Published online Jan 27, 2025. doi: 10.4254/wjh.v17.i1.98658
Revised: November 25, 2024
Accepted: December 13, 2024
Published online: January 27, 2025
Processing time: 188 Days and 6.7 Hours
Hepatitis B virus (HBV) infection is a global health concern. The current sequen
Core Tip: Elimination of hepatitis B virus (HBV) infection by 2030 is a common global goal proposed by the World Health Organization, and the challenge of HBV cure is the persistence of viral replication driven by the covalently closed circular DNA (cccDNA) pool. Currently, clinical markers of HBV DNA and hepatitis B surface antigen have limited utility in predicting HBV cure, either alone or in combination. In this review, we concentrate on the serum hepatitis B core-related antigen, an emerging serologic marker that correlates well with cccDNA, the continued monitoring of which during long-term therapy may aid in clinical management and facilitate the achievement of a complete cure in patients with chronic hepatitis B.
- Citation: Qiu Y, Tang Q, Liu XQ, Xue YL, Zeng Y, Hu P. Hepatitis B core-related antigen as a promising serological marker for monitoring hepatitis B virus cure. World J Hepatol 2025; 17(1): 98658
- URL: https://www.wjgnet.com/1948-5182/full/v17/i1/98658.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i1.98658
Hepatitis B virus (HBV) infection causes chronic hepatitis B (CHB) worldwide, which affects approximately 304 million patients and poses a serious public health hazard[1]. Patients with CHB are at risk of developing liver diseases with poor prognoses, such as cirrhosis, liver failure, and hepatocellular carcinoma (HCC)[2]. Despite the availability of effective hepatitis B vaccines, CHB continues to pose an important global health problem and is associated with high mortality rate[3,4].
Current treatments are unable to completely eradicate covalently closed circular DNA (cccDNA) from infected hepatocytes, because relaxed circular DNA (rcDNA) is continually converted to cccDNA in the nucleus of infected hepatocytes during HBV infection[5]. For the clinical treatment and management of patients, it is crucial to monitor the amount and transcriptional activity of cccDNA throughout the course of the disease. However, the process of detecting cccDNA is complicated, time-consuming, and expensive, and conventional methods require invasive experiments that are not always clinically applicable[6]. HBV serological markers are crucial for predicting CHB progression and indicating intrahepatic HBV replicative activity. It can be used as a minimally invasive substitute for liver biopsy.
Although hepatitis B surface antigen (HBsAg) is one of the oldest serological indicators of HBV infection and represents the state of HBV infection, it gradually loses its correlation with cccDNA after therapy due to its numerous mutations and integrated origin[7]. Hepatitis B core-related antigen (HBcrAg) and pregenomic RNA (pgRNA) are emerging serological markers of cccDNA. The lack of an international standard for quantitative RNA to validate labo
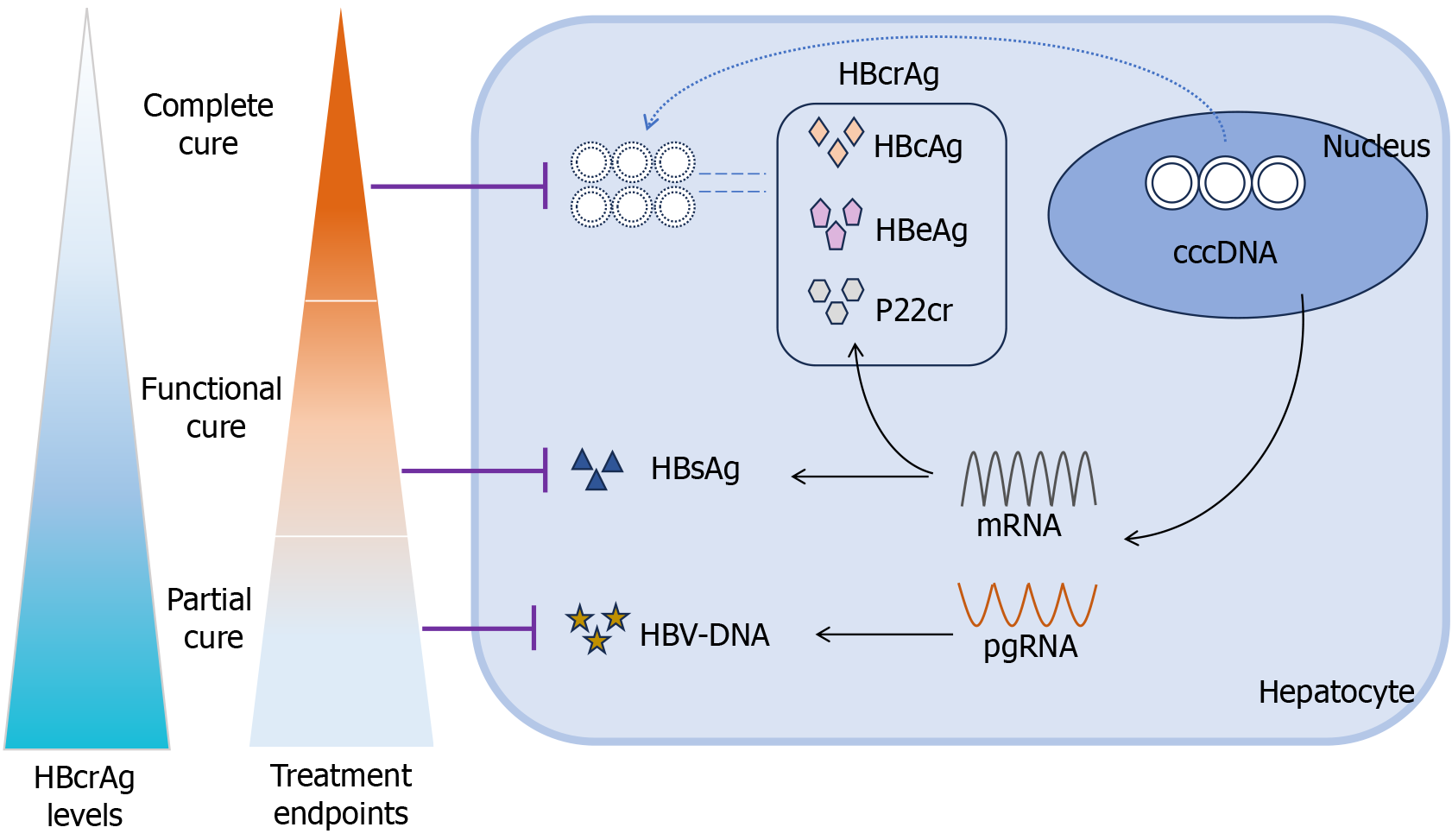
The current endpoints of antiviral therapy for chronic HBV infection can be divided into four stages. The initial goal is to achieve sustained viral suppression, indicated by undetectable HBV DNA, also known as a partial cure. The next step is to achieve serological conversion of HBeAg. The third stage involves the loss of HBsAg, which represents a functional cure. The ultimate goal is to achieve viral clearance, meaning the elimination of cccDNA, also called a complete cure[11]. Numerous studies have demonstrated that the baseline HBcrAg levels and changes in HBcrAg levels during treatment are indicative of treatment outcomes in patients with CHB. However, to date, no review has comprehensively summarized the role of HBcrAg as a sequential cure target for CHB.
In this review, we provide a thorough assessment of the contribution of serum HBcrAg in monitoring the cure of CHB. Therefore, long-term continuous monitoring of serum HBcrAg levels enhances the choice of treatment regimen and aids in the clinical management of patients, making it a promising indicator of treatment efficacy in patients with CHB.
Hepatocytes are target cells susceptible to HBV infection[12]. HBV delivers its 3.2-kb rcDNA genome to the nucleus of host hepatocytes after internalization through the cellular receptor sodium taurocholate cotransporting polypeptide[13]. In the presence of host cell enzymes such as nucleic acid endonucleases, DNA polymerases, and various ligases, the rcDNA is converted into cccDNA[14-16]. cccDNA is the sole transcriptional template for viral RNA and produces all relevant viral RNAs, including pre-genomic and messenger RNAs[17]. Pre-genomic RNA is transcribed into core and polymerase proteins and serves as a template for reverse transcription into new rcDNA. Messenger RNA can be translated into various HBV-associated structural or non-structural proteins[18]. HBcAg is a structural protein derived from the core gene that acts as a nucleocapsid component, wrapping viral DNA in Dane particles[19]. HBeAg, on the other hand, is also derived from the core gene but is a circulating protein that is modified and secreted by infected liver cells. p22Cr is a truncated 22-kDa precore protein that contains the N- and C-terminal processed product[20]. Fur
HBcrAg was first identified in 2002 by Kimura et al[8], who defined HBcAg and HBeAg as HBcrAg, using a sensitive enzyme immunoassay (EIA)[8]. The Lumipulse G HBcrAg assay, a commercial assay for HBcrAg detection, was later developed by Fujirebio (Japan). This test not only identified p22cr but also HBeAg and HBcAg serologically. Recently, iTACT-HBcrAg, a chemiluminescent EIA, has been used to detect HBcrAg[22]. Compared to the conventional G-HBcrAg assay, the iTACT-HBcrAg assay has higher sensitivity and efficiency, lower cost, and shorter detection time. The limit of detection of the iTACT-HBcrAg assay was 2.1 LogU/mL, which is approximately 10 times greater than that of the tra
A recent detailed study characterizing the components of HBcrAg suggested that serum HBeAg and PreC proteins have similar buoyant densities and size distributions, and both show density and size heterogeneity[23]. HBcAg, but not HBeAg or PreC antigen, was the main component of capsids in DNA-containing or empty virions. Neither HBeAg nor PreC can form capsids in cells or in vitro under physiological conditions[23]. Several studies have characterized the kinetics of HBcrAg in patients with HBV infection, revealing that its natural history differs between HBeAg-positive and HBeAg-negative individuals. Studies on cohorts from Europe and Asia have revealed that HBcrAg titers were substantially lower in HBeAg-negative individuals, particularly in inactive carriers (median titer 2.00 Log10 U/mL)[24,25]. In HBeAg-positive patients, HBeAg accounts for approximately 72% of the HBcrAg, whereas the contributions of HBcAg and p22cr cannot be quantified in HBeAg-negative patients[26]. Interestingly, during antiviral therapy, serum HBcrAg levels decreased progressively, regardless of the HBeAg status. After three years of therapy, Fan et al[27] reported that 67% of 66 HBeAg-negative patients, the majority of whom were non-Asians with genotype A or D infections, had undetectable HBcrAg. A two-by-two comparison of HBV DNA, HBsAg, and HBcrAg levels in a different study revealed that HBcrAg levels were higher in patients with the C genotype (n = 19) than in those with the E genotype (n = 16), with a difference of 1.3 Log10 U/mL, P = 0.0165, indicating that there may also be a correlation between HBcrAg, race and genotype[28]. Moreover, it has been found that the rate of decrease of HBcrAg is significantly slower compared to the decrease of HBV DNA during nucleos(t)ide analogue (NA) treatment. This is attributed to the fact that antiviral therapy does not directly inhibit transcription or translation processes[29]. Another cohort study demonstrated that HBcrAg declines early and reaches undetectable levels (< 1000 U/mL) approximately 10-14 years prior to the seroclearance of HBsAg in patients who achieve serologic clearance. In contrast, HBcrAg levels remain high in patients who did not achieve serologic clearance[30]. Furthermore, as evidenced by additional research, HBcrAg can be found in individuals with undetectable HBV DNA or HBsAg, indicating that cccDNA may still exist in individuals with occult HBV infection or HBsAg clearance[31,32].
HBV DNA levels are the visual representation of the level of HBV replication. The inhibition of HBV replication to render HBV DNA undetectable is the initial correlation between baseline HBcrAg and HBV DNA, regardless of HBeAg status, and has been demonstrated in multiple studies[33-36]. HBcrAg > 3.6 Log IU/mL was significantly associated with HBV DNA > 2000 IU/mL and HBcrAg > 5.3 Log IU/mL was diagnostic of HBV DNA > 200000 IU/mL in a Gambian cohort[37]. A study comprising 130 patients with untreated CHB revealed a strong correlation between the prevalence of HBcrAg and HBV DNA levels in the general population (r = 0.82, P < 0.0001). In contrast to previous research, the correlation between HBcrAg and HBV DNA in this investigation was dependent on patient status, with a strong correlation observed in HBeAg+ (r = 0.46, P = 0.005) and HBeAg-CH (r = 0.57, P < 0.001) patients, but not in HBeAg-CI patients[32]. Additionally, genotype influences the correlation between serum HBcrAg and HBV DNA levels. An independent analysis of each genotype in a prospective study involving 202 HBeAg-negative, treatment-naïve patients with CHB revealed that the correlation between HBcrAg and HBV DNA was only observed in genotype A-infected patients (r = 0.43, P < 0.001)[38]. Furthermore, a notable association was observed between the decline in HBcrAg levels and reduction in HBV DNA levels during the course of treatment. The correlation coefficients for this relationship were R = 0.366, P = 0.001 for HBeAg-positive patients, and R = 0.626, P < 0.001 for HBeAg-negative patients[39]. A subsequent trial conducted in a prospective randomized cohort revealed that individuals who received treatment continued to exhibit detectable levels of HBcrAg, although having undetectable levels of HBV DNA. The association between HBcrAg and HBV DNA was no longer observed after 48 weeks of entecavir treatment. However, it remained significant following interferon (IFN) treatment, with a correlation coefficient of 0.702 and a P value less than 0.0001[40]. These findings are consistent with those of a prior investigation[41]. In summary, a positive association was observed between baseline HBcrAg and HBV DNA levels, which progressively declined throughout the course of treatment.
Sustained viral suppression is a critical prerequisite for HBV partial cure. Prior research has demonstrated that serum HBcrAg is a reliable predictor of persistent viral remission (PVR; defined as persistent HBV DNA levels < 2000 IU/mL), and that low HBcrAg levels are linked to a favorable virological response[42]. Regression analyses revealed that baseline and decreasing HBcrAg levels after 12 weeks of treatment were both correlated with PVR in a retrospective study involving 121 Thai patients with CHB who tested negative for HBeAg[43]. In predicting PVR, the area under the receiver operating characteristic (ROC) curve (AUROC) of the log10 HBcrAg declines following 12-week treatment with pegylated IFN (PEG-IFN) alone or in combination with entecavir was 0.706 (0.604-0.808; P < 0.001). According to the ROC analysis, the optimal cutoff values for predicting PVR based on the reduction of HBcrAg from baseline were 0.45 Log10 U/mL[43]. Multivariate logistic regression analysis revealed that serum HBcrAg at week 12 was an independent predictor of virological response in patients with HBeAg-positive CHB receiving PEG-IFN therapy, according to another Thai study that defined virological remission (VR) as HBeAg clearance and HBV DNA < 2000 IU/mL[44]. With a sensitivity of 93.3%, specificity of 54.8%, and positive and negative predictive values of 50.0% and 94.4%, respectively, the ROC analysis revealed that a serum HBcrAg level of 8.0 Log10 U/mL at week 12 was the optimal threshold for predicting VR. This threshold may be useful for the early identification of patients who are insensitive to PEG-IFN therapy[44]. In another study on sequential NAs/PEG-IFN therapy, the response was determined in accordance with the recommendations of the Japanese Society of Hepatology: Alanine transaminase (ALT) < 31 IU/L, negative HBeAg result, HBV DNA < 4.0 Log copies/mL, and no recommendation for NAs[45,46]. As indicated by the multivariate analyses of patient factors associated with switching from NA to PEG-IFN, responders were significantly associated with lower levels of HBsAg and HBcrAg. ROC analysis revealed that a 3.9 LogU/mL HBcrAg level accurately predicted responders, with an area under the curve (AUC) of 0.692 (P = 0.005)[45]. During tenofovir therapy, HBV DNA levels in all individuals were undetectable within 10 months, regardless of whether HBcrAg was undetectable in 10 years in a median follow-up of 12 years. Another study showed that HBcrAg levels at the end of treatment (EOT) were < 4 Log U/L or not, and 90% of patients showed virological relapse (HBV-DNA > 2000 IU/mL) in the 96-week follow-up period without treatment[47]. More interesting, a multicenter cohort showed that the incidence of relapse in 96-week was significantly higher with EOT HBcrAg ≥ 1000 U/mL than with HBcrAg < 1000 U/mL (P = 0.002)[48]. There is a significant association between HBcrAg and HBV DNA, and alterations in HBV DNA levels during therapy are linked to changes in HBcrAg levels. Moreover, the limited extent of HBcrAg fluctuation throughout the treatment period may indicate a reduced likelihood of sustained VR. Importantly, incorporating HBcrAg into the considerations for drug discontinuation in clinical applications may be beneficial for patient prognosis, reducing clinical recurrence, and ultimately achieving a partial cure.
HBeAg seroconversion, which is defined as the loss of HBeAg and the production of anti-HBe antibodies, is a clinically desirable therapeutic endpoint because it reflects partial immune control. HBeAg seroconversion is usually accompanied by sustained suppression of HBV DNA, normalization of ALT levels, and long-term remission of hepatic inflammation as confirmed by liver biopsy. It indicates a positive long-term result throughout the duration of the condition[49-51]. HBeAg may spontaneously disappear during the immunological clearance phase in patients with HBeAg-positive CHB without the need for antiviral medication, with a likelihood of 5%-10% annually[50]. NA analogs such as tenofovir disoproxil fumarate and entecavir have been shown to result in HBeAg seroconversion rates of 40%-53% after prolonged treatment for 5 years[52,53].
HBeAg is a component of the HBcrAg. HBeAg constitutes the majority of HBcrAgs in HBeAg-positive individuals, and the two proteins have a strong association over time[54]. According to previous studies, HBcrAg can distinguish between patients with active disease who are HBeAg-positive and those who are HBeAg-negative[33]. Additionally, HBcrAg is crucial for determining the serological conversion of HBeAg.
Previous research has demonstrated that HBcrAg is an excellent predictor of spontaneous HBeAg seroconversion in patients with CHB. Eighteen participants with spontaneous HBeAg seroconversion and 35 patients with non-spontaneous HBeAg seroconversion were randomly matched in a Beijing cohort[55]. One patient was excluded owing to sample contamination. According to multivariate logistic regression findings, spontaneous HBeAg seroconversion was associated with both HBcrAg levels at week 28 and HBcrAg reduction at week 28. The HBcrAg levels and the reduction in HBcrAg levels at week 28 had respective AUROCs of 0.913 (P = 0.001, 95%CI: 0.804-1.000) and 0.922 (P = 0.001, 95%CI: 0.847-0.997). This indicated that the predictive values were adequate. At week 28, the optimal cutoffs for HBcrAg and the reduction in the HBcrAg levels were 4.90 and 2.00 Log10 kU/mL, respectively, with Youden's index values of 0.77 and 0.75, respectively. HBcrAg levels below 4.90 Log10 kU/mL, and HBcrAg levels > 2.00 Log10 kU/mL at week 28 appeared to be associated with the greatest likelihood of spontaneous HBeAg seroconversion in patients with CHB[55]. After 76 weeks of follow-up during the immune clearance phase, Xie et al[56] increased the sample size in their most recent study. They consistently found that a significant decline in HBcrAg from baseline to 28 weeks was a good predictor of spontaneous HBeAg seroconversion (AUROC: 0.93; 95%CI: 0.74-1.08; P = 0.001).
Several studies have indicated that HBcrAg levels may decrease before NA-induced HBeAg conversion[33,57]. According to a report by Sonneveld et al[58], patients who attained HBeAg seroconversion had lower levels of HBcrAg than those who did not. However, multifactorial analysis did not reveal this association[58]. In a study conducted by Lee et al[59], 194 patients who were administered NA following liver biopsy were included, with a median treatment duration of 48.3 months. Cox regression analysis revealed that the sole independent predictor of NA-induced HBeAg seroconversion was high HBcrAg level [hazard ratio (HR) = 1.285, P = 0.028]. With a median follow-up of 11 years, the most recent Korean study retrospectively enrolled 70 patients with HBeAg-positive CHB, who were treated with entecavir or tenofovir. Of these patients, 21 (30%) experienced HBeAg seroconversion at 28 months after antiviral therapy[60]. In multivariate analysis, HBeAg seroconversion was predicted by serum HBcrAg levels at baseline and two years after antiviral therapy (HR = 0.326; 95%CI: 0.111-0.958; P = 0.042 and HR = 0.4555; 95%CI: 0.211-0.984; P = 0.045). The AUROCs of HBcrAg levels ≤ 6.5 Log10 U/mL at baseline and levels ≤ 5.3 Log10 U/mL at 2 years after antiviral therapy for predicting HBeAg seroconversion were 0.712 (95%CI: 0.596-0.830) and 0.745 (95%CI: 0.599-0.891), respectively. Furthermore, a study assessing the predictive power of HBcrAg for seroconversion in IFN-treated HBeAg-positive patients with CHB discovered that an HBcrAg level < 34225 kU/mL at the 12-week post-treatment follow-up was a strong indicator of HBeAg serological conversion at the conclusion of the follow-up (AUROC = 0.896; 95%CI: 0.766-1.000; P = 0.013)[54]. Another retrospective study of 31 patients with HBeAg-positive CHB who were followed-up for 6 months after 12 months of PEG-IFN treatment found that the absolute quantitative HBcrAG (qHBcrAg) level and extent of qHBcrAg decline at month 1 were better predictors of HBeAg seroconversion at month 6 after treatment than qHBsAg[61]. Moreover, in another randomized controlled trial, it was discovered that baseline qHBcrAg did not predict treatment response and that the optimal time for qHBcrAg to predict HBeAg seroconversion was 24 and 48 weeks, with areas under the curve measuring 0.794 (P < 0.0001) and 0.825 (P < 0.0001), respectively[40].
In summary, HBcrAg remains concordant in patients with CHB who test positive for HBeAg, and plays a significant role in anticipating HBeAg seroconversion, either naturally or during antiviral treatment.
HBsAg is one of the most significant serological markers of HBV infection. qHBsAg can be used to predict various clinically relevant outcomes, whereas qualitative HBsAg is currently regarded as a therapeutic endpoint for functional cure. The correlation between HBsAg and HBcrAg levels has been documented in numerous studies. A study comprising 2666 patients (both HBeAg-positive and -negative) employing Pearson's correlation analysis revealed a moderate but notable correlation between baseline HBcrAg and HBsAg levels (r = 0.59; P < 0.001)[62]. In a study conducted in North America involving 373 individuals with HBeAg positivity and 978 with HBeAg negativity, the correlation between baseline HBcrAg and qHBsAg was significant but diminished during the HBeAg-negative period[63]. Consistent with this, in a study by Mak et al[41], the correlation between baseline HBcrAg and qHBsAg was greater in HBeAg-positive patients than in HBeAg-negative patients, with correlation coefficients of 0.686 and 0.436, respectively. Thus, in patients lacking HBeAg, there was no correlation between HBsAg and HBcrAg levels. This may be due to the fact that the biological pathway of HBsAg differs from that of HBcrAg. HBsAg is primarily derived from recombinant HBV DNA in HBeAg-negative patients, whereas HBcrAg is produced via intrahepatic cccDNA transcription[64]. Unsurprisingly, the correlation between HBsAg and HBcrAg decreases further in both HBeAg-positive and HBeAg-negative patients[39,43,44].
Numerous studies have investigated the predictive value of HBcrAg levels for HBsAg clearance. Numerous studies have demonstrated a correlation between lower baseline HBcrAg levels and higher rates of HBsAg loss[43]. Owing to the heterogeneity of individual studies, the optimal cutoff values that have been established vary. A recent retrospective study conducted in Taiwan examined the relationship between HBcrAg levels, kinetics, and HBsAg seroclearance over time in 2614 patients with primary CHB without cirrhosis. Both lower HBcrAg levels at baseline and a decrease in HBcrAg levels were associated with an increase in HBsAg seroclearance over time in primigravida patients, according to the findings. Furthermore, when the study population was restricted to individuals with HBsAg levels exceeding 1000 IU/mL, the only independent viral marker associated with HBsAg seroclearance over time was a baseline HBcrAg < 10000 U/mL[30].
Similarly, several studies have revealed the prognostic impact of on-treatment alterations in HBcrAg levels on HBsAg loss. Ma et al[65] conducted a study that revealed that the extent of reduction in HBcrAg levels at the beginning of treatment and throughout the treatment process was linked to HBsAg loss. However, the effectiveness of using the reduction in the HBcrAg levels at 12 months as a predictor for HBsAg loss was significantly lower compared to the reduction in the HBsAg levels (AUC = 0.939; 95%CI: 0.868-1.000 vs AUC = 0.521; 95%CI: 0.344-0.698). In addition, Mak et al[66] found that the reduction in serum HBcrAg levels at week 4 following NA treatment in patients with HBeAg-negative CHB was a strong indicator of a favorable HBsAg response (FHR; defined as HBsAg < 100 IU/mL or HBsAg seroclearance) at the end of follow-up (EOFU). The performance characteristics of the week 4 change in serum HBcrAg levels to predict FHR at EOFU were evaluated using ROC analysis, with an AUC of 0.789 (95%CI: 0.596-0.982; P = 0.003). The study also found a significant association between the composite variable "biomarker response at week 4" and FHR. This composite variable includes a decrease in pgRNA of ≥ 5.32 Log copies/mL for individuals with HBeAg+ at week 4, or a decrease in HBcrAg of ≥ 2.05 Log U/mL for individuals with HBeAg- at week 4. The hazard ratio for this association was 10.378 (95%CI: 2.312-46.589; P = 0.002), indicating that a rapid and substantial decrease in HBcrAg levels after antiviral therapy is a more favorable treatment outcome. In contrast, patients who do not exhibit an initial reduction in biomarkers may have a diminished likelihood of attaining a functional cure after prolonged nucleoside analog therapy[66].
The significance of the HBcrAg level at EOT as a prognostic factor for HBsAg loss has been the subject of contradictory findings in multiple studies. According to the findings of Kuo and Huang, the EOT HBcrAg level at EOT following NA cessation of NA did not constitute a significant predictor of HBsAg loss[67,68]. According to two recent multicenter studies conducted in the Netherlands and China, patients with a higher likelihood of experiencing HBsAg loss following discontinuation of NA may be identified by low or undetectable levels of HBcrAg at EOT[69,70]. In a prospective study in Wuhan, researchers discovered that patients who received peg-IFN treatment and had HBcrAg levels < 4 Log10 U/mL at the EOT experienced long-lasting suppression of the virus, as well as lower levels of HBV DNA, HBsAg, and HBeAg compared to patients with HBcrAg levels ≥ 4 Log10 U/mL at EOT. Subsequently, a composite model was developed by combining the variables HBcrAg < 4 Log10 U/mL and HBsAb > 2 Log10 IU/L. This model, known as “HBV Cure crAb” accurately identified patients who were likely to acquire a durable functional cure, with a positive rate of up to 100%[71]. These findings indicate that stratifying patient management based on HBcrAg levels < 4 Log U/mL, in combination with existing serological indicators such as HBsAg and HBV DNA levels, serves as a better basis for determining treatment cessation. This approach may more effectively achieve long-term viral suppression and facilitate functional cure.
A crucial factor in sustained viral infection caused by HBV is the presence of intracellular HBV replication intermediates, specifically cccDNA. cccDNA is present as a non-integrating plasmid-like molecule within the nucleus of an infected cell and consistently produces offspring viruses[72,73]. cccDNA is freely present in the nuclei of hepatocytes as a reverse transcription template for all HBV RNAs, which can be transcribed into HBV DNA and translated into seven viral proteins[74]. The long half-life and dual origin of cccDNA (imported viral particles and intracellular nuclear capsid recycling) render its eradication challenging even after protracted treatment[75]. Permanent cccDNA silencing or elimination is crucial for preventing viral reactivation; therefore, cccDNA is regarded as the ultimate target for HBV treatment[76].
HBcrAg contains three proteins that are transcription products of cccDNA, and several prior studies have demonstrated a positive correlation between HBcrAg levels and liver cccDNA[77,78]. Subsequent research demonstrated that HBcrAg is significantly correlated with the transcriptional activity of cccDNA (pgRNA/cccDNA) in addition to cccDNA levels[32]. Notably, this correlation was considerably stronger than that between qHBsAg and serum HBV DNA levels, indicating that HBcrAg is a more accurate biological indicator of cccDNA levels than HBsAg or HBV DNA[32].
Despite the challenges of directly quantifying cccDNA, HBcrAg levels may accurately reflect cccDNA in vivo[79]. This suggests that HBcrAg levels may serve as an alternative indicator of cccDNA elimination therapy. There are currently two major obstacles to the removal of cccDNA from infected cells: First, disease replication and cccDNA replenishment must be completely inhibited, and second, preexisting cccDNA must be depleted within a reasonable time window[80]. Currently, medications that have been conceived produce target cccDNA by impeding the production of cccDNA and expediting its disintegration via either cytotoxic or noncytotoxic methods. Phase II clinical trials have been initiated for drugs that target cccDNA, including entry inhibitors, transcription inhibitors, RNA silencers, core protein allosteric modulators, noncompetitive polymerase inhibitors, and viral protein export inhibitors, among others[81]. During drug development and efficacy validation, HBcrAg was utilized as a non-invasive yet effective method for monitoring treatment-induced modification levels of intrahepatic HBV cccDNA. Vonafexor, an agonist of the nuclear farnesoid X receptor, in conjunction with peg-IFN-2a combination therapy, decreased the levels of HBcrAg and pgRNA has been reported[82]. Recent randomized human trials have demonstrated that RG6346 as an RNA inhibitor is safe and well tolerated, with moderate to significant sustained reductions in HBeAg, HBV DNA, and HBcrAg levels observed during treatment[83]. Although HBcrAg is an effective serological replacement for cccDNA, its predictive or monitoring function in cccDNA clearance remains unclear.
Hepatitis B continues to be a significant public health concern worldwide. The proposition put up by the World Health Organization suggests that the eradication of acquired immunodeficiency syndrome, viral hepatitis (specifically, CHB and chronic hepatitis C), and sexually transmitted illnesses is attainable by the year 2030[84]. The sequential objective of HBV treatment for cancer prevention involves three key milestones: Conversion of HBV DNA and HBsAg, and elimination of HBV cccDNA[85]. Nevertheless, the eradication of cccDNA poses significant challenges, and the suppression of cccDNA transcriptional activity holds promise as a viable therapeutic strategy. An accurate reflection of cccDNA transcriptional activity was observed using HBcrAg. When serum HBcrAg clearance occurs, specifically HBcrAg negativity, as determined by high-sensitivity testing, HBV is in a state of epigenetic quiescence. This may serve as a potential therapeutic target.
HBV DNA is also considered a significant serological indicator of HBV replication; however, it is typically undetectable following NA treatment[86]. Treatment with NA can efficiently inhibit HBV DNA replication but cannot clear cccDNA. Patients who receive IFN have a greater chance of achieving HBsAg clearance but are more susceptible to adverse effects[87]. Certain patients with CHB infection may require ongoing antiviral therapy. Serum HBcrAg levels at baseline and at the conclusion of treatment were more accurate predictors of serological conversion after NA therapy in patients with CHB. Additionally, the trend of HBcrAg alterations throughout treatment can be used to identify patients who are insensitive to PEG-IFN therapy[44]. Following treatment, certain conventional serological and histological markers may exhibit partial restoration to normal levels and HBsAg may become undetectable. However, it is important to note that some patients may still test positive for HBcrAg, indicating that complete remission of their condition has not been achieved. Furthermore, the liver may harbor cccDNA, which has the potential to advance the development of hepatocellular lesions. Hence, serum HBcrAg level serves as a surrogate test for HBV cccDNA levels to comprehensively evaluate antiviral efficacy and predict treatment endpoints. This also enables the provision of individualized guidance for clinical anti-HBV treatment, appropriate timing of drug discontinuation, and prognostic analysis. However, to enhance the sensitivity and specificity of the prediction, the incorporation of additional serological markers may be necessary, despite their potential in predicting treatment outcomes and future complications. Moreover, extensive studies involving large cohorts are essential to establish a precise threshold value for accurate prediction.
However, clinical application of HBcrAg quantification remains challenging. Current evidence demonstrates that it is not a substitute for HBV DNA levels in predicting the development of HCC or as a criterion for starting antiviral therapy; however, whether HBcrAg levels can be used as a biomarker to optimize the management of HBeAg-negative patients in the gray zone, either with high HBV DNA levels or mildly elevated ALT levels, warrants further investigation[85]. Given that serum HBcrAg remains detectable in certain patients with undetectable HBeAg and HBsAg levels, it is crucial to conduct future research comparing the long-term outcomes of patients with and without HBcrAg to enhance the prognosis of patients with CHB. Furthermore, most previous reports on HBcrAg were published in Asia. Large prospective clinical trials in the United States and Europe are required to investigate and justify the use of HBcrAg in order to improve its clinical application.
In summary, HBcrAg is a promising HBV marker that maintains excellent concordance with intrahepatic cccDNA, and predicts the treatment response and prognosis of patients with CHB well. Observing the rational and dynamic changes in HBcrAg levels may benefit the clinical management of patients with CHB and optimize treatment selection.
| 1. | Jeng WJ, Papatheodoridis GV, Lok ASF. Hepatitis B. Lancet. 2023;401:1039-1052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 264] [Article Influence: 132.0] [Reference Citation Analysis (0)] |
| 2. | Polaris Observatory Collaborators. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3:383-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1260] [Cited by in RCA: 1215] [Article Influence: 173.6] [Reference Citation Analysis (2)] |
| 3. | Piermatteo L, D'Anna S, Bertoli A, Bellocchi M, Carioti L, Fabeni L, Alkhatib M, Frazia S, Lichtner M, Mastroianni C, Sanctis G, Marignani M, Pasquazzi C, Iapadre N, Parruti G, Cappiello G, Vecchiet J, Malagnino V, Grelli S, Ceccherini-Silbertein F, Andreoni M, Sarmati L, Svicher V, Salpini R. Unexpected rise in the circulation of complex HBV variants enriched of HBsAg vaccine-escape mutations in HBV genotype-D: potential impact on HBsAg detection/quantification and vaccination strategies. Emerg Microbes Infect. 2023;12:2219347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 4. | Cooke GS, Andrieux-Meyer I, Applegate TL, Atun R, Burry JR, Cheinquer H, Dusheiko G, Feld JJ, Gore C, Griswold MG, Hamid S, Hellard ME, Hou J, Howell J, Jia J, Kravchenko N, Lazarus JV, Lemoine M, Lesi OA, Maistat L, McMahon BJ, Razavi H, Roberts T, Simmons B, Sonderup MW, Spearman CW, Taylor BE, Thomas DL, Waked I, Ward JW, Wiktor SZ; Lancet Gastroenterology & Hepatology Commissioners. Accelerating the elimination of viral hepatitis: a Lancet Gastroenterology & Hepatology Commission. Lancet Gastroenterol Hepatol. 2019;4:135-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 397] [Article Influence: 66.2] [Reference Citation Analysis (0)] |
| 5. | Eller C, Heydmann L, Colpitts CC, El Saghire H, Piccioni F, Jühling F, Majzoub K, Pons C, Bach C, Lucifora J, Lupberger J, Nassal M, Cowley GS, Fujiwara N, Hsieh SY, Hoshida Y, Felli E, Pessaux P, Sureau C, Schuster C, Root DE, Verrier ER, Baumert TF. A genome-wide gain-of-function screen identifies CDKN2C as a HBV host factor. Nat Commun. 2020;11:2707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Cai D, Nie H, Yan R, Guo JT, Block TM, Guo H. A southern blot assay for detection of hepatitis B virus covalently closed circular DNA from cell cultures. Methods Mol Biol. 2013;1030:151-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 118] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 7. | Iannacone M, Guidotti LG. Immunobiology and pathogenesis of hepatitis B virus infection. Nat Rev Immunol. 2022;22:19-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 291] [Article Influence: 97.0] [Reference Citation Analysis (0)] |
| 8. | Kimura T, Rokuhara A, Sakamoto Y, Yagi S, Tanaka E, Kiyosawa K, Maki N. Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load. J Clin Microbiol. 2002;40:439-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 191] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 9. | Inoue T, Tanaka Y. Novel biomarkers for the management of chronic hepatitis B. Clin Mol Hepatol. 2020;26:261-279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 92] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 10. | Lam YF, Seto WK, Wong D, Cheung KS, Fung J, Mak LY, Yuen J, Chong CK, Lai CL, Yuen MF. Seven-Year Treatment Outcome of Entecavir in a Real-World Cohort: Effects on Clinical Parameters, HBsAg and HBcrAg Levels. Clin Transl Gastroenterol. 2017;8:e125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 11. | You H, Wang F, Li T, Xu X, Sun Y, Nan Y, Wang G, Hou J, Duan Z, Wei L, Jia J, Zhuang H; Chinese Society of Hepatology, Chinese Medical Association; Chinese Society of Infectious Diseases, Chinese Medical Association. Guidelines for the Prevention and Treatment of Chronic Hepatitis B (version 2022). J Clin Transl Hepatol. 2023;11:1425-1442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 82] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 12. | Yang F, Wu L, Xu W, Liu Y, Zhen L, Ning G, Song J, Jiao Q, Zheng Y, Chen T, Xie C, Peng L. Diverse Effects of the NTCP p.Ser267Phe Variant on Disease Progression During Chronic HBV Infection and on HBV preS1 Variability. Front Cell Infect Microbiol. 2019;9:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1280] [Cited by in RCA: 1600] [Article Influence: 123.1] [Reference Citation Analysis (1)] |
| 14. | Menéndez-Arias L, Sebastián-Martín A, Álvarez M. Viral reverse transcriptases. Virus Res. 2017;234:153-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 81] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 15. | Kitamura K, Que L, Shimadu M, Koura M, Ishihara Y, Wakae K, Nakamura T, Watashi K, Wakita T, Muramatsu M. Flap endonuclease 1 is involved in cccDNA formation in the hepatitis B virus. PLoS Pathog. 2018;14:e1007124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 16. | Qi Y, Gao Z, Xu G, Peng B, Liu C, Yan H, Yao Q, Sun G, Liu Y, Tang D, Song Z, He W, Sun Y, Guo JT, Li W. DNA Polymerase κ Is a Key Cellular Factor for the Formation of Covalently Closed Circular DNA of Hepatitis B Virus. PLoS Pathog. 2016;12:e1005893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 159] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 17. | Tsukuda S, Watashi K. Hepatitis B virus biology and life cycle. Antiviral Res. 2020;182:104925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 232] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 18. | Nair S, Li L, Francis S, Turner WW, VanNieuwenhze M, Zlotnick A. Use of a Fluorescent Analogue of a HBV Core Protein-Directed Drug To Interrogate an Antiviral Mechanism. J Am Chem Soc. 2018;140:15261-15269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 19. | Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. Structural organization of the hepatitis B virus minichromosome. J Mol Biol. 2001;307:183-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 280] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 20. | Kimura T, Ohno N, Terada N, Rokuhara A, Matsumoto A, Yagi S, Tanaka E, Kiyosawa K, Ohno S, Maki N. Hepatitis B virus DNA-negative dane particles lack core protein but contain a 22-kDa precore protein without C-terminal arginine-rich domain. J Biol Chem. 2005;280:21713-21719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 128] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 21. | Mak LY, Hui RW, Fung J, Seto WK, Yuen MF. The role of different viral biomarkers on the management of chronic hepatitis B. Clin Mol Hepatol. 2023;29:263-276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 22. | Inoue T, Kusumoto S, Iio E, Ogawa S, Suzuki T, Yagi S, Kaneko A, Matsuura K, Aoyagi K, Tanaka Y. Clinical efficacy of a novel, high-sensitivity HBcrAg assay in the management of chronic hepatitis B and HBV reactivation. J Hepatol. 2021;75:302-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 83] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 23. | Hong X, Luckenbaugh L, Mendenhall M, Walsh R, Cabuang L, Soppe S, Revill PA, Burdette D, Feierbach B, Delaney W, Hu J. Characterization of Hepatitis B Precore/Core-Related Antigens. J Virol. 2021;95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 24. | Maasoumy B, Wiegand SB, Jaroszewicz J, Bremer B, Lehmann P, Deterding K, Taranta A, Manns MP, Wedemeyer H, Glebe D, Cornberg M. Hepatitis B core-related antigen (HBcrAg) levels in the natural history of hepatitis B virus infection in a large European cohort predominantly infected with genotypes A and D. Clin Microbiol Infect. 2015;21:606.e1-606.10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 25. | Seto WK, Wong DK, Fung J, Huang FY, Liu KS, Lai CL, Yuen MF. Linearized hepatitis B surface antigen and hepatitis B core-related antigen in the natural history of chronic hepatitis B. Clin Microbiol Infect. 2014;20:1173-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 116] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 26. | Adraneda C, Tan YC, Yeo EJ, Kew GS, Khakpoor A, Lim SG. A critique and systematic review of the clinical utility of hepatitis B core-related antigen. J Hepatol. 2023;78:731-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 27. | Fan R, Peng J, Xie Q, Tan D, Xu M, Niu J, Wang H, Ren H, Chen X, Wang M, Sheng J, Tang H, Bai X, Wu Y, Zhou B, Sun J, Hou J; Chronic Hepatitis B Study Consortium. Combining Hepatitis B Virus RNA and Hepatitis B Core-Related Antigen: Guidance for Safely Stopping Nucleos(t)ide Analogues in Hepatitis B e Antigen-Positive Patients With Chronic Hepatitis B. J Infect Dis. 2020;222:611-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 65] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 28. | Brunetto MR, Carey I, Maasoumy B, Marcos-Fosch C, Boonstra A, Caviglia GP, Loglio A, Cavallone D, Scholtes C, Ricco G, Smedile A, Riveiro-Barciela M, van Bömmel F, van der Eijk A, Zoulim F, Berg T, Cornberg M, Lampertico P, Agarwal K, Buti M. Incremental value of HBcrAg to classify 1582 HBeAg-negative individuals in chronic infection without liver disease or hepatitis. Aliment Pharmacol Ther. 2021;53:733-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 29. | Ma G, Lou B, Lv F, Zhao D, Zhang Z, Chen Y. HBcrAg and pg RNA and the therapeutic effect in HBeAg-positive patients receiving anti-viral therapy, baseline serum HBV-RNA is a powerful predictor of response. J Viral Hepat. 2020;27:837-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 30. | Tseng TC, Chiang C, Liu CJ, Hong CM, Su TH, Yang HC, Yang WT, Liu CH, Chen PJ, Kao JH. Low Hepatitis B Core-Related Antigen Levels Correlate Higher Spontaneous Seroclearance of Hepatitis B Surface Antigen in Chronic Hepatitis B Patients With High Hepatitis B Surface Antigen Levels. Gastroenterology. 2023;164:669-679.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 31. | Tseng TN, Hu TH, Wang JH, Kuo YH, Hung CH, Lu SN, Jeng WJ, Chen CH. Incidence and Factors Associated With HBV Relapse After Cessation of Entecavir or Tenofovir in Patients With HBsAg Below 100 IU/mL. Clin Gastroenterol Hepatol. 2020;18:2803-2812.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 32. | Testoni B, Lebossé F, Scholtes C, Berby F, Miaglia C, Subic M, Loglio A, Facchetti F, Lampertico P, Levrero M, Zoulim F. Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J Hepatol. 2019;70:615-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 217] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 33. | Chan HLY, Yasuda S, Wong GLH, Tada T, Chan CKM, Kumada T, Tse YK, Wong VWS, Toyoda H. Use of hepatitis B virus core-related antigen to evaluate natural history of chronic hepatitis B. J Gastroenterol Hepatol. 2020;35:2202-2209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 34. | Loggi E, Vukotic R, Conti F, Grandini E, Gitto S, Cursaro C, Galli S, Furlini G, Re MC, Andreone P. Serum hepatitis B core-related antigen is an effective tool to categorize patients with HBeAg-negative chronic hepatitis B. J Viral Hepat. 2019;26:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | van Campenhout MJH, Rijckborst V, Brouwer WP, van Oord GW, Ferenci P, Tabak F, Akdogan M, Pinarbasi B, Simon K, de Knegt RJ, Boonstra A, Janssen HLA, Hansen BE. Hepatitis B core-related antigen monitoring during peginterferon alfa treatment for HBeAg-negative chronic hepatitis B. J Viral Hepat. 2019;26:1156-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 36. | Zhang ZQ, Lu W, Wang YB, Weng QC, Zhang ZY, Yang ZQ, Feng YL. Measurement of the hepatitis B core-related antigen is valuable for predicting the pathological status of liver tissues in chronic hepatitis B patients. J Virol Methods. 2016;235:92-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 37. | Shimakawa Y, Ndow G, Njie R, Njai HF, Takahashi K, Akbar SMF, Cohen D, Nayagam S, Jeng A, Ceesay A, Sanneh B, Baldeh I, Imaizumi M, Moriyama K, Aoyagi K, D'Alessandro U, Mishiro S, Chemin I, Mendy M, Thursz MR, Lemoine M. Hepatitis B Core-related Antigen: An Alternative to Hepatitis B Virus DNA to Assess Treatment Eligibility in Africa. Clin Infect Dis. 2020;70:1442-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 38. | Riveiro-Barciela M, Bes M, Rodríguez-Frías F, Tabernero D, Ruiz A, Casillas R, Vidal-González J, Homs M, Nieto L, Sauleda S, Esteban R, Buti M. Serum hepatitis B core-related antigen is more accurate than hepatitis B surface antigen to identify inactive carriers, regardless of hepatitis B virus genotype. Clin Microbiol Infect. 2017;23:860-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 39. | Li J, Wu Z, Wang GQ, Zhao H. Hepatitis B core-related antigen reflects viral replication and protein production in chronic hepatitis B patients. Chin Med J (Engl). 2021;134:1160-1167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 40. | Chi XM, Wang XM, Wang ZF, Wu RH, Gao XZ, Xu HQ, Ding YH, Niu JQ. Serum hepatitis B core-related antigen as a surrogate marker of hepatitis B e antigen seroconversion in chronic hepatitis B. World J Gastroenterol. 2021;27:6927-6938. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 41. | Mak LY, Cloherty G, Wong DK, Gersch J, Seto WK, Fung J, Yuen MF. HBV RNA Profiles in Patients With Chronic Hepatitis B Under Different Disease Phases and Antiviral Therapy. Hepatology. 2021;73:2167-2179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 63] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 42. | Okuhara S, Umemura T, Joshita S, Shibata S, Kimura T, Morita S, Komatsu M, Matsumoto A, Yoshizawa K, Katsuyama Y, Ota M, Tanaka E. Serum levels of interleukin-22 and hepatitis B core-related antigen are associated with treatment response to entecavir therapy in chronic hepatitis B. Hepatol Res. 2014;44:E172-E180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 43. | Chuaypen N, Posuwan N, Chittmittraprap S, Hirankarn N, Treeprasertsuk S, Tanaka Y, Shinkai N, Poovorawan Y, Tangkijvanich P. Predictive role of serum HBsAg and HBcrAg kinetics in patients with HBeAg-negative chronic hepatitis B receiving pegylated interferon-based therapy. Clin Microbiol Infect. 2018;24:306.e7-306.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 44. | Chuaypen N, Posuwan N, Payungporn S, Tanaka Y, Shinkai N, Poovorawan Y, Tangkijvanich P. Serum hepatitis B core-related antigen as a treatment predictor of pegylated interferon in patients with HBeAg-positive chronic hepatitis B. Liver Int. 2016;36:827-836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 45. | Matsumoto A, Nishiguchi S, Enomoto H, Kang JH, Tanaka Y, Shinkai N, Kurosaki M, Enomoto M, Kanda T, Yokosuka O, Yatsuhashi H, Nagaoka S, Okuse C, Kagawa T, Mine T, Takaguchi K, Saito S, Hino K, Ikeda F, Sakisaka S, Morihara D, Miyase S, Tsuge M, Chayama K, Hiramatsu N, Suzuki Y, Murata K, Tanaka E. Combinational use of hepatitis B viral antigens predicts responses to nucleos(t)ide analogue/peg-interferon sequential therapy. J Gastroenterol. 2018;53:247-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 46. | Kumada H, Okanoue T, Onji M, Moriwaki H, Izumi N, Tanaka E, Chayama K, Sakisaka S, Takehara T, Oketani M, Suzuki F, Toyota J, Nomura H, Yoshioka K, Seike M, Yotsuyanagi H, Ueno Y; Study Group for the Standardization of Treatment of Viral Hepatitis Including Cirrhosis, Ministry of Health, Labor and Welfare of Japan. Guidelines for the treatment of chronic hepatitis and cirrhosis due to hepatitis B virus infection for the fiscal year 2008 in Japan. Hepatol Res. 2010;40:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 47. | Hume SJ, Wong DK, Yuen MF, Jackson K, Bonanzinga S, Vogrin S, Hall SAL, Burns GS, Desmond PV, Sundararajan V, Ratnam D, Levy MT, Lubel JS, Nicoll AJ, Strasser SI, Sievert W, Ngu MC, Sinclair M, Meredith C, Matthews G, Revill PA, Littlejohn M, Bowden S, Visvanathan K, Holmes JA, Thompson AJ. High end-of-treatment hepatitis B core-related antigen levels predict hepatitis flare after stopping nucleot(s)ide analogue therapy. Liver Int. 2024;44:2605-2614. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 48. | Tsai YN, Wu JL, Tseng CH, Chen TH, Wu YL, Chen CC, Fang YJ, Yang TH, Nguyen MH, Lin JT, Hsu YC. Hepatitis B core-related antigen dynamics and risk of subsequent clinical relapses after nucleos(t)ide analog cessation. Clin Mol Hepatol. 2024;30:98-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 49. | Bortolotti F, Guido M, Bartolacci S, Cadrobbi P, Crivellaro C, Noventa F, Morsica G, Moriondo M, Gatta A. Chronic hepatitis B in children after e antigen seroclearance: final report of a 29-year longitudinal study. Hepatology. 2006;43:556-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 162] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 50. | Hui CK, Leung N, Shek TW, Yao H, Lee WK, Lai JY, Lai ST, Wong WM, Lai LS, Poon RT, Lo CM, Fan ST, Lau GK; Hong Kong Liver Fibrosis Study Group. Sustained disease remission after spontaneous HBeAg seroconversion is associated with reduction in fibrosis progression in chronic hepatitis B Chinese patients. Hepatology. 2007;46:690-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 51. | Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, Huang GT, Iloeje UH; REVEAL-HBV Study Group. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2309] [Cited by in RCA: 2365] [Article Influence: 124.5] [Reference Citation Analysis (0)] |
| 52. | Arends P, Sonneveld MJ, Zoutendijk R, Carey I, Brown A, Fasano M, Mutimer D, Deterding K, Reijnders JG, Oo Y, Petersen J, van Bömmel F, de Knegt RJ, Santantonio T, Berg T, Welzel TM, Wedemeyer H, Buti M, Pradat P, Zoulim F, Hansen B, Janssen HL; VIRGIL Surveillance Study Group. Entecavir treatment does not eliminate the risk of hepatocellular carcinoma in chronic hepatitis B: limited role for risk scores in Caucasians. Gut. 2015;64:1289-1295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 167] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 53. | Marcellin P, Gane E, Buti M, Afdhal N, Sievert W, Jacobson IM, Washington MK, Germanidis G, Flaherty JF, Aguilar Schall R, Bornstein JD, Kitrinos KM, Subramanian GM, McHutchison JG, Heathcote EJ. Regression of cirrhosis during treatment with tenofovir disoproxil fumarate for chronic hepatitis B: a 5-year open-label follow-up study. Lancet. 2013;381:468-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1228] [Cited by in RCA: 1369] [Article Influence: 114.1] [Reference Citation Analysis (0)] |
| 54. | Ma H, Yang RF, Li XH, Jin Q, Wei L. HBcrAg Identifies Patients Failing to Achieve HBeAg Seroconversion Treated with Pegylated Interferon Alfa-2b. Chin Med J (Engl). 2016;129:2212-2219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 55. | Song G, Yang R, Rao H, Feng B, Ma H, Jin Q, Wei L. Serum HBV core-related antigen is a good predictor for spontaneous HBeAg seroconversion in chronic hepatitis B patients. J Med Virol. 2017;89:463-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 56. | Xie Y, Ma H, Feng B, Song G. Combining the HBcrAg decline and HBV mutations predicts spontaneous HBeAg seroconversion in chronic hepatitis B patients during the immune clearance phase. J Med Virol. 2022;94:2694-2701. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 57. | Wang B, Carey I, Bruce M, Montague S, Dusheiko G, Agarwal K. HBsAg and HBcrAg as predictors of HBeAg seroconversion in HBeAg-positive patients treated with nucleos(t)ide analogues. J Viral Hepat. 2018;25:886-893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 58. | Sonneveld MJ, van Oord GW, van Campenhout MJ, De Man RA, Janssen HLA, de Knegt RJ, Boonstra A, van der Eijk AA. Relationship between hepatitis B core-related antigen levels and sustained HBeAg seroconversion in patients treated with nucleo(s)tide analogues. J Viral Hepat. 2019;26:828-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 59. | Lee HA, Lee HW, Park Y, Kim HS, Seo YS. Hepatitis B Core-Related Antigen Is Useful for Predicting Phase and Prognosis of Hepatitis B e Antigen-Positive Patients. J Clin Med. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 60. | Hwang SY, Yoo SH, Chang HY, Kim S, Lee JI, Lee KS, Cho YY, Joon KH, Lee HW. Baseline and on-treatment HBcrAg levels as predictors of HBeAg seroconversion in chronic hepatitis B patients treated with antivirals. J Viral Hepat. 2023;30:39-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 61. | Wang ML, Liao J, Wei B, Zhang DM, He M, Tao MC, Chen EQ, Tang H. Comparison of hepatitis B virus core-related antigen and hepatitis B surface antigen for predicting HBeAg seroconversion in chronic hepatitis B patients with pegylated interferon therapy. Infect Dis (Lond). 2018;50:522-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 62. | Tseng TC, Liu CJ, Hsu CY, Hong CM, Su TH, Yang WT, Chen CL, Yang HC, Huang YT, Fang-Tzu Kuo S, Liu CH, Chen PJ, Chen DS, Kao JH. High Level of Hepatitis B Core-Related Antigen Associated With Increased Risk of Hepatocellular Carcinoma in Patients With Chronic HBV Infection of Intermediate Viral Load. Gastroenterology. 2019;157:1518-1529.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 63. | Ghany MG, King WC, Lisker-Melman M, Lok ASF, Terrault N, Janssen HLA, Khalili M, Chung RT, Lee WM, Lau DTY, Cloherty GA, Sterling RK. Comparison of HBV RNA and Hepatitis B Core Related Antigen With Conventional HBV Markers Among Untreated Adults With Chronic Hepatitis B in North America. Hepatology. 2021;74:2395-2409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 64. | Gan W, Gao N, Gu L, Mo Z, Pang X, Lei Z, Gao Z. Reduction in Intrahepatic cccDNA and Integration of HBV in Chronic Hepatitis B Patients with a Functional Cure. J Clin Transl Hepatol. 2023;11:314-322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 65. | Ma G, Lou B, Lv F, Zhao D, Chen H, Ye X, Chen Y. HBcrAg, pg RNA and HBsAg dynamically supervise the seroconversion of HBsAg with anti-viral therapy: "Loss of HBsAg" maybe not a good end-point of anti-viral therapy. Clin Chim Acta. 2020;501:264-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 66. | Mak LY, Wong D, Kuchta A, Hilfiker M, Hamilton A, Chow N, Mao X, Seto WK, Yuen MF. Hepatitis B virus pre-genomic RNA and hepatitis B core-related antigen reductions at week 4 predict favourable hepatitis B surface antigen response upon long-term nucleos(t)ide analogue in chronic hepatitis B. Clin Mol Hepatol. 2023;29:146-162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 67. | Huang PY, Wang JH, Hung CH, Lu SN, Hu TH, Chen CH. The role of hepatitis B virus core-related antigen in predicting hepatitis B virus relapse after cessation of entecavir in hepatitis B e antigen-negative patients. J Viral Hepat. 2021;28:1141-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 68. | Kuo YH, Wang JH, Hung CH, Lu SN, Hu TH, Chen CH. Combining end-of-treatment HBsAg and baseline hepatitis B core-related antigen reduce HBV relapse rate after tenofovir cessation. Hepatol Int. 2021;15:301-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 69. | Sonneveld MJ, Chiu SM, Park JY, Brakenhoff SM, Kaewdech A, Seto WK, Tanaka Y, Carey I, Papatheodoridi M, Colombatto P, van Bömmel F, Berg T, Zoulim F, Ahn SH, Dalekos GN, Erler NS, Brunetto M, Wedemeyer H, Cornberg M, Yuen MF, Agarwal K, Boonstra A, Buti M, Piratvisuth T, Papatheodoridis G, Chen CH, Maasoumy B; CREATE study group. Lower pretreatment HBV DNA levels are associated with better off-treatment outcomes after nucleo(s)tide analogue withdrawal in patients with HBeAg-negative chronic hepatitis B: A multicentre cohort study. JHEP Rep. 2023;5:100790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Reference Citation Analysis (0)] |
| 70. | Xie Y, Li M, Ou X, Zheng S, Gao Y, Xu X, Yang Y, Ma A, Li J, Nan Y, Zheng H, Liu J, Wei L, Feng B. Lower end of treatment HBsAg and HBcrAg were associated with HBsAg loss after nucleos(t)ide analog cessation. BMC Gastroenterol. 2023;23:224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 71. | Huang D, Wu D, Wang P, Wang Y, Yuan W, Hu D, Hu J, Wang Y, Tao R, Xiao F, Zhang X, Wang X, Han M, Luo X, Yan W, Ning Q. End-of-treatment HBcrAg and HBsAb levels identify durable functional cure after Peg-IFN-based therapy in patients with CHB. J Hepatol. 2022;77:42-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 55] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 72. | Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 695] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 73. | Hong X, Kim ES, Guo H. Epigenetic regulation of hepatitis B virus covalently closed circular DNA: Implications for epigenetic therapy against chronic hepatitis B. Hepatology. 2017;66:2066-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 156] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 74. | Martinez MG, Boyd A, Combe E, Testoni B, Zoulim F. Covalently closed circular DNA: The ultimate therapeutic target for curing HBV infections. J Hepatol. 2021;75:706-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 104] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 75. | Boyd A, Lacombe K, Lavocat F, Maylin S, Miailhes P, Lascoux-Combe C, Delaugerre C, Girard PM, Zoulim F. Decay of ccc-DNA marks persistence of intrahepatic viral DNA synthesis under tenofovir in HIV-HBV co-infected patients. J Hepatol. 2016;65:683-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 76. | Zoulim F, Testoni B. Eliminating cccDNA to cure hepatitis B virus infection. J Hepatol. 2023;78:677-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 77. | Chen EQ, Feng S, Wang ML, Liang LB, Zhou LY, Du LY, Yan LB, Tao CM, Tang H. Serum hepatitis B core-related antigen is a satisfactory surrogate marker of intrahepatic covalently closed circular DNA in chronic hepatitis B. Sci Rep. 2017;7:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 78. | Wang X, Chi X, Wu R, Xu H, Gao X, Yu L, Liu L, Zhang M, Tan Y, Niu J, Jin Q. Serum HBV RNA correlated with intrahepatic cccDNA more strongly than other HBV markers during peg-interferon treatment. Virol J. 2021;18:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 79. | Mak LY, Seto WK, Fung J, Yuen MF. New Biomarkers of Chronic Hepatitis B. Gut Liver. 2019;13:589-595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 80. | Huang Q, Zhou B, Cai D, Zong Y, Wu Y, Liu S, Mercier A, Guo H, Hou J, Colonno R, Sun J. Rapid Turnover of Hepatitis B Virus Covalently Closed Circular DNA Indicated by Monitoring Emergence and Reversion of Signature-Mutation in Treated Chronic Hepatitis B Patients. Hepatology. 2021;73:41-52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 81. | Jin Y, Wang S, Xu S, Zhao S, Xu X, Poongavanam V, Menéndez-Arias L, Zhan P, Liu X. Targeting hepatitis B virus cccDNA levels: Recent progress in seeking small molecule drug candidates. Drug Discov Today. 2023;28:103617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 82. | Erken R, Andre P, Roy E, Kootstra N, Barzic N, Girma H, Laveille C, Radreau-Pierini P, Darteil R, Vonderscher J, Scalfaro P, Tangkijvanich P, Flisiak R, Reesink H. Farnesoid X receptor agonist for the treatment of chronic hepatitis B: A safety study. J Viral Hepat. 2021;28:1690-1698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 83. | Gane EJ, Kim W, Lim TH, Tangkijvanich P, Yoon JH, Sievert W, Sukeepaisarnjaroen W, Thompson AJ, Pavlovic V, Surujbally B, Wat C, Brown BD, Achneck HE, Yuen MF. First-in-human randomized study of RNAi therapeutic RG6346 for chronic hepatitis B virus infection. J Hepatol. 2023;79:1139-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 84. | World Health Organization. Hepatitis B. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b. |
| 85. | Nguyen MH, Wong G, Gane E, Kao JH, Dusheiko G. Hepatitis B Virus: Advances in Prevention, Diagnosis, and Therapy. Clin Microbiol Rev. 2020;33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 310] [Article Influence: 62.0] [Reference Citation Analysis (0)] |
| 86. | Luo H, Zhang XX, Cao LH, Tan N, Kang Q, Xi HL, Yu M, Xu XY. Serum hepatitis B virus RNA is a predictor of HBeAg seroconversion and virological response with entecavir treatment in chronic hepatitis B patients. World J Gastroenterol. 2019;25:719-728. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (2)] |
| 87. | de Niet A, Jansen L, Stelma F, Willemse SB, Kuiken SD, Weijer S, van Nieuwkerk CMJ, Zaaijer HL, Molenkamp R, Takkenberg RB, Koot M, Verheij J, Beuers U, Reesink HW. Peg-interferon plus nucleotide analogue treatment versus no treatment in patients with chronic hepatitis B with a low viral load: a randomised controlled, open-label trial. Lancet Gastroenterol Hepatol. 2017;2:576-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |