Published online Apr 27, 2025. doi: 10.4254/wjh.v17.i4.102027
Revised: February 21, 2025
Accepted: April 9, 2025
Published online: April 27, 2025
Processing time: 200 Days and 21.3 Hours
The Streptococcus salivarius (S. salivarius) group, which produces the enzyme urease has been identified as a potential contributor to ammonia production in the gut. Researchers have reported that patients with minimal HE had an increased abundance of the S. salivarius group, which is a specific change in the gut mi
To quantify S. salivarius using digital PCR (dPCR) as a liver fibrosis marker of CLD.
This study retrospectively analysed 52 patients with CLD. To quantify S. salivarius in patients with CLD using dPCR, we evaluated the specificity and sensitivity of S. salivarius bacterial load using dPCR for a type strain. Next, we evaluated the clinical usefulness of dPCR for S. salivarius load quantification for detecting liver fibrosis in patients with CLD. The liver fibrosis stage was categorized into mild and advanced fibrosis based on pathological findings.
The dPCR assay revealed that S. salivarius was highly positive for the tnpA gene. The lower limit of quantification for dPCR using the tnpA gene with a 1 μL template comprising 1.28 × 102 CFU/mL was 4.3 copies. After considering the detection range in dPCR, we adjusted the extracted DNA concentration to 5.0 × 10-4 ng/μL from 200 mg stool samples. The median bacterial loads of S. salivarius in stool sample from patients with mild and advanced fibrosis were 1.9 and 7.4 copies/μL, respectively. The quantification of S. salivarius load was observed more frequently in patients with advanced fibrosis than in those with mild fibrosis (P = 0.032).
Quantifying of S. salivarius load using digital PCR is a useful biomarker for liver fibrosis in patients with CLD.
Core Tip: This is a retrospective single-centre observational study and the first to quantify Streptococcus salivarius (S. salivarius) load in stool samples as a liver fibrosis marker for patients with chronic liver disease (CLD) Using digital PCR (dPCR). DPCR is a high precision technique for measuring the absolute quantity of the bacterial species. The quantification of S. salivarius using dPCR is proposed as a potentially useful biomarker for liver fibrosis in patients with CLD.
- Citation: Iwasaki S, Také A, Uojima H, Horio K, Sakaguchi Y, Gotoh K, Satoh T, Hidaka H, Tanaka Y, Hayashi S, Kusano C. Quantification of Streptococcus salivarius using the digital polymerase chain reaction as a liver fibrosis marker. World J Hepatol 2025; 17(4): 102027
- URL: https://www.wjgnet.com/1948-5182/full/v17/i4/102027.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i4.102027
The human microbiome comprises a vast community of microorganisms that inhabit various internal and external parts of the human body[1]. The gut microbiome, which is one of the most influential microbial communities in the human body, plays a crucial role in various physiological functions, including digestion, metabolism, immune system deve
Advancements in molecular biology have revealed that an imbalanced gut microbiome can contribute to chronic inflammation in the liver, leading to the development of conditions such as liver fibrosis[3-5]. Changes in the balance and diversity of the gut microbiota, known as dysbiosis, can cause an imbalance in microbial metabolites. Some bacteria produce metabolites and toxins that can be harmful to the host. The portal vein connects the gastrointestinal tract, including the intestines where the gut microbiota resides, to the liver. The influx of harmful substances from the gut into the liver through the portal vein promotes liver inflammation and the progression of chronic liver disease (CLD)[6,7].
Some researchers have identified the ammonia-contributing factors of gut microbiota dysbiosis in patients with CLD[8,9]. It is an important biological reaction commonly associated with certain bacteria, including those that inhabit the gastrointestinal tract. The ability of streptococci to produce urease and amplify its activity is a significant biological function with potential implications for the gut environment. The hydrolysis of urea by urease produces ammonia, which is basic, leading to an increase in local pH. Therefore, Streptococci with amplified urease activity could potentially contribute to alterations in gut pH. Changes in gut pH can impact the growth and survival of different bacterial species. Consequently, dysbiosis of the gut microbiota contributes significantly to liver inflammation, fibrosis, and other CLD-related complications[10]. The association between liver cirrhosis and an increased abundance of Streptococcus salivarius
Modulation of the gut microbiota, often referred to as microbiota-targeted therapy, holds promise for treating CLD. Interventions to modulate the composition of the gut microbiota, including the control of specific microbial strains such as S. salivarius, offer a novel strategy for treating CLD. However, metagenomic analysis, which involves sequencing the entire gut microbiota genetic material o, is expensive and resource-intensive. This process involves multiple steps including DNA extraction, library preparation, high-throughput sequencing, and extensive computational analyses. Therefore, concentrated efforts have been made to optimize and streamline metagenomic workflows[12,13]. Of these, molecular biology techniques such as digital PCR (dPCR) used for the quantification of specific microbes can reduce the cost and complexity associated with studying the gut microbiome[14-17]. However, the correlation between the aggregation of specific bacterial species of the genus Streptococcus and fibrosis progression in CLD is complex and yet to be fully elucidated using screening approaches such as dPCR. Therefore, the present study aimed to quantify S. salivarius as a marker of liver fibrosis in patients with CLD using dPCR.
This study protocol was approved by the Institutional Review Board Ethics Committees of Kitasato University School of Medicine (approval number: C21-212). Informed consent was obtained from all participants. This study was registered with the UMIN Clinical Trials Registry (UMIN 000048201).
The study retrospectively analysed 70 patients with CLD (diagnosed by liver biopsy) based on faecal samples collected in a prospective cohort study at the Kitasato University School of Medicine between January 2019 and August 2023. Of these, 18 were excluded from the study because of a history of malignancies other than hepatocellular carcinoma (HCC) or antibiotic use within three months. Patients with CLD cases include those with metabolic dysfunction-associated fatty liver disease (MAFLD) and non-MAFLD. The definition of MAFLD is provided in Supplementary material[18].
To quantify S. salivarius as a marker of liver fibrosis in patients with CLD using dPCR, we evaluated the analytical specificity, sensitivity, and repeatability of the dPCR assay in quantifying the bacterial load of S. salivarius for the type strain obtained from the Japan Collection of Microorganisms (JCM). Analytical specificity was evaluated using primers and probes specific for S. salivarius among 16 common microorganisms (Supplementary Table 1). Sensitivity was evaluated using a limit of quantification (LOQ). Repeatability was evaluated using intra-assay coefficients of variability (CV), which were used to express the relative variability of a set of data points. Furthermore, quantification of the S. salivarius bacterial load using dPCR was validated or cross-verified using quantitative PCR (qPCR) with the same set of primers and probes.
Next, we evaluated the clinical usefulness of t S. salivarius load quantification using dPCR for detecting liver fibrosis in patients with CLD. The pathological assessment of liver fibrosis was conducted by a pathologist at our institution to ensure the accuracy and reliability of fibrosis staging. The fibrosis stages were classified based on steatosis, activity, and fibrosis scoring[19]. Furthermore, these stages were categorised into mild fibrosis or advanced fibrosis groups, depending on the presence or absence of bridging fibrosis, respectively, based on pathological findings. We evaluated the clinical factors influencing of S. salivarius load quantification in patients with CLD, except at the fibrosis stage.
Faecal samples were collected from the enrolled patients using collection kits. To preserve the microbial community and prevent degradation, the samples were storedat 4 °C (refrigeration) and -80 °C (freezing) for short- and long-term storage, respectively.
Microbial DNA was extracted using the DNeasy PowerSoil Kit (QIAGEN). This kit was designed to efficiently extracts DNA from environmental samples, including soil and faecal samples.
The selected strains of the genus Streptococcus were inoculated into Todd-Hewitt broth (Difco, Becton, Dickinson and Company, MD, United States) with 0.3% yeast extract (THY) medium, and the others were inoculated in brain heart infusion (BHI, Difco) medium (Supplementary Table 1). Bifidobacterium bifidum JCM 1255T, Bacteroides uniformis JCM 5828T, Enterococcus faecalis JCM 5803T, Clostridium ramosum JCM 1298T, Eggerthella lenta JCM 9979T, and Blautia coccoides JCM 1395T were cultured overnight at 35 °C under anaerobic conditions, and other bacteria were cultured overnight at 37 °C under aerobic conditions. The cultures were collected and centrifuged, and bacterial DNAs samples were extracted from the pellet using a NucleoSpin® Microbial DNA Kit (TaKaRa Bio).
The primers and probes were designed to target conserved regions within the 16S rRNA for Streptococcus spp. (Stc16S primer set) and tnpA gene specific to S. salivarius (Sal-tnpA primer set)[20-22].
dPCR was performed using a QX200 Droplet Digital PCR system (Bio-Rad, Hercules, CA, United States). The mastermix used in the dPCR assay comprised 1 × dPCR multiplex Supermix for Probes (Bio-Rad), 900 nM target primers, and 250 nM target probes with sample DNA. Subsequent amplification was performed in a Veriti 96-well thermal cycler (Applied Biosystems) under the following PCR conditions: 95 °C for 10 minutes enzyme activation followed by 45 cycles of denaturation at 94 °C for 30 seconds, 59 °C for 2 minutes and enzyme deactivation at 98 °C for 10 minutes[23].
qPCR assays for the target S. salivarius were conducted using LightCycler® 96 (Roche). The total reaction volume for qPCRs was 20 μL, which included each template DNA, 500 nM target primers, 200 nM target probes, 10 μL of Probe FastStart Essential DNA ProbeMaster (Roche), and 9 μL of PCR grade water. The qPCR conditions were as follows: 95°C for 10 minutes, followed by 40 cycles at 95 °C for 30 seconds and at 60°C.
To validate qPCR and dPCR results, the S. salivarius strain was cultivated in Todd-Hewitt broth (Difco, Becton, Dickinson and Company, MD, United States) with 0.3% THY broth overnight at 37 °C. The cultured broth was dispensed into 0.1 mL portions for DNA extraction and colony counting. To extract DNA, the cultured broth was centrifuged for 1 min at 10000 × g, and washed with Tris-EDTA buffer TE (pH = 8.0). The cells were resuspended in TE buffer and heated at 95 °C for 10 minutes. The suspensions were centrifuged, and the supernatants containing the DNA templates were collected. To calculate the colony forming units (CFUs) of the DNA template, 0.1 mL of the cultured broth was diluted with PBS and plated on BHI (Difco) agar medium. The CFU of the DNA template was calculated from the number of colonies grown on the plate.
IBM SPSS Statistics software (version 23.0 New York, NY, United States) was used for statistical analyses. P ≤ 0.05 indicated statistical significance. Continuous variables were expressed as the median and interquartile range in statistical reporting. The test statistic of χ2 was calculated based on the observed frequencies in the contingency table. The Mann-Whitney U test was used to compare distributions of dPCR quantification between the presence of advanced fibrosis in load.
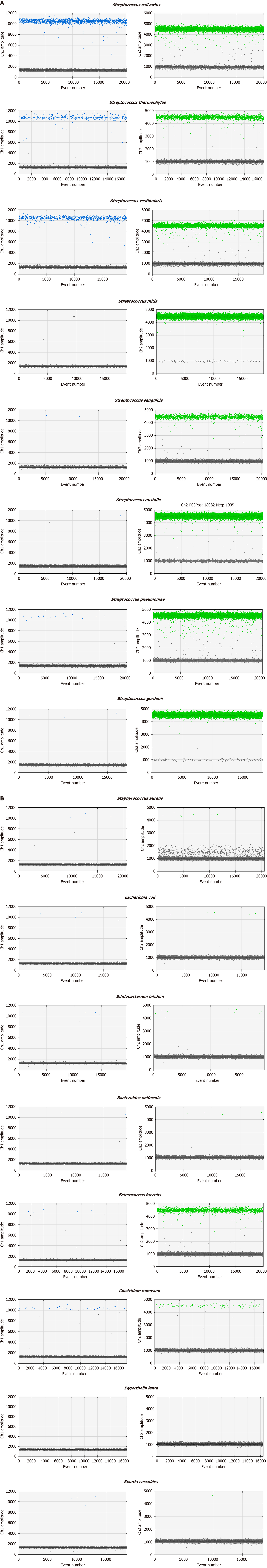
Figure 1 shows the positive droplets of 16 common microorganisms using the primer sets of Stc16S and Sal-tnpA with channel amplitude signals. The dPCR assay revealed that the S. salivarius group, including S. salivarius, Streptococcus thermophilus (S. thermophilus), and Streptococcus vestibularis (S. vestibularis) type strains, were highly positive for the Sal-tnpA primer set. Streptococcus spp. and Enterococcus faecalis (E. faecalis) were positive for the Stc16S primer set. Clostridium ramosum was mildly positive for the primer sets of Stc16S and Sal-tnpA. dPCR using the Sal-tnpA primer set showed excellent specificity, with no cross-reactivity to 13 different microorganisms.
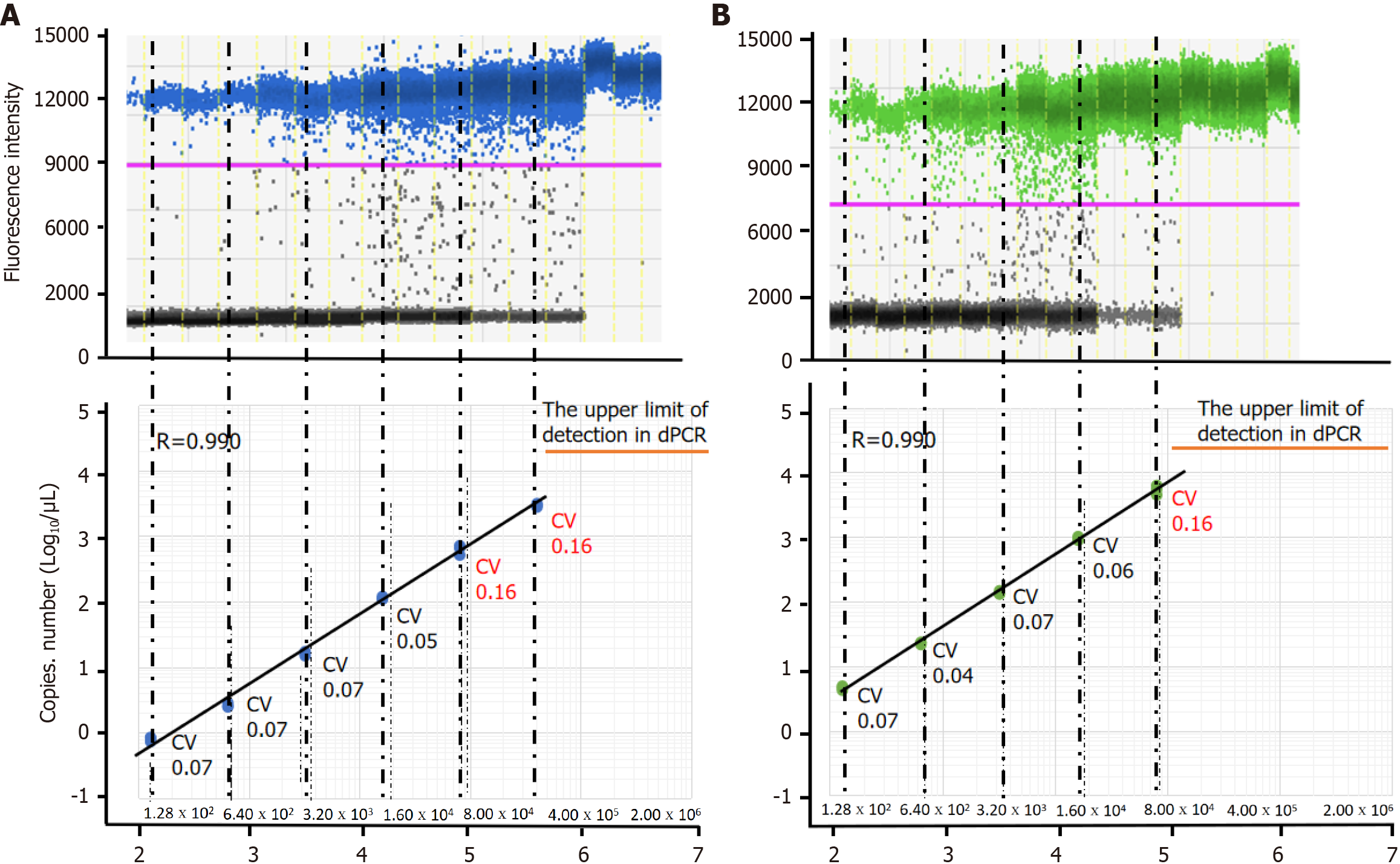
Figure 2 shows a comparison of bacterial load measured using dPCR (copy number/μL) and bacterial CFU measured using the dilution plate method (CFU/mL). The dPCR assay targeting both the tnpA and 16S rRNA genes reflected the change in DNA concentration. A 5-fold dilution of DNA resulted in a 5-fold decrease in the final readouts indicating unbiased detection of S. salivarius (Figure 2, Supplementary Figure 1).
The lower limit of detections with dPCR with the 1 μL template comprising 1.28 × 102 CFU/mL were 6 copies of the 16S rRNA and 4.3 copies of the tnpA gene. However, analysis using substantialdroplets was complex. The upper LOQ for the dPCR assay was about 20000 copies/mL. Beyond this limit, the assay may become less reliable, and the risk of false positives due to overlapping signals or background noise may increase.
The intra-assay CV values of the bacterial load for S. salivarius using the Sal-tnpA primer set in each 5-fold dilution of input microorganisms, were 0.07, 0.07, 0.07, 0.05, 0.16, and 0.16, respectively. The intra-assay CV values of the bacterial load for S. salivarius using the Stc16S primer set in each 5-fold dilution of input microorganisms were 0.07, 0.04, 0.07, 0.06, and 0.16, respectively. The dPCR assay for two different target genes exhibited excellent intra-assay coefficients of CV. A high intra-assay CV in dPCR was observed at template DNA concentration of 8.00 × 104 and 4.00 × 105 CFU/mL.
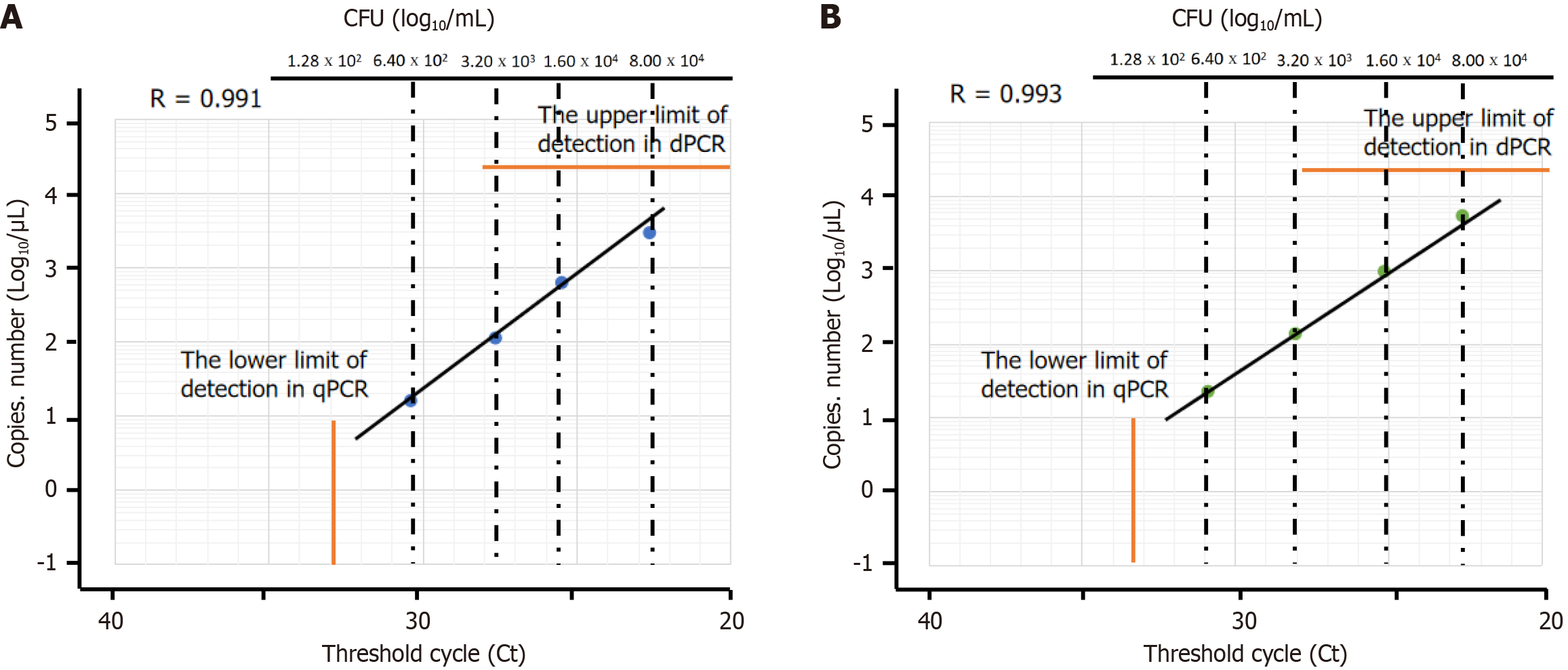
Figure 3 shows a comparison of bacterial load measured using dPCR (copy number/μL) and qPCR (Ct value). A statistically significant correlation was observed between the results obtained using dPCR and qPCR assays targeting the tnpA and 16S rRNA genes.
The lower LOQs for qPCR using the1 μL template 6.4 × 102 CFU/mL were 31 Ct of the 16S rRNA and 33 Ct of the tnpA gene. dPCR was more sensitive than qPCR in detecting low concentrations of bacteria.
After considering the dPCR range of detection, we adjusted the extracted DNA concentration to 5 × 10-4 ng/μL from 200 mg stool samples (Supplementary Figure 2). The median bacterial load for S. salivarius in stool samples based on the primer sets of Sal-tnpA and Stc16S were 2.5 (0.5-160) and 32.8 (0.9-1257) copies/mL, respectively. The correlation coefficient of the detected bacterial load of S. salivarius using the primer sets of Sal-tnpA and streptococci using the primer sets of Stc16S was 0.944. These measurements showed strong correlations for all ranges.
When a threshold of > 1 copy/μL load for S. salivarius in stool samples based on the dPCR using tnpA was defined as positive, 36 patients met this criterion. An association between a positive group based on the defined threshold using the primer sets of Sal-tnpA and a higher frequency of advanced fibrosis among the patients (P = 0.023). was observed. Furthermore, the age was higher in the positive group compared to the negative group (P = 0.006). The presence of HCC was higher in the positive group compared to the negative group (P = 0.017). The proportion of patients using gastric acid secretion inhibitors was higher in the positive group than in the negative group; however, this difference was not statistically significant (P = 0.146). No significant differences were observed between the two groups in terms of sex, weight, the frequency of MAFLD, hepatic encephalopathy, ascites, white blood cells, platelets count, aspartate aminotransferase, alanine aminotransferase, and total bilirubin (Table 1).
| Clinical characteristics | All patients | Positive (≤ 1 copy/ μL) | Negative (> 1 copy/ μL) | P value |
| Number of patients | 52 | 36 | 16 | |
| Age (year) | 65.0 (24-84) | 68.5 (24-84) | 55.0 (41-74) | 0.006 |
| Gender: Male [n (%)] | 31 (54.3) | 23 (63.8) | 8 (50.0) | 0.450 |
| Weight(kg) | 64 (41-121) | 64 (41-121) | 70 (47-92) | 0.234 |
| Body mass index (kg/m2) | 25.1 (16.1-46.1) | 24.9 (16.0-46.1) | 25.3 (16.1-32.1) | 0.706 |
| MAFLD/non-MAFLD (n) | 28/24 | 19/17 | 9/7 | 0.772 |
| Alcohol [n (%)] | 4 (7.7) | 2 (5.6) | 2 (12.5) | 0.578 |
| Hepatocellular carcinoma [n (%)] | 10 (19.2) | 10 (27.8) | 0 | 0.017 |
| Fibrosis stage 0/1/2/3/4 (n) | 4/20/9/13/6 | 1/12/7/11/5 | 3/8/2/2/1 | 0.023 |
| Mild/advance fibrosis (n) | 33/19 | 20/16 | 13/3 | |
| Hepatic encephalopathy [n (%)] | 2 (3.8) | 2 (5.6) | 0 | 0.587 |
| Hepatic ascites [n (%)] | 2 (3.8) | 2 (5.6) | 0 | 0.587 |
| Gastric acid secretion inhibitor [n (%)] | 13 (25.0) | 11 (30.6) | 2 (12.5) | 0.146 |
| White blood cells (/μL) | 5400 (2400-12200) | 5250 (3100-12200) | 6250 (2400-9300) | 0.201 |
| Platelets (× 104/μL) | 18.7 (6.3-47.0) | 17.3 (6.3-40.7) | 19.9 (10.6-30.0) | 0.204 |
| Prothrombin time (%) | 94 (40-107) | 99.0 (40-114) | 100 (61-107) | 0.571 |
| Serum albumin (g/dL) | 4.2 (2.5-5.0) | 4.2 (2.5-5.0) | 4.3 (3.4-4.8) | 0.068 |
| Aspartate aminotransferase (IU/L) | 36.5 (16-138) | 35.5 (16-138) | 37.5 (18-127) | 0.684 |
| Alanine aminotransferase (IU/L) | 46.0 (11-179) | 36.5 (11-179) | 48.5 (14-146) | 0.252 |
| Total bilirubin (mg/dL) | 0.7 (0.4-4.9) | 0.7 (0.4-4.9) | 0.8 (0.4-2.0) | 0.114 |
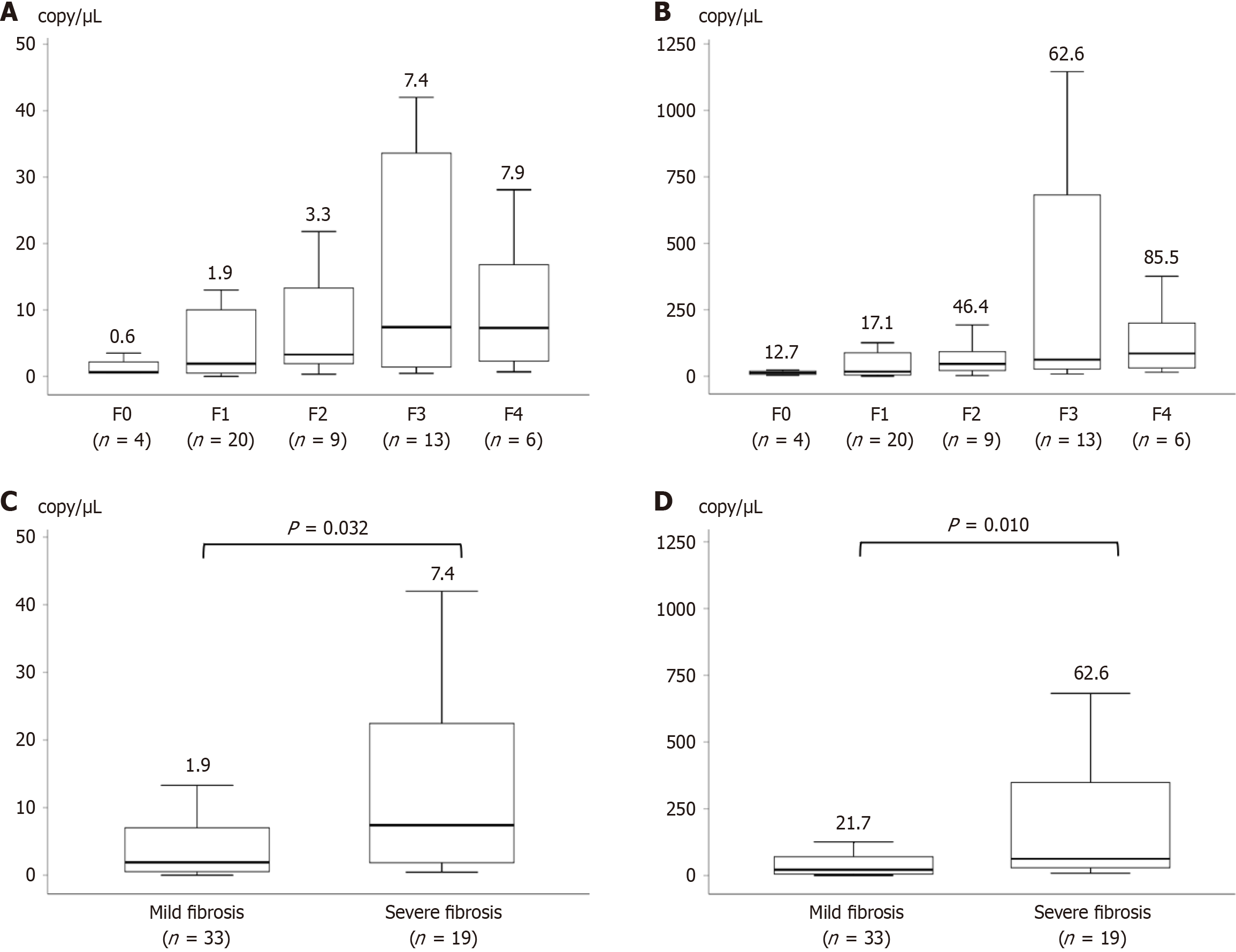
An association was observed between increasing S. salivarius load based on the dPCR and liver fibrosis progression in patients with CLD. The median bacterial loads of S. salivarius from stool sample using the primer sets of Sal-tnpA in patients with mild and severe fibrosis were 1.9 and 7.4 copies/μL, respectively. The median bacterial loads of streptococci using the primer sets of Stc16S in patients with mild and severe fibrosis were 21.7 and 62.6 copies/μL, respectively. The bacterial load of S. salivarius was observed more frequently in patients with advanced fibrosis than in those with mild fibrosis (Stc16S, P = 0.010 and Sal-tnpA, P = 0.032; Figure 4).
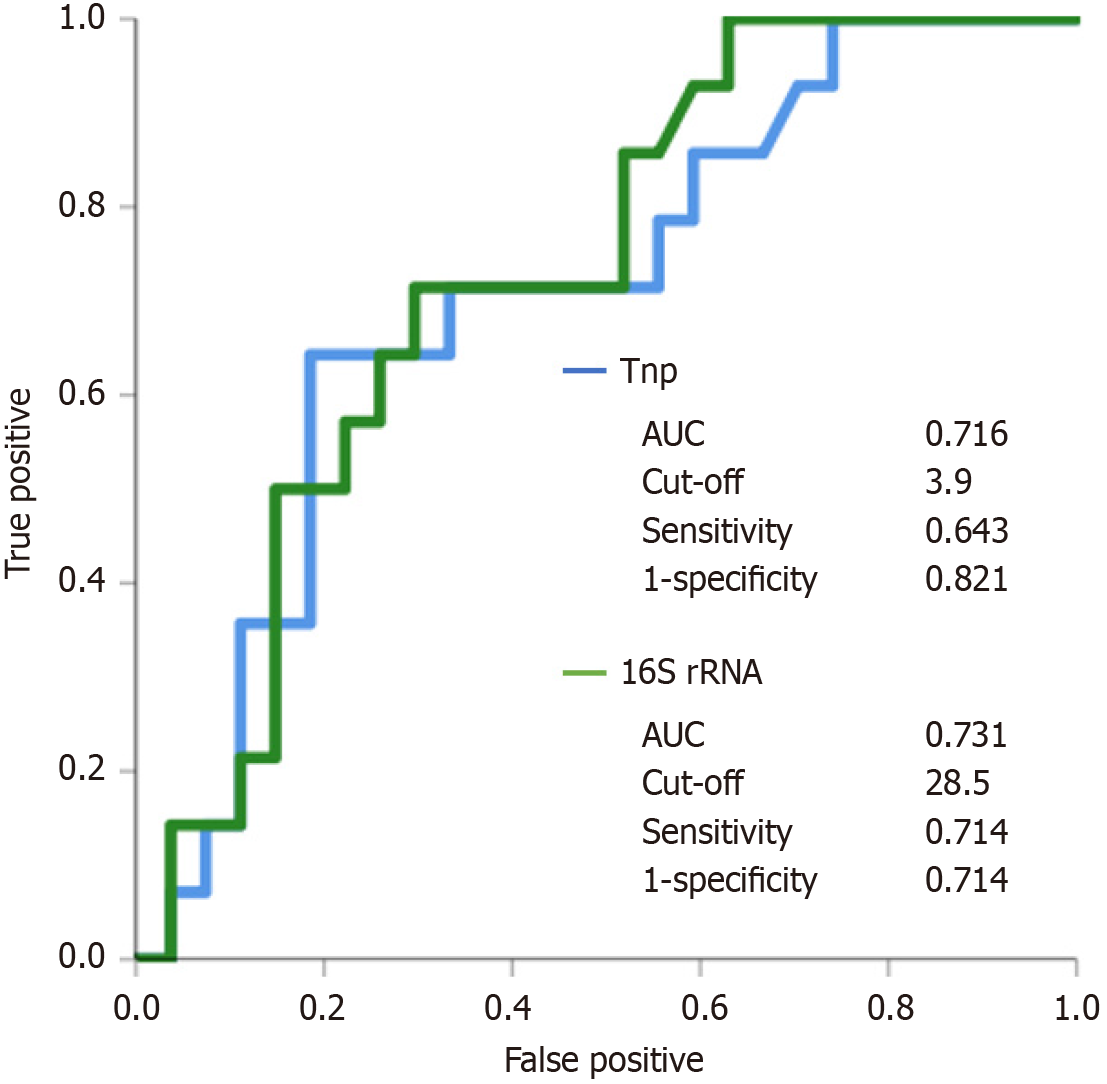
We performed a receiver operating characteristic (ROC) analysis to assess advanced fibrosis in patients with CLD. The cut-off points for advanced fibrosis were estimated using ROC curves of S. salivarius and streptococci bacterial loads using the primer sets Sal-tnpA and Stc16S, respectively, by identifying the threshold value that optimally differentiatedbetween patients with and without advanced fibrosis. Using a cut-off value of 3.9 for the primer sets of Sal-tnpA to, predict the presence of advanced liver fibrosis resulted ina sensitivity of 64.3% and a specificity of 82.1%. Using a cut-off value of 28.5 for the primer sets of Stc16S to, predict the presence of advanced fibrosis resulted in a sensitivity of 71.4% and a specificity of 71.4% (Figure 5).
This is the first report to quantify S. saribarius load in stool samples using dPCR as a liver fibrosis marker for patients with CLD. Certain Streptococcus species, such as S. salivarius, contribute to oral health by competing with pathogenic bacteria and producing antimicrobial substances[24,25]. However, imbalances in the gut microbial community, including an overgrowth of S. salivarius, can disrupt the normal functioning of the microbiota and contribute to CLD progression[26]. Monitoring changes in the S. salivarius in the gut could potentially serve as diagnostic or prognostic markers for liver disease progression. dPCR is an appropriate method for the detection and quantification of infectious disease research and clinical practice.
The dPCR method divides the adjusted sample into 20000 droplets. By dividing the sample into droplets, target DNA and background DNA are randomly distributed in each droplet, and the PCR is initiated in each droplet[27,28]. Counting the number of positive and negative droplets enables absolute quantification of the DNA target. To quantify of S. salivarius load in stool samples, the dPCR assay targeted both the tnpA and 16S rRNA genes, which were previously used to detect and quantify S. salivarius[11]. The present study showed that the dPCR assay targeting the tnpA was only positive for S. salivarius, S. vestibularis, and S. thermophilus (a group of bacteria classified into three genetically similar species). However, the dPCR assay targeting the 16S rRNA was positive for E. faecalis, in addition to the Streptococcus species. E. faecalis and various Streptococcus species are members of the phylum Firmicutes and the class Bacilli, and they are similar regardingtheir gram-positive cell wall structure and certain physiological characteristics[29]. The 16S rRNA gene is commonly used for bacterial identification and phylogenetic studies owing to its conserved nature across bacterial species. Furthermore, dPCR has high sensitivity and is appropriate for determining relative bacterial load[30]. The amplification of individual DNA molecules in separate droplets enhances the sensitivity of dPCR, facilitating the detection of low-abundance targets[31]. The lower LOQs for qPCR and dPCR with a 1 μL template were 6.40 × 102 and 1.28 × 102 CFU/mL, respectively. These results suggested that dPCR is more sensitive than qPCR in detecting low concentrations of bacteria. Contrarily, the upper LOQ for dPCR was observed. The intra-assay CV in dPCR was relatively high for template DNA concentrations over 8.00 × 104 CFU/mL. The risk of false positives may increase as the upper LOQ is exceeded owing to overlapping signals, background noise, or saturation effects in the detection system[32].
After the bacterial load of S. salivarius from the type strain was quantified using the dPCR, we quantified the S. salivarius in the stool using dPCR for patients with CLD. The bacterial load for S. salivarius was more frequently observed in patients with advanced fibrosis compared to those with mild fibrosis suggesting a potential association between the presence of S. salivarius and the progression of CLD. However, the copy numbers of any bacterial species in stool samples can be influenced by various factors related to the host environment at the time of sample collection[33]. The role of pH and age in influencing gut microbiota is notable[34]. The pH level within the gastrointestinal tract is crucial in shaping the gut microbiota. Age-related physiological changes or medication use, including gastric acid secretion inhibitors, can alter the pH environment within the GI tract, potentially leading to dysbiosis and bacterial overgrowth. Therefore, careful consideration and monitoring are essential when prescribing PPIs, particularly in older adults.
Our final objective is to develop microbiome-targeted interventions to mitigate fibrosis progression. By identifying bacteria associated with fibrosis severity, personalized therapeutic strategies can be tailored to individual microbial profiles and disease stages. For example, S. salivarius in oral saliva, which has been linked to fibrosis progression, may serve as a modifiable factor through oral microbiome-targeted interventions. Preliminary evidence suggests that reducing urease-positive S. salivarius via oral care could attenuate fibrosis progression[11]. If validated in clinical trials, this approach could provide a non-invasive, cost-effective strategy for fibrosis management. DPCR enables precise microbial quantification, reducing the cost and complexity of microbiome analysis[35]. Targeting bacteria or microbial pathways involved in fibrosis progression may facilitate the development of novel therapeutics, including probiotics, prebiotics, antimicrobial agents, and other microbiome-modulating interventions.
This study has certain limitations. First, prospective studies aimed at validating dPCR performance in patients with liver cirrhosis diagnosis remain scarce. Second, this was a single-centre prospective study to evaluate the correlation between dPCR and fibrosis progression in patients with CLD. Third, this study had a relatively small sample size and was conducted at a single centre. Therefore, more research is needed to further validate and generalise the findings.
In conclusion, dPCR is a high precision technique to-quantify bacterial species. The quantification of S. salivarius using digital PCR is a potentially useful biomarker for liver fibrosis in patients with CLD.
In conclusion, S. salivarius quantification using dPCR is a useful biomarker for liver fibrosis in patients with CLD.
We thank STATISTA Corporation for assistance with the statistical analyses in this study.
| 1. | Chen Y, Zhou J, Wang L. Role and Mechanism of Gut Microbiota in Human Disease. Front Cell Infect Microbiol. 2021;11:625913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 302] [Article Influence: 75.5] [Reference Citation Analysis (0)] |
| 2. | Zhang X, Li L, Butcher J, Stintzi A, Figeys D. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome. 2019;7:154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 188] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 3. | Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 674] [Cited by in RCA: 797] [Article Influence: 56.9] [Reference Citation Analysis (3)] |
| 4. | Acharya C, Bajaj JS. Altered Microbiome in Patients With Cirrhosis and Complications. Clin Gastroenterol Hepatol. 2019;17:307-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 108] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 5. | Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Chen Y, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1230] [Cited by in RCA: 1534] [Article Influence: 139.5] [Reference Citation Analysis (40)] |
| 6. | Bajaj JS, Khoruts A. Microbiota changes and intestinal microbiota transplantation in liver diseases and cirrhosis. J Hepatol. 2020;72:1003-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 129] [Article Influence: 25.8] [Reference Citation Analysis (1)] |
| 7. | Lee NY, Suk KT. The Role of the Gut Microbiome in Liver Cirrhosis Treatment. Int J Mol Sci. 2020;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 8. | Ikegami T, Honda A. Reciprocal interactions between bile acids and gut microbiota in human liver diseases. Hepatol Res. 2018;48:15-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 9. | Bajaj JS, Sikaroodi M, Shamsaddini A, Henseler Z, Santiago-Rodriguez T, Acharya C, Fagan A, Hylemon PB, Fuchs M, Gavis E, Ward T, Knights D, Gillevet PM. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut. 2021;70:1162-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 10. | Kaci G, Goudercourt D, Dennin V, Pot B, Doré J, Ehrlich SD, Renault P, Blottière HM, Daniel C, Delorme C. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl Environ Microbiol. 2014;80:928-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 148] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 11. | Také A, Uojima H, Sakaguchi Y, Gotoh K, Satoh T, Hidaka H, Horio K, Mizokami M, Hayashi S, Kusano C. Impact of liver fibrosis on the relative abundance of a urease-positive Streptococcus salivarius group from saliva in patients with chronic liver disease. Hepatol Res. 2023;53:998-1007. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 12. | Yoshiji H, Nagoshi S, Akahane T, Asaoka Y, Ueno Y, Ogawa K, Kawaguchi T, Kurosaki M, Sakaida I, Shimizu M, Taniai M, Terai S, Nishikawa H, Hiasa Y, Hidaka H, Miwa H, Chayama K, Enomoto N, Shimosegawa T, Takehara T, Koike K. Evidence-based clinical practice guidelines for liver cirrhosis 2020. Hepatol Res. 2021;51:725-749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 125] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 13. | Pombert JF, Sistek V, Boissinot M, Frenette M. Evolutionary relationships among salivarius streptococci as inferred from multilocus phylogenies based on 16S rRNA-encoding, recA, secA, and secY gene sequences. BMC Microbiol. 2009;9:232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 14. | Aralar A, Yuan Y, Chen K, Geng Y, Ortiz Velez D, Sinha M, Lawrence SM, Fraley SI. Improving Quantitative Power in Digital PCR through Digital High-Resolution Melting. J Clin Microbiol. 2020;58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 15. | O'Sullivan DM, Laver T, Temisak S, Redshaw N, Harris KA, Foy CA, Studholme DJ, Huggett JF. Assessing the accuracy of quantitative molecular microbial profiling. Int J Mol Sci. 2014;15:21476-21491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 16. | Willemse D, Kaushal D. Using genomic DNA copies to enumerate Mycobacterium tuberculosis load in macaque tissue samples. Tuberculosis (Edinb). 2021;129:102102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 17. | Shin J, Shina S, Jung SH, Park C, Cho SY, Lee DG, Chung YJ. Duplex dPCR System for Rapid Identification of Gram-Negative Pathogens in the Blood of Patients with Bloodstream Infection: A Culture-Independent Approach. J Microbiol Biotechnol. 2021;31:1481-1489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 18. | Eslam M, Newsome PN, Sarin SK, Anstee QM, Targher G, Romero-Gomez M, Zelber-Sagi S, Wai-Sun Wong V, Dufour JF, Schattenberg JM, Kawaguchi T, Arrese M, Valenti L, Shiha G, Tiribelli C, Yki-Järvinen H, Fan JG, Grønbæk H, Yilmaz Y, Cortez-Pinto H, Oliveira CP, Bedossa P, Adams LA, Zheng MH, Fouad Y, Chan WK, Mendez-Sanchez N, Ahn SH, Castera L, Bugianesi E, Ratziu V, George J. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J Hepatol. 2020;73:202-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2883] [Cited by in RCA: 2819] [Article Influence: 563.8] [Reference Citation Analysis (1)] |
| 19. | Tamaki N, Munaganuru N, Jung J, Yonan AQ, Bettencourt R, Ajmera V, Valasek MA, Behling C, Loomba R. Clinical Utility of Change in Nonalcoholic Fatty Liver Disease Activity Score and Change in Fibrosis in NAFLD. Clin Gastroenterol Hepatol. 2021;19:2673-2674.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 20. | Abdelbary MMH, Wilms G, Conrads G. A New Species-Specific Typing Method for Salivarius Group Streptococci Based on the Dephospho-Coenzyme A Kinase (coaE) Gene Sequencing. Front Cell Infect Microbiol. 2021;11:685657. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 21. | Seow WK, Lam JH, Tsang AK, Holcombe T, Bird PS. Oral Streptococcus species in pre-term and full-term children - a longitudinal study. Int J Paediatr Dent. 2009;19:406-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 22. | Matsumoto Y. Method for measuring the number of oral streptococci and a PCR primers-probe set used for the same. United States patent US 20080182265 A1. 2008 July 31. |
| 23. | Pomari E, Piubelli C, Perandin F, Bisoffi Z. Digital PCR: a new technology for diagnosis of parasitic infections. Clin Microbiol Infect. 2019;25:1510-1516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 24. | Zhang Z, Zhai H, Geng J, Yu R, Ren H, Fan H, Shi P. Large-scale survey of gut microbiota associated with MHE Via 16S rRNA-based pyrosequencing. Am J Gastroenterol. 2013;108:1601-1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 25. | Mouries J, Brescia P, Silvestri A, Spadoni I, Sorribas M, Wiest R, Mileti E, Galbiati M, Invernizzi P, Adorini L, Penna G, Rescigno M. Microbiota-driven gut vascular barrier disruption is a prerequisite for non-alcoholic steatohepatitis development. J Hepatol. 2019;71:1216-1228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 230] [Cited by in RCA: 456] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 26. | Yukawa-Muto Y, Kamiya T, Fujii H, Mori H, Toyoda A, Sato I, Konishi Y, Hirayama A, Hara E, Fukuda S, Kawada N, Ohtani N. Distinct responsiveness to rifaximin in patients with hepatic encephalopathy depends on functional gut microbial species. Hepatol Commun. 2022;6:2090-2104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 27. | Hsu C, Caussy C, Imajo K, Chen J, Singh S, Kaulback K, Le MD, Hooker J, Tu X, Bettencourt R, Yin M, Sirlin CB, Ehman RL, Nakajima A, Loomba R. Magnetic Resonance vs Transient Elastography Analysis of Patients With Nonalcoholic Fatty Liver Disease: A Systematic Review and Pooled Analysis of Individual Participants. Clin Gastroenterol Hepatol. 2019;17:630-637.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 301] [Article Influence: 50.2] [Reference Citation Analysis (0)] |
| 28. | Milosevic I, Vujovic A, Barac A, Djelic M, Korac M, Radovanovic Spurnic A, Gmizic I, Stevanovic O, Djordjevic V, Lekic N, Russo E, Amedei A. Gut-Liver Axis, Gut Microbiota, and Its Modulation in the Management of Liver Diseases: A Review of the Literature. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 393] [Cited by in RCA: 355] [Article Influence: 59.2] [Reference Citation Analysis (0)] |
| 29. | Patterson MJ. Streptococcus. Medical Microbiology. 4th ed. Baron S, editor. Galveston (TX): University of Texas Medical Branch at Galveston, 1996. |
| 30. | Huang Y, Pan H, Xu X, Lv P, Wang X, Zhao Z. Droplet digital PCR (ddPCR) for the detection and quantification of Ureaplasma spp. BMC Infect Dis. 2021;21:804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 31. | Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003-1005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 927] [Cited by in RCA: 1094] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 32. | Lunde TM, Roberts AP, Al-Haroni M. Determination of copy number and circularization ratio of Tn916-Tn1545 family of conjugative transposons in oral streptococci by droplet digital PCR. J Oral Microbiol. 2019;11:1552060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 33. | DeJong EN, Surette MG, Bowdish DME. The Gut Microbiota and Unhealthy Aging: Disentangling Cause from Consequence. Cell Host Microbe. 2020;28:180-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 235] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 34. | Mangiola F, Nicoletti A, Gasbarrini A, Ponziani FR. Gut microbiota and aging. Eur Rev Med Pharmacol Sci. 2018;22:7404-7413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 48] [Reference Citation Analysis (0)] |
| 35. | Ding M, Lang Y, Shu H, Shao J, Cui L. Microbiota-Gut-Brain Axis and Epilepsy: A Review on Mechanisms and Potential Therapeutics. Front Immunol. 2021;12:742449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (0)] |