaa AI, Khan SA, Gedroyc W, Taylor-Robinson SD. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev Gastroenterol Hepatol. 2009;3:353-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 226] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 2. | Daniels D, Grytdal S, Wasley A. Surveillance for acute viral hepatitis - United States, 2007. MMWR Surveill Summ. 2009;58:1-27. [PubMed] |
| 3. | Papatheodoridis GV, Manolakopoulos S, Dusheiko G, Archimandritis AJ. Therapeutic strategies in the management of patients with chronic hepatitis B virus infection. Lancet Infect Dis. 2008;8:167-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 157] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 4. | Zanetti AR, Van Damme P, Shouval D. The global impact of vaccination against hepatitis B: a historical overview. Vaccine. 2008;26:6266-6273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 5. | Fontana RJ. Side effects of long-term oral antiviral therapy for hepatitis B. Hepatology. 2009;49:S185-S195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 217] [Article Influence: 13.6] [Reference Citation Analysis (1)] |
| 6. | Dienstag JL. Benefits and risks of nucleoside analog therapy for hepatitis B. Hepatology. 2009;49:S112-S121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 89] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 7. | Tacke F, Amini-Bavil-Olyaee S, Heim A, Luedde T, Manns MP, Trautwein C. Acute hepatitis B virus infection by genotype F despite successful vaccination in an immune-competent German patient. J Clin Virol. 2007;38:353-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 8. | Kim W, Oe Lim S, Kim JS, Ryu YH, Byeon JY, Kim HJ, Kim YI, Heo JS, Park YM, Jung G. Comparison of proteome between hepatitis B virus- and hepatitis C virus-associated hepatocellular carcinoma. Clin Cancer Res. 2003;9:5493-5500. [PubMed] |
| 9. | Kim MY, Park E, Park JH, Park DH, Moon WS, Cho BH, Shin HS, Kim DG. Expression profile of nine novel genes differentially expressed in hepatitis B virus-associated hepatocellular carcinomas. Oncogene. 2001;20:4568-4575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 10. | Iizuka N, Oka M, Yamada-Okabe H, Mori N, Tamesa T, Okada T, Takemoto N, Hashimoto K, Tangoku A, Hamada K. Differential gene expression in distinct virologic types of hepatocellular carcinoma: association with liver cirrhosis. Oncogene. 2003;22:3007-3014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 61] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 11. | Chan DW, Ng IO. Knock-down of hepatitis B virus X protein reduces the tumorigenicity of hepatocellular carcinoma cells. J Pathol. 2006;208:372-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 12. | Raimondo G, Burk RD, Lieberman HM, Muschel J, Hadziyannis SJ, Will H, Kew MC, Dusheiko GM, Shafritz DA. Interrupted replication of hepatitis B virus in liver tissue of HBsAg carriers with hepatocellular carcinoma. Virology. 1988;166:103-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 54] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 13. | Wang Y, Wu MC, Sham JS, Tai LS, Fang Y, Wu WQ, Xie D, Guan XY. Different expression of hepatitis B surface antigen between hepatocellular carcinoma and its surrounding liver tissue, studied using a tissue microarray. J Pathol. 2002;197:610-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 14. | Otsuka M, Aizaki H, Kato N, Suzuki T, Miyamura T, Omata M, Seki N. Differential cellular gene expression induced by hepatitis B and C viruses. Biochem Biophys Res Commun. 2003;300:443-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Giladi H, Ketzinel-Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther. 2003;8:769-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 236] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 16. | Hu Z, Zhang Z, Kim JW, Huang Y, Liang TJ. Altered proteolysis and global gene expression in hepatitis B virus X transgenic mouse liver. J Virol. 2006;80:1405-1413. [PubMed] |
| 17. | Hajjou M, Norel R, Carver R, Marion P, Cullen J, Rogler LE, Rogler CE. cDNA microarray analysis of HBV transgenic mouse liver identifies genes in lipid biosynthetic and growth control pathways affected by HBV. J Med Virol. 2005;77:57-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 64] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 18. | Guo Y, Guo H, Zhang L, Xie H, Zhao X, Wang F, Li Z, Wang Y, Ma S, Tao J. Genomic analysis of anti-hepatitis B virus (HBV) activity by small interfering RNA and lamivudine in stable HBV-producing cells. J Virol. 2005;79:14392-14403. [PubMed] |
| 19. | Suda T, Gao X, Stolz DB, Liu D. Structural impact of hydrodynamic injection on mouse liver. Gene Ther. 2007;14:129-137. [PubMed] |
| 20. | Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 880] [Cited by in RCA: 868] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 21. | Igarashi M, Suda T, Hara H, Takimoto M, Nomoto M, Takahashi T, Okoshi S, Kawai H, Mita Y, Waguri N. Interferon can block telomere erosion and in rare cases result in hepatocellular carcinoma development with telomeric repeat binding factor 1 overexpression in chronic hepatitis C. Clin Cancer Res. 2003;9:5264-5270. [PubMed] |
| 22. | Ueda H, Ullrich SJ, Gangemi JD, Kappel CA, Ngo L, Feitelson MA, Jay G. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat Genet. 1995;9:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 255] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 23. | McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, Wieland SF, Marion PL, Kay MA. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 458] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 24. | Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, Chen J, Shankar P, Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 839] [Cited by in RCA: 814] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 25. | Whither RNAi? Nat Cell Biol. 2003;5:489-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 148] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 26. | Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 622] [Cited by in RCA: 649] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 27. | Maeda K, Okubo K, Shimomura I, Funahashi T, Matsuzawa Y, Matsubara K. cDNA cloning and expression of a novel adipose specific collagen-like factor, apM1 (AdiPose Most abundant Gene transcript 1). Biochem Biophys Res Commun. 1996;221:286-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1483] [Cited by in RCA: 1444] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 28. | Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 710] [Cited by in RCA: 816] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 29. | Prasad NB, Biankin AV, Fukushima N, Maitra A, Dhara S, Elkahloun AG, Hruban RH, Goggins M, Leach SD. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells. Cancer Res. 2005;65:1619-1626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 178] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 30. | Onda M, Emi M, Nagai H, Nagahata T, Tsumagari K, Fujimoto T, Akiyama F, Sakamoto G, Makita M, Kasumi F. Gene expression patterns as marker for 5-year postoperative prognosis of primary breast cancers. J Cancer Res Clin Oncol. 2004;130:537-545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 31. | Kang HC, Kim IJ, Park JH, Shin Y, Ku JL, Jung MS, Yoo BC, Kim HK, Park JG. Identification of genes with differential expression in acquired drug-resistant gastric cancer cells using high-density oligonucleotide microarrays. Clin Cancer Res. 2004;10:272-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 148] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 32. | Scott BJ, Qutob S, Liu QY, Ng CE. APM2 is a novel mediator of cisplatin resistance in a variety of cancer cell types regardless of p53 or MMR status. Int J Cancer. 2009;125:1193-1204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 33. | Norton PA, Gong Q, Mehta AS, Lu X, Block TM. Hepatitis B virus-mediated changes of apolipoprotein mRNA abundance in cultured hepatoma cells. J Virol. 2003;77:5503-5506. [PubMed] |
| 34. | Tong A, Wu L, Lin Q, Lau QC, Zhao X, Li J, Chen P, Chen L, Tang H, Huang C. Proteomic analysis of cellular protein alterations using a hepatitis B virus-producing cellular model. Proteomics. 2008;8:2012-2023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 35. | Niu D, Sui J, Zhang J, Feng H, Chen WN. iTRAQ-coupled 2-D LC-MS/MS analysis of protein profile associated with HBV-modulated DNA methylation. Proteomics. 2009;9:3856-3868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 36. | Narayan R, Gangadharan B, Hantz O, Antrobus R, García A, Dwek RA, Zitzmann N. Proteomic analysis of HepaRG cells: a novel cell line that supports hepatitis B virus infection. J Proteome Res. 2009;8:118-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 37. | Tong A, Gou L, Lau QC, Chen B, Zhao X, Li J, Tang H, Chen L, Tang M, Huang C. Proteomic profiling identifies aberrant epigenetic modifications induced by hepatitis B virus X protein. J Proteome Res. 2009;8:1037-1046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |