Published online Jan 27, 2024. doi: 10.4254/wjh.v16.i1.75
Peer-review started: November 3, 2023
First decision: December 1, 2023
Revised: December 11, 2023
Accepted: January 2, 2024
Article in press: January 2, 2024
Published online: January 27, 2024
Processing time: 80 Days and 10.8 Hours
Prevalence of hepatocellular carcinoma (HCC) is increasing, especially in patients with metabolic dysfunction-associated steatotic liver disease (MASLD).
To investigate rifaximin (RIF) effects on epigenetic/autophagy markers in animals.
Adult Sprague-Dawley rats were randomly assigned (n = 8, each) and treated from 5-16 wk: Control [standard diet, water plus gavage with vehicle (Veh)], HCC [high-fat choline deficient diet (HFCD), diethylnitrosamine (DEN) in drinking water and Veh gavage], and RIF [HFCD, DEN and RIF (50 mg/kg/d) gavage]. Gene expression of epigenetic/autophagy markers and circulating miRNAs were obtained.
All HCC and RIF animals developed metabolic-dysfunction associated steatohepatitis fibrosis, and cirrhosis, but three RIF-group did not develop HCC. Comparing animals who developed HCC with those who did not, miR-122, miR-34a, tubulin alpha-1c (Tuba-1c), metalloproteinases-2 (Mmp2), and metalloproteinases-9 (Mmp9) were significantly higher in the HCC-group. The opposite occurred with Becn1, coactivator associated arginine methyltransferase-1 (Carm1), enhancer of zeste homolog-2
RIF might have a possible beneficial effect on preventing/delaying liver carcinogenesis through epigenetic modulation in a rat model of MASLD-HCC.
Core Tip: Managing metabolic dysfunction-associated steatotic liver disease (MASLD)-hepatocellular carcinoma (HCC) is a clinical challenge, with many unanswered questions, as autophagy and epigenetics appear to contribute to drug resistance. Additionally, the broad-spectrum oral antibiotic Rifaximin influences inflammation, energy metabolism, and fat storage. Utilizing animal models for MASLD-HCC is crucial in understanding pathophysiological mechanisms and potential therapeutic targets. Furthermore, Rifaximin may have a beneficial effect in rats by possibly preventing or delaying hepatic carcinogenesis through epigenetic modulation.
- Citation: Michalczuk MT, Longo L, Keingeski MB, Basso BS, Guerreiro GTS, Ferrari JT, Vargas JE, Oliveira CP, Uribe-Cruz C, Cerski CTS, Filippi-Chiela E, Álvares-da-Silva MR. Rifaximin on epigenetics and autophagy in animal model of hepatocellular carcinoma secondary to metabolic-dysfunction associated steatotic liver disease. World J Hepatol 2024; 16(1): 75-90
- URL: https://www.wjgnet.com/1948-5182/full/v16/i1/75.htm
- DOI: https://dx.doi.org/10.4254/wjh.v16.i1.75
Metabolic dysfunction-associated steatotic liver disease (MASLD) comprises a spectrum of histological abnormalities, ranging from isolated steatosis to steatohepatitis, characterized by inflammation, necrosis, and hepatocellular ballooning, and progression to fibrosis, cirrhosis, liver failure and/or hepatocellular carcinoma (HCC)[1,2]. Along with diet and sedentary lifestyle, many other factors often determine the progression of MASLD and the development of MASLD-associated carcinogenesis[3]. Although only 2.4% to 12.4% of cirrhotic MASLD patients develop HCC, it is expected that it will become the leading HCC cause by 2030[4,5].
Several MASLD-associated oncogenesis mechanisms, such as structural genomic defects, epigenetic alterations and autophagy promote significant changes in regulatory and signaling pathways, creating a favorable hepatic microenvironment for lesion progression and HCC development, which can occur with or without cirrhosis[6-8]. Among the epigenetic mechanisms, microRNAS act on the expression or suppression of genes responsible for the worsening of liver damage, that is, they have the potential to be used as new biomarkers for early diagnosis and/or therapeutic targets to HCC[8-10]. Autophagy is recognized for playing a beneficial role in the initial liver injury by contributing to the removal of protein aggregates, damaged organelles, and lipid droplets, preventing the formation of pre-tumor cells[6,7]. However, during tumor promotion, autophagy predominantly acts on the adaptive mechanism, contributing to tumor maintenance and growth through the supply of energy substrates and metabolic adaptation, which improves their survival ability in hypoxic and low-nutrient environments, promoting tumor progression[6,11]. Autophagy tends to play a complicit role in HCC treatment resistance, the interface between these two processes being multifactorial and therefore crosstalk can occur in different target proteins. In this sense, therapies for the epigenetic control of autophagy are promising targets for treatment of liver injury[6,11].
Currently, there is no approved pharmacological therapy for steatohepatitis, and several studies are being conducted with different targets. Dietary management, such as flavonoids, has shown beneficial effects on the epigenetic mechanisms of MASLD-HCC[12]. On the other hand, although microbiota plays an important role in MASLD patho
Twenty-four adult (60-d-old) male Sprague Dawley rats weighing 250–400 g was included for this study. The animals were group-housed in polypropylene cages with sawdust-covered floors. Rats were maintained on a standard 12-h light/dark cycle, in a temperature-controlled environment (22 ± 2 °C). The Institutional Ethics Committee approved all experiments and procedures for the Use of Animals, No. 2021-0105. The procedures for scientific animal’s use were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (8th ed, 2011) and law No. 11.794 (Brazil, 2008).
After acclimatization to the environment, the animals were randomized by weight into a control group (n = 8) that received a standard diet, water free of diethylnitrosamine (DEN, catalog number N0756, Sigma-Aldrich, United States) and gavage with vehicle (Veh) solution from the 5th week of the experiment until the 16th week; HCC-group (n = 8) that received a high-fat and choline-deficient diet (HFCD, catalog number RH19576, Rhoster, Brazil), 135 mg/L DEN in drinking water and gavage with Veh solution, during the same period previously described; and the RIF-group (n = 8) that received HFCD diet plus DEN and RIF (catalog number R9904, Sigma-Aldrich, United States) by gavage from the 5th week of the experiment until the 16th week of the experiment. The DEN dose and the experimental period for the development of this protocol were based on a previous study, which combine a dietary model of fatty liver based on high trans-fat with exposure to a known hepatic carcinogen as a means of provoking and accelerating more severe injury. The model replicated many features of MASLD including steatohepatitis with ballooning, fibrosis, cirrhosis, and HCC[17]. The experimental study design is presented in Figure 1. The animals were weighed twice a week during the experimental period. After 16 wk of the experiment, all the animals were euthanized by cardiac exsanguination. Serum samples and liver fragments were collected aseptically, frozen in liquid nitrogen, and stored in an ultra-freezer at -80 °C until the experimental procedures were carried out. A portion of each liver sample was fixed in 10% formalin for histological analysis.
Animals in the control group received a standard rodent diet (Nuvilab CR-1; Quimtia SA, Brazil) with an energy value of 2.93 kcal/g. The diet consisted of 55.0% carbohydrates, 22.0% protein, 4.5% fat, and 18.5% of other nutrients such as fibers and vitamins. Animals in the intervention groups received an HFCD diet with an energy value of 4.3 kcal/g. This product consisted of 54.5% carbohydrates, 14.0% protein, and 31.5% fat (enriched with 54.0% trans fatty acids). The diet of the intervention group was chosen as such because it mirrors many of the phenotypes observed in humans with MASLD, as previously demonstrated by our research group[18]. The diet offered to the animals in the control and intervention groups was replaced every two days. Both the groups received water and food ad libitum during the experimental period.
The therapeutic intervention through the administration of daily gavage with RIF or the offer of Veh solution, also by gavage, in the respective experimental groups occurred daily from the 5th week of the experiment until the date of euthanasia. The animals in the control group and HCC-group received daily gavage with a Veh solution (0.5 mL/kg distilled water) in order to undergo the same stress conditions as those in the RIF-group. The RIF-group received a daily dose of 50 mg/kg/d of RIF by daily gavage from the 5th week of the experiment until the 16th week of the experiment. The dose of RIF used was in accordance with a previous study published in the literature[19].
The total RNA was extracted from fragments of the liver tissue using TRIzol (catalog number 15596026, Invitrogen, United States). The cDNA conversion, from 2 μg of RNA, was performed using the High-capacity cDNA Reverse Transcription kit (catalog number 4368814, Applied Biosystems, United States). Quantitative (q) real-time polymerase chain reaction (RT-PCR) with TaqMan assay (Applied Biosystems, United States) was used to assess the gene expression of aldolase-B (Aldob), tubulin alpha-1c (Tuba1c), alpha-fetoprotein (Afp), metalloproteinases-2 (Mmp2), Mmp9, autophagy-related factor LC3A/B (Map1 Lc3b), p62/sequestosome-1 (p62/Sqstm1), beclin-1 (Becn1), enhancer of zeste homolog-2
To analyze the circulating microRNAs from serum, total RNA was extracted using the miRNeasy serum/plasma kit (catalog number 217184, Qiagen, United States). Then, cel-miR-39 (1.6 × 108 copies) spike in control (catalog number 21961, Qiagen, United States) was added to provide an internal reference. cDNA conversion was performed from 10 ng of total RNA using the TaqMan microRNA Reverse Transcription kit (catalog number 4366597, Applied Biosystems, United States). Analysis of the gene expression of miR-122, miR-34a, miR-26b, miR-224, miR-33a, miR-143, miR-155, miR-375 and miR-21, together with the cell-miR-39 normalizer, was performed by RT-qPCR using TaqMan assay (Applied Biosystems, United States). The sequences and codes of the assessed microRNAs are described in Supplementary Table 1. Values were calculated by formula 2-ΔΔCt.
For western blotting analysis, protein extraction was performed on samples of liver tissue from rats. The samples were homogenized in a solution containing Triton X-100, β-mercaptoethanol, tris-buffered saline (TTBS), ethylenediaminetetraacetic acid and proteases inhibitor cocktail (catalog number 29131, Santa Cruz Biotechnology, United States). The samples were then normalized to 40 µg of protein. Subsequently, proteins were separated by electrophoresis using a 12% w/v polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was washed with TTBS, and then incubated for one hour in a blocking solution containing 3% bovine serum albumin in TTBS. Following the blocking step, the membrane was washed three times with TTBS and incubated overnight at 4 °C in a blocking solution containing the following primary antibodies: Anti-actin (catalog number A5060, Sigma-Aldrich, United States), anti-LC3B (catalog number ab128025, Abcam, United Kingdom), and anti-SQSTM1 (catalog number ab56416, Abcam, United Kingdom). The primary antibodies were used at a dilution of 1:1000. After the overnight incubation, the membrane was washed three times with TTBS and then incubated for two hours in a solution containing a horseradish peroxidase-conjugated anti-IgG secondary antibody in TBBS, at a concentration of 1:2000. For band detection, Clarity Western ECL Substrate (catalog number 1705062, BioRad, United States) was used, and the resulting image was captured using an ImageQuant LAS 500 (GE Healthcare Lifesciences) imaging system. Band intensities were quantified using the ImageJ software, and actin was used as a constitutive protein reference.
For this analysis, in relation to markers related to hepatocarcinogenesis we selected Mmp2 Mmp9, Afp, Tuba1c and Aldob and associated with the autophagy and epigenetic process we selected Becn1, p62/Sqstm1, Map1 Lc3b, Ezh2, Carm1, p62 (protein ratio), and LC3B (protein ratio). Regarding microRNAs, we selected miR-122, miR-26b, miR-224 and miR-34a, which are markers related to liver disease and which showed a significant difference between the experimental groups.
Formalin-fixed liver tissue samples were embedded in paraffin and stained with hematoxylin and eosin (H&E) and picrosirius red. Histopathological lesions of the different evolutionary stages of nonalcoholic fatty liver disease were assessed according to the score by Liang et al[20], which is a highly reproducible scoring system applicable to experimental rodent models[20]. This scoring system was developed through the assessment of various experimental models, aiming to establish generic criteria for analysis. To validate the proposed scoring, biological material from rodents was evaluated by blinded pathologists in two separate assessments, with an interval of over 3 mo between them. In this validation, observers estimated the percentage of macrovesicular steatosis, microvesicular steatosis, hypertrophy, and the number of inflammatory foci per field[20]. The degree of fibrosis was evaluated using the slides stained with picrosirius red, and cancerous lesions were graded according to the Edmondson & Steiner classification[21]. The analysis was performed by an experienced pathologist, who was blinded to the experimental groups.
The sample size estimation was performed using the WINPEPI 11.20 software (Brixton Health, Israel), based on a previously published study by the research group demonstrating HCC development in an experimental model[17]. Considering a power of 80% and a significance level of 5%, it was determined that 8 animals per experimental group would be required for conducting this study. The outcome used for the calculation was the prevention of HCC.
Normality was verified for all variables using Shapiro-Wilk test and histograms. Nonparametric data were analyzed using the Kruskal-Wallis followed by Dunn test. Quantitative variables were expressed as median and interquartile ranges (25th–75th) and percentage. Spearman's correlation coefficient was performed, with a moderate (0.3 < r < 0.6), strong (0.6 < r < 0.9) or very strong (0.9 < r < 1.0) correlation were adopted. Statistical significance was set at P < 0.05. Data were analyzed using the Statistical Package for Social Sciences (version 28.0; SPSS Inc., United States).
During the 16 wk of the experiment, one animal from the HCC-group and another from the RIF-group died, totaling 7 animals per experimental group. There were no deaths in the control group (n = 8) during the study period.
The data of gene expression of hepatocarcinogenesis markers are shown in Table 1. There was no difference between the experimental groups for the gene expression of Tuba1c (P = 0.839), Aldob (P = 0.595), Afp (P = 0.837) and Mmp2 (P = 0.101). There was a significant difference between the experimental groups in the gene expression of Mmp9 (P = 0.035). For this marker, there was a significant increase in its expression in the HCC-group compared to the control group (P = 0.013). However, there was no significant difference in the expression of Mmp9 between the HCC-group and RIF-group (P = 0.071).
| Variables1 | Control (n = 8) | HCC (n = 7) | RIF (n = 7) | P value |
| Tuba1c | 1.28 (0.18-2.76) | 4.27 (0.00-9.69) | 1.38 (0.29-19.8) | 0.839 |
| Aldob | 2.46 (0.01-5.78) | 4.24 (0.00-9.92) | 0.12 (0.00-2.69) | 0.595 |
| Afp | 1.28 (0.18-2.76) | 2.33 (0.00-9.69) | 1.38 (0.29-19.8) | 0.837 |
| Mmp2 | 1.81 (0.03-5.15) | 31.4 (0.25-62.2) | 0.92 (0.16-18.4) | 0.101 |
| Mmp9 | 2.36 (0.02-4.25)a | 23.6 (2.69-76.8)b | 14.6 (1.32-28.7)a,b | 0.035 |
| miR-122 | 1.14 (0.34-2.34)a | 7.07 (1.41-11.8)b | 2.62 (1.20-6.00)a,b | 0.005 |
| miR-34a | 0.94 (0.63-1.70)b | 0.39 (0.33-0.44)a | 0.33 (0.13-0.89)a | 0.007 |
| miR-26b | 1.04 (0.61-1.57)a, b | 0.36 (0.21-1.85)a | 2.22 (1.46-2.96)b | 0.026 |
| miR-224 | 1.05 (0.41-2.45)a, b | 2.97 (1.96-5.94)b | 0.58 (0.13-0.85)a | 0.005 |
| miR-33a | 0.98 (0.53 – 2.16) | 0.60 (0.54-1.17) | 1.4 (0.70-1.60) | 0.445 |
| miR-143 | 1.18 (0.25-3.67) | 1.40 (0.54-5.16) | 0.62 (0.17-2.54) | 0.471 |
| miR-155 | 1.46 (0.32-1.96) | 0.63 (0.01-1.18) | 0.34 (0.04-1.46) | 0.074 |
| miR-375 | 0.79 (0.61-2.42) | 0.85 (0.17-1.09) | 0.38 (0.14-1.76) | 0.216 |
| miR-21 | 1.12 (0.34-2.13) | 1.49 (0.91-6.08) | 1.69 (0.84-3.68) | 0.188 |
Data obtained for the gene expression of markers of autophagy are shown in Figure 2. There was a significant decrease in Map1 Lc3b gene expression in the HCC and RIF-groups compared to the control group (P = 0.002 and P = 0.001, respectively). There was no significant difference in p62/Sqstm1 expression in the RIF-group compared to the control (P = 0.078) and HCC-group (P = 0.890); however, the HCC-group showed a significant decrease in its expression compared to the control (P = 0.010). Becn1 and Ezh2 showed a significant decrease in their expression in the HCC-group (P = 0.001 and P < 0.001, respectively) and RIF-group (P = 0.009 and P = 0.010, respectively) compared to the control. There was a significant reduction in Carm1 expression in the HCC-group compared to the RIF-group (P = 0.004).
The results obtained from the gene expression of the circulating microRNAs related to liver damage are demonstrated in Table 1. There was a significant increase in miR-122 gene expression in the HCC-group compared to the control group (P < 0.001), however the RIF-group did not differ significantly from the control (P = 0.118). There was a significant difference between the experimental groups in the gene expression of miR-34a (P = 0.007). The expression of miR-34a was significantly lower in the RIF-group compared to the control (P = 0.005). There was a significant difference between the experimental groups in the gene expression of miR-26b (P = 0.026). The HCC-group showed a significant decrease in miR-26b expression compared to the RIF-group (P = 0.008), which was like the control (P = 0.469). The gene expression of miR-224 was significantly higher in the HCC-group compared to the RIF-group (P < 0.001), which is like the control group (P = 0.099). There was no significant difference between groups in the gene expression of miR-33a (P = 0.445), miR-143 (P = 0.471), miR-155 (P = 0.074), miR-375 (P = 0.216) and miR-21 (P = 0.188).
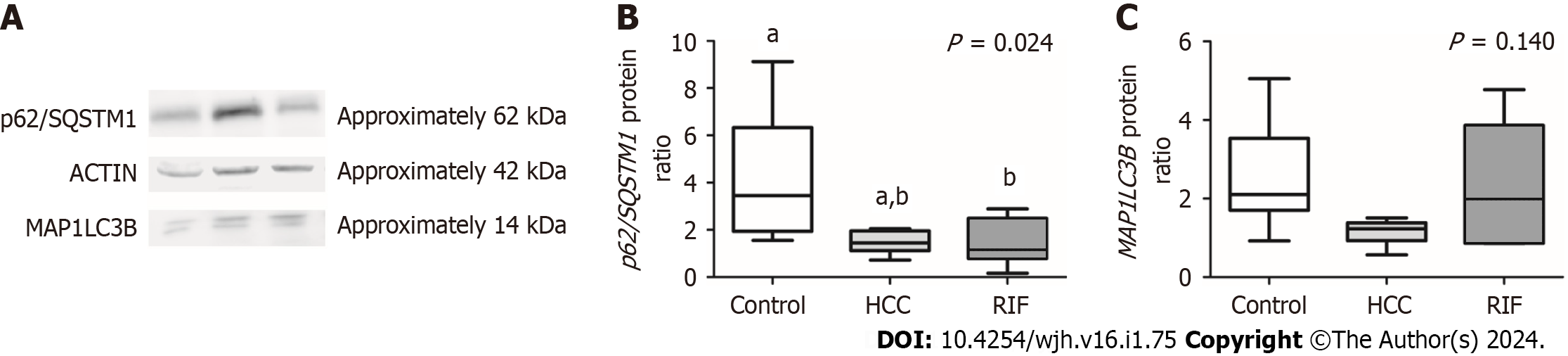
The data obtained from the protein expression analysis of p62/SQSTM1 and MAP1LC3B, conducted through the Western blot technique, are represented in Figure 3. A difference in the expression of the MAP1LC3B protein between the experimental groups was observed (P = 0.024). A significant decrease in its expression was noted in the RIF-group animals compared to the control group (P = 0.039). No significant differences in MAP1LC3B expression were found between the animals receiving RIF treatment and the animals in the HCC-group (P > 0.05). Regarding the protein expression of p62/SQSTM1, no significant differences were observed between the experimental groups (P = 0.140).
The values obtained from the correlations between markers of hepatocarcinogenesis, autophagy and epigenetic are described in Table 2. There was a negative correlation between the expression of miR-122 and miR-34a, both related to the severity of liver injury compared to the autophagy markers Map1 Lc3b, p62/Sqstm1 and Becn1 that act in the development of the autophagosome. Additionally, this negative correlation was also demonstrated between these microRNAs and Ezh2, an enzyme that modulates the expression of autophagy genes. There was a positive correlation between the three autophagy indicators (Map1 Lc3b, p62/Sqstm1 and Becn1), however there was no correlation between Ezh2 and Carm1, demonstrating that these enzymes that perform epigenetic control of autophagy act in different ways. Ezh2 positively correlated with Map1 Lc3b, p62/Sqstm1 and Becn1. While the correlations were strong between the autophagy markers Map1 Lc3b, p62/Sqstm1 and Ezh2, it was only moderate with Becn1. As Ezh2 can catalyze the methylation of lysine to histone (H3K27), which can decrease the expression of target genes, an attempt was made to assess whether Becn1 would be on the target of Ezh2-H3K27 axis. The result was described in Supplementary Figure 1. Metalloproteinases were positively correlated with the expression of Afp and Tuba1c, markers related to hepatocarcinogenesis. Aldob expression was positively correlated with Mmp2, Afp and Tuba1c. The protein expression of p62 correlated positively with the gene expression of the markers Becn1, p62/Sqstm1, and Map1 Lc3b.
| Variables1 | microRNAs | Hepatocarcinogenesis | Autophagy and Epigenetic | |||||||||||||
| miR-26b | miR-224 | miR-34a | Mmp2 | Mmp9 | Afp | Tuba1c | Aldob | Becn1 | p62/Sqstm1 | Map1lc3b | Ezh2 | Carm1 | p62 protein | MAP1LC3B protein | ||
| microRNAs | miR-122 | -0.235 | 0.075 | 0.400a | 0.195 | 0.422 | -0.189 | 0.085 | 0.030 | -0.502a | -0.673b | -0.554a | -0.760b | -0.375 | -0.245 | 0.017 |
| miR-26b | -0.360 | 0.538a | -0.071 | 0.203 | 0.199 | -0.090 | -0.113 | -0.089 | 0.164 | -0.047 | -0.005 | 0.525a | -0.124 | 0.186 | ||
| miR-224 | -0.195 | 0.041 | -0.241 | -0.201 | 0.025 | 0.153 | -0.298 | -0.022 | 0.032 | -0.061 | -0.335 | -0.007 | -0.393 | |||
| miR-34a | 0.110 | 0.426 | 0.123 | 0.110 | -0.127 | -0.461a | -0.340 | -0.612b | -0.584b | 0.181 | -0.400 | -0.191 | ||||
| Hepatocarcinogenesis | Mmp2 | 0.742b | 0.704b | 0.957b | 0.548b | -0.321 | -0.133 | -0.321 | -0.361 | -0.286 | -0.318 | -0.368 | ||||
| Mmp9 | 0.521a | 0.703b | 0.292 | -0.398 | -0.276 | -0.351 | -0.586b | -0.278 | -0.547a | -0.229 | ||||||
| Afp | 0.702b | 0.468a | -0.086 | 0.049 | -0.173 | 0.038 | -0.073 | -0.328 | -0.078 | |||||||
| Tuba1c | 0.644b | -0.324 | -0.058 | -0.281 | -0.350 | -0.282 | -0.258 | -0.246 | ||||||||
| Aldob | -0.137 | -0.042 | 0.186 | 0.111 | -0.314 | -0.032 | 0.174 | |||||||||
| Autophagy and Epigenetic | Becn1 | 0.541b | 0.771b | 0.597b | 0.433 | 0.631b | 0.302 | |||||||||
| p62/Sqstm1 | 0.657b | 0.609b | 0.459 | 0.554a | 0.265 | |||||||||||
| Map1lc3b | 0.718b | 0.230 | 0.512a | 0.343 | ||||||||||||
| Ezh2 | 0.288 | 0.342 | 0.253 | |||||||||||||
| Carm1 | 0.185 | -0.044 | ||||||||||||||
| p62 protein | 0.406 | |||||||||||||||
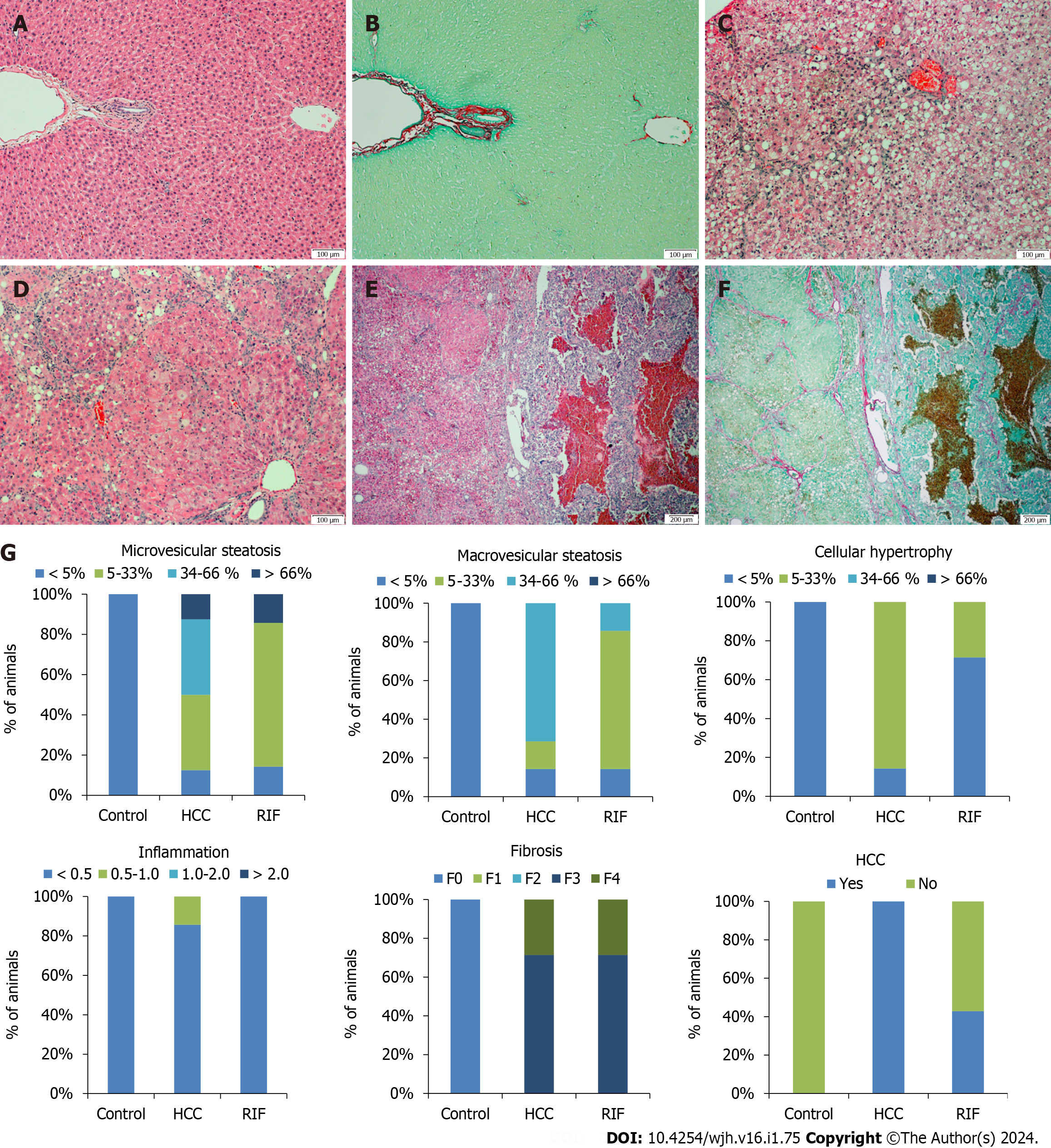
No hepatic histopathological changes were observed in the liver tissue of the control group (Figure 4A and B). The animals in the HCC-group had predominantly macrovesicular steatosis along with microvesicular steatosis of moderate intensity, with inflammatory activity and mild hypertrophy (Figure 4C and D). The RIF-group presented mild macrovesicular and microvesicular steatosis, no inflammation was observed and in only two animals the presence of hypertrophy was evidenced. In the staging of the histopathological lesion, seven animals in the HCC-group and RIF-group developed steatohepatitis (Table 3). In the evaluation of hepatic fibrosis, through H&E and picrosirius red staining respectively, five animals from the HCC-group and RIF-group developed fibrosis with multiple septa without the presence of cirrhosis and two animals from both experimental groups developed liver cirrhosis (Figure 4E and F). The breakdown by experimental group observed for microvesicular steatosis, macrovesicular steatosis, hypertrophy, inflammation, fibrosis, and HCC as a percentage is shown in Figure 4G.
| Variable1 | Control (n = 8) | HCC (n = 7) | RIF (n = 7) |
| No MASLD | 8 (100) | 0 (0.0) | 0 (0.0) |
| MASLD | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| MASH | 0 (0.0) | 7 (100) | 7 (100) |
In the evaluation of the tumor classification by the Edmondson & Steiner score, we reported a lesion grade three and four, corresponding to the presence of poorly differentiated and undifferentiated cancer, respectively, in the HCC-group. This finding was replicated for some animals in the RIF-group; however, three animals did not develop cancer.
Additionally, to better detail the results obtained between the autophagy and epigenetics markers with the hepatic histopathological results, we performed the subdivision of the experimental groups. Initially, we stratified the RIF-group, and performed a comparison analysis between the animals that developed HCC (n = 4) and those that did not develop HCC (n = 3). No significant differences (P > 0.05) were observed in gene expression of microRNAs, markers of hepatocarcinogenesis and autophagy between animals that developed or did not develop HCC in the RIF-group, as detailed in Table 4.
| Variables1 | Developed HCC (n = 4), median (min–max) | Did not developed HCC (n = 3), median (min–max) | P value |
| miR-122 | 2.27 (1.20–4.57) | 2.97 (1.51–5.98) | 0.571 |
| miR-375 | 0.42 (0.14–1.76) | 0.33 (0.30–0.67) | 1.000 |
| miR-26b | 1.97 (1.10–3.38) | 2.41 (1.97–3.99) | 0.571 |
| miR-224 | 0.46 (0.13–0.85) | 0.58 (0.46–0.82) | 0.857 |
| miR-34 | 11.0 (7.17–18.9) | 10.0 (8.64–10.5) | 0.786 |
| miR-21 | 2.36 (0.84–3.68) | 1.68 (1.00–1.69) | 0.700 |
| miR-143 | 0.76 (0.17–2.53) | 0.48 (0.25–1.96) | 1.000 |
| miR-155 | 0.69 (0.06–1.46) | 0.13 (0.03–0.36) | 0.250 |
| miR-33a | 0.88 (0.52–1.86) | 1.55 (1.40–1.58) | 0.400 |
| Mm2 | 7.95 (0.58–18.4) | 0.71 (0.16–0.92) | 0.229 |
| Mm9 | 24.0 (1.32–28.7) | 8.48 (1.73–14.6) | 0.400 |
| Afp | 6.99 (1.38–19.8) | 0.46 (0.29–0.48) | 0.057 |
| Tuba1c | 4.63 (0.30–7.12) | 0.26 (0.25–0.30) | 0.057 |
| Aldob | 2.32 (0.11–2.69) | 0.07 (0.00–0.12) | 0.114 |
| Becn1 | 0.70 (0.67–0.85) | 0.74 (0.66–0.76) | 1.000 |
| p62/Sqstm1 | 0.84 (0.54–0.98) | 0.72 (0.64–0.79) | 0.629 |
| Map1lc3b | 0.24 (0.22–0.35) | 0.27 (0.21–0.29) | 1.000 |
| Ezh2 | 29.0 (28.0–30.1) | 27.5 (27.2–28.0) | 0.114 |
| Carm1 | 0.79 (0.77–1.67) | 1.23 (1.08–1.68) | 0.400 |
| p62/SQSTM1 protein | 1.07 (0.16–2.89) | 1.78 (1.19–2.36) | 0.355 |
| MAP1LC3B protein | 2.00 (0.87–4.78) | 2.20 (0.86–3.57) | 0.643 |
Subsequently, we carried out a new stratification of the experimental groups to compare the results obtained between the animals that developed HCC and those that did not develop HCC. The comparison of results is detailed in Table 5. In total, eleven animals developed HCC and eleven did not develop this clinical condition at the end of the study. Regarding epigenetic markers, we reported a significant increase in the gene expression of miR-122 (P = 0.029) and miR-34a (P = 0.012) in animals with HCC compared to animals without HCC. There was a significant increase in the gene expression of Mm2 (P = 0.017), Mm9 (P = 0.013) and Tuba1c (P = 0.017), markers related to hepatic hepatocarcinogenesis, in animals with HCC compared to those that did not develop HCC. Regarding autophagy markers, there was a significant reduction in the gene expression of Becn1 (P = 0.004), Map1 Lc3b (P = 0.004), Ezh2 (P = 0.010) and Carm1 (P = 0.026) in animals with HCC compared to animals who did not develop this clinical condition. Additionally, we demonstrated a significant decrease in the protein expression of p62/SQSTM1 (P = 0.013) in the animals with HCC compared to the animals without HCC in the RIF group.
| Variables1 | HCC (n = 11), median (min–max) | No HCC (n = 11), median (min–max) | P value |
| miR-122 | 4.49 (1.20–11.8) | 1.41 (0.34–5.98) | 0.029a |
| miR-375 | 0.64 (0.14–1.76) | 0.72 (0.30–2.42) | 0.512 |
| miR-26b | 1.10 (0.46–3.38) | 1.23 (0.59–3.99) | 0.656 |
| miR-224 | 1.40 (0.13–5.94) | 0.82 (0.41–2.45) | 0.545 |
| miR-34a | 7.73 (2.29–18.9) | 1.79 (0.14–10.5) | 0.012a |
| miR-21 | 1.92 (0.84–6.08) | 1.46 (0.34–2.13) | 0.109 |
| miR-143 | 1.08 (0.17–5.16) | 1.13 (0.25–3.67) | 0.756 |
| miR-155 | 0.66 (0.01–1.46) | 0.60 (0.03–1.95) | 0.349 |
| miR-33a | 0.66 (0.52–1.86) | 1.40 (0.34–2.64) | 0.200 |
| Mm2 | 18.4 (0.25–62.2) | 1.17 (0.03–5.15) | 0.017a |
| Mm9 | 23.6 (1.32–76.8) | 2.76 (0.02–14.6) | 0.013a |
| Afp | 3.23 (0.00–19.8) | 0.62 (0.18–2.76) | 0.190 |
| Tuba1c | 7.12 (0.12–27.8) | 0.76 (0.12–3.71) | 0.017a |
| Aldob | 2.32 (0.00–9.92) | 1.35 (0.00–5.78) | 0.549 |
| Becn1 | 0.68 (0.44–0.87) | 0.88 (0.66–1.39) | 0.004a |
| p62/Sqstm1 | 0.65 (0.32–1.06) | 0.92 (0.59–1.31) | 0.052 |
| Map1lc3b | 0.25 (0.19–0.37) | 0.86 (0.21–1.53) | 0.004a |
| Ezh2 | 28.4 (25.8–30.1) | 30.2 (27.2–31.8) | 0.010a |
| Carm1 | 0.69 (0.37–1.67) | 1.10 (0.56–1.68) | 0.026a |
| p62/SQSTM1 protein | 1.27 (0.16–2.39) | 2.87 (1.19–9.12) | 0.013a |
| MAP1LC3B protein | 1.29 (0.57–4.78) | 2.10 (0.86–5.05) | 0.183 |
In this study, carried out in an experimental model of HCC secondary to MASLD, with the objective of evaluating the effect of treatment with RIF in relation to autophagy and epigenetic markers, we demonstrated: (1) A positive correlation between Map1cl3b, p62/Sqstm1 and Becn1 in HCC, however, there was no significant difference in the expression of these markers between the HCC-group and RIF-group; (2) a lack of correlation between Ezh2 and Carm1, demonstrating that these enzymes regulate different signaling pathways, although RIF promoted a significant increase in Carm1 expression; (3) a significant increase in miR-26b expression in the RIF-group compared to the HCC-group; the inverse was observed for miR-224; (4) a negative correlation between the expression of miR-122 and miR-34a related to liver injury with Map1 Lc3b, p62/Sqstm1 and Becn1; and (5) finally, steatohepatitis developed in all animals of the HCC-group and RIF-group, but three animals treated with RIF did not develop HCC.
RIF, a semisynthetic derivative of rifamycin, has an intestinal absorption of approximately 0.007%, which prevents it from exerting systemic or adverse effects[13,16]. It has broad-spectrum in vitro activity against aerobic and anaerobic enteric bacteria and has been used in the treatment of hepatic encephalopathy, traveler's diarrhea, and irritable bowel syndrome[13,22]. Recent studies report that RIF inhibited hepatic fibrosis in rodent models of alcoholic liver injury and steatohepatitis[23,24]. In clinical practice, its use seems to be effective and safe in MASLD, promoting the reduction of serum endotoxemia, improvement of insulin resistance, inflammatory process, and histopathological score[13,16]. To our knowledge, no studies evaluate the effect of RIF on the progression from MASLD to HCC in relation to autophagy and epigenetic markers. We emphasize that the treatment with RIF in the animals started from the fifth week of the experiment when the animals probably already had steatohepatitis, as demonstrated in previous studies[17]. In the histopathological evaluation, all animals in the HCC and RIF groups developed steatohepatitis; however, three animals treated with RIF did not develop HCC. This interesting finding continues to be investigated by our research group and we seek to understand the role of RIF in this process. A multicenter, double-blind, randomized, placebo-controlled study in patients with biopsy-proven steatohepatitis investigated the effects of daily administration of RIF for six months[13]. The results obtained demonstrated that patients treated with RIF showed a significant decline in cytokeratin-18 Levels, a biomarker capable of predicting the histopathological manifestations of steatohepatitis[13]. However, Cobbold et al[14] reported that RIF therapy in patients with steatohepatitis for six weeks caused no changes in liver triglyceride content, insulin sensitivity, or systemic inflammation[14]. Due to the lack of studies evaluating the effect of RIF on HCC secondary to steatohepatitis, further studies are needed in the search for new therapies for the prevention of disease progression and future clinical application.
It is known that MASLD-HCC is a tumor of specific molecular characteristics compared to other etiologies of HCC. In the tumor microenvironment, the adaptive response caused by cellular hypoxia promotes the activation of pathways that intensify the process of angiogenesis, inflammation, fibrogenesis, and autophagy[9,25]. This response is dependent on hypoxia-inducible transcriptional factors, such as Aldob, Tuba1c, and metalloproteinases, among other markers. In this study animals of the HCC-group showed a significant increase in the expression of Mmp9 in relation to the control group, and the treatment with RIF caused a reduction of this expression, obtaining values like the control. Additionally, we reported an increase in the gene expression of Mm2, Mm9 and Tuba1c in animals that developed HCC compared to animals that did not develop this clinical condition. The increased expression of these markers is linked to the process of hepatic hepatocarcinogenesis, as it is known that metalloproteinases are master regulators in the process of cell proliferation and migration, and play a role in cell apoptosis, tissue regeneration, and immune response[26,27]. It is known that inflammation drives the progression of MASLD; however, the mechanisms associated with this process are not fully elucidated. In clinical practice, assessing the dietary inflammatory index and the systemic immunological inflammation index are tools that can be used to predict and evaluate the prognosis of liver disease, considering the associated genetic, environmental, and dietary factors[28,29].
Autophagy contributes with cellular homeostasis, thus avoiding cell transformation and tumor initiation. However, in the advanced stages of the tumor, autophagy acts mainly as a suppressor of cell death, allowing the adaptation of cancer cells to stressful conditions[30,31]. The autophagy process is regulated by different markers, including Map1 Lc3b, p62/Sqstm1, and Becn1[30]. In this study, we demonstrated a significant decrease in the expression of these genes in the HCC-group and RIF-group compared to healthy animals. It is known that there is no pattern in the expression of these markers in different cell types and tissues of origin[10,30]. An example of this process is the expression of Becn1 which is reduced in glioblastoma, ovarian, lung, and esophageal cancer, but increased in colorectal and gastric cancer cells[32-34]. The results obtained in this study have a similar pattern of autophagy markers in HCC and hepatic fibrosis that was found in previous studies[10,35-38]. Becn1 acts as an initiator of autophagy and its deregulation increase the susceptibility of cells to transformation, that is, its lower expression is predictive of inferior survival in HCC[10,33,37,38]. Becn1 deficiency is also associated with increased angiogenesis, which indirectly corroborates the results obtained in this study[39]. Al-Shenawy[10] reports that autophagy and apoptosis in the liver are interrelated processes, in which elevated levels of Becn1 in patients with chronic hepatitis may limit liver damage and interact with progression to cancer, where Becn1 Later it is suppressed in aggressive cases of HCC[10]. The absence of microtubule-associated protein-1 Light chain-3 expression, an essential component in the formation of autophagosomes, is predictive of immediate mortality from HCC[35]. Duran et al[36] report that p62/Sqstm1 deficiency increased the activation phenotype of hepatic stellate cells, inhibiting their anti-fibrotic and anti-inflammatory functions[36]. Although we reported a significant decrease in the expression of these autophagy markers in the HCC and RIF groups, we did not observe a significant difference with the treatment of RIF. It is known that the greater the difference in the levels of autophagy between cancer and normal tissue, the worse the prognosis of the lesion, a result observed in this study.
Epigenetics is involved in autophagy “turn on, turn off” along the carcinogenesis, through the activity of metyltransferases such as Ezh2 and Carm1[40-42]. In this study, there was a significant decrease in Ezh2 expression in the HCC-group and RIF-group compared to the control. Treatment with RIF promoted a significant increase in Carm1 expression. No significant correlation was observed between the expression of Ezh2 and Carm1, probably because they act in different ways of epigenetic control of autophagy. Ezh2 represses the expression of genes related to the mammalian target of rapamycin pathway and Carm1 acts on transcription factors such as p53, and factor nuclear-κB[40,41]. Inhibition of Ezh2 activity induces autophagy, through the formation of LC3B and consequently the formation of the autophagosome, which corroborates with this study, given the lower expression of Map1 Lc3b[40,42]. The function of Carm1, like other epigenetic controllers, is dependent on the stage of lesion development, acting as a tumor repressor or promoter[41].
Epigenetic regulation is also carried out by microRNAs, which are short RNA sequences that function as modulators of mRNA expression, by either impairing translation or promoting its degradation[43,44]. Subtle dysregulation of anyone step in microRNAs biogenesis may lead to tumorigenesis. The miR-122 acts in the balance of proliferation and differentiation of hepatocytes, however its physiological role in carcinogenesis is variable and the mechanism by which it contributes to the progression of the lesion is undetermined[43]. In this study, the HCC-group showed a significant increase in miR-122 expression compared to healthy animals, a result that corroborates the literature[43,45]. We emphasize that this increase in gene expression was maintained in animals that developed HCC compared to animals that did not develop this clinical condition, in the stratified analysis of experimental groups. However, contradictory data are also reported, a possible explanation being the heterogeneity of the samples under different environmental conditions[44]. We did not observe a significant difference in the expression of miR-34a between the HCC-group and RIF-group, however there was a significant increase in its expression in the RIF-group compared to healthy animals. This increase in expression may represent a beneficial effect of RIF, as miR-34 inhibits the process of carcinogenesis through the regulation of p53, promoting apoptosis, cell cycle arrest, and senescence[46,47]. This data corroborates the increase in the expression of Carm1 regulator of autophagy through the expression of p53. Additionally, we reported a significant increase in miR-26b expression in RIF treated animals. We infer that it is a beneficial effect of the treatment since the lower expression of this microRNA is associated with a worse prognosis of HCC[48,49]. The suppression of miR-26 may result in an increase in the expression of Ezh2, in opposite to our results. However, we know that the expression of these epigenetic markers is variable according to the tissue and stage of the lesion[50]. Another possible beneficial effect of RIF treatment was the decrease in the expression of miR-224, which acts as an oncomiR in HCC cells, and its upregulation promotes the proliferation and migration of malignant hepatocytes[51,52]. Carm1 regulates the expression of transcription factors such as p53 and nuclear factor-κB which regulate the expression of miR-224. Possibly this signaling mechanism is activated and RIF acts in its modulation, however further studies are necessary to be carried out for the elucidation of the process[51,52]. Additionally, we compared the results of animals that developed or not HCC, and we found no difference within them in the RIF-group, probably due to the small number of animals. However, analyzing the entire group of animals, there was a difference between miR-122, miR-34a, Tuba1c, metalloproteinases and autophagy markers between the groups that developed cancer vs those who did not present HCC. These interesting findings also may demonstrate the influence of epigenetic and autophagy markers in the development of HCC in this scenario.
So far, there is no specific medication approved by the Food and Drug Administration in clinical practice for the treatment of MASLD; therefore, counseling focuses on lifestyle to prevent progression to HCC. Biological aging and the progression of liver disease occur through the interference of various factors, including environmental, genetic, and dietary issues[28]. The assessment of these mechanisms can contribute to the search for preventive strategies in the development of MASLD-HCC. In this study, animals were subjected to a HFCD with the aim of developing the liver condition, allowing the study of the use of RIF as a preventive measure. Since dietary management tools are essential for controlling this clinical condition, one can speculate if including additional dietary intervention in the model, such as high fiber diet or flavonoids, for instance, could alleviate liver damage[12,53,54].
In summary, MASLD-HCC management is a clinical challenge, and many questions need to be addressed, including the response to the new immunotherapy agents. The process of autophagy and epigenetics tends to play a complicit role in drug resistance and the interface between the two is multifactorial and crosstalk occurs in different proteins of each process[6,55]. In this study, we observed, mainly in relation to the epigenetic markers evaluated, a possible beneficial effect of the treatment with RIF in rats with MASLD-HCC, suggesting it could be useful to prevent or delay carcinogenesis. On the other hand, the study has some weaknesses to be considered, like the small number of animals and the gene expression markers analysis. Thus, new preclinical studies are needed to evaluate the epigenetic and autophagy mechanisms in MASLD-HCC for a better understanding of the role of RIF, as there are factors that need to be better explored, such as the variability of the course of the disease, the complexity of the autophagy mechanism and the individualized treatment requirements.
Metabolic-dysfunction associated steatotic liver disease (MASLD) incidence is increasing worldwide. Hepatocellular carcinoma (HCC) is a complex and heterogeneous neoplasm, and there’s evidence showing MASLD-related HCC has some unique features, including gut microbiota (GM). However, current treatment does not take this heterogeneity into account, dealing with viral and non-viral HCC in the same way. This study is intended to characterize autophagy and epigenetics in experimental MASLD-HCC and its response to rifaximin (RIF), a minimally-absorbed broad-spectrum oral antibiotic, that may interfere in GM-derived inflammation.
Epigenetic changes, autophagy and GM are involved in hepatocarcinogenesis, but there is no definite evidence of a positive effect of its modulation in human steatohepatitis. RIF may influence in these complex mechanisms. Understanding GM influence on epigenetics and autophagy can help not only as a diagnostic tool but also as a target for new therapies.
The main objective was to investigate rifaximin (RIF) effects on epigenetic and autophagy markers in experimental HCC secondary to MASLD. Future research in humans with MASLD can open a new therapeutic pathway to decrease HCC burden in this setting.
We conducted an innovative RIF experiment in a MASLD-HCC model with 24 adult Sprague-Dawley rats, randomly assigned in three groups (n = 8, each) and treated from 5-16 wk. We compared the results of control animals to RIF group and MASLD (animals in the last two groups received a high-fat choline deficient diet plus diethylnitrosamine in drinking water. Gene expression of epigenetic and autophagy markers was obtained at the end of experiment.
All animals in RIF and MASLD groups developed steatohepatitis, fibrosis, and cirrhosis. All MASLD animals also presented HCC, but in RIF group three rats did not develop tumor. Some microRNAs, metalloproteinases and aggressivity markers were higher in rats that developed HCC comparing with those that not developed, and the opposite occurred with the autophagy markers.
The results suggest that autophagy and epigenetics could exert influence on MASLD-HCC via GM interference with RIF and support clinical studies in the area.
RIF may have effect on autophagy and epigenetic markers as shown in this study. These initial results in animals shall be confirmed in other preclinical and clinical studies before recommending its use in high-risk patients with MASLD cirrhosis.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Liu M, China S-Editor: Li L L-Editor: A P-Editor: Zhao S
| 1. | Rinella ME, Lazarus JV, Ratziu V, Francque SM, Sanyal AJ, Kanwal F, Romero D, Abdelmalek MF, Anstee QM, Arab JP, Arrese M, Bataller R, Beuers U, Boursier J, Bugianesi E, Byrne CD, Narro GEC, Chowdhury A, Cortez-Pinto H, Cryer DR, Cusi K, El-Kassas M, Klein S, Eskridge W, Fan J, Gawrieh S, Guy CD, Harrison SA, Kim SU, Koot BG, Korenjak M, Kowdley KV, Lacaille F, Loomba R, Mitchell-Thain R, Morgan TR, Powell EE, Roden M, Romero-Gómez M, Silva M, Singh SP, Sookoian SC, Spearman CW, Tiniakos D, Valenti L, Vos MB, Wong VW, Xanthakos S, Yilmaz Y, Younossi Z, Hobbs A, Villota-Rivas M, Newsome PN; NAFLD Nomenclature consensus group. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Ann Hepatol. 2024;29:101133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 318] [Article Influence: 318.0] [Reference Citation Analysis (0)] |
| 2. | Kuchay MS, Choudhary NS, Mishra SK. Pathophysiological mechanisms underlying MAFLD. Diabetes Metab Syndr. 2020;14:1875-1887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 3. | Guo F, Estévez-Vázquez O, Benedé-Ubieto R, Maya-Miles D, Zheng K, Gallego-Durán R, Rojas Á, Ampuero J, Romero-Gómez M, Philip K, Egbuniwe IU, Chen C, Simon J, Delgado TC, Martínez-Chantar ML, Sun J, Reissing J, Bruns T, Lamas-Paz A, Moral MGD, Woitok MM, Vaquero J, Regueiro JR, Liedtke C, Trautwein C, Bañares R, Cubero FJ, Nevzorova YA. A Shortcut from Metabolic-Associated Fatty Liver Disease (MAFLD) to Hepatocellular Carcinoma (HCC): c-MYC a Promising Target for Preventative Strategies and Individualized Therapy. Cancers (Basel). 2021;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 4. | White DL, Kanwal F, El-Serag HB. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin Gastroenterol Hepatol. 2012;10:1342-1359.e2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 586] [Cited by in RCA: 574] [Article Influence: 44.2] [Reference Citation Analysis (2)] |
| 5. | Estes C, Razavi H, Loomba R, Younossi Z, Sanyal AJ. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2018;67:123-133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1028] [Cited by in RCA: 1701] [Article Influence: 243.0] [Reference Citation Analysis (0)] |
| 6. | Wu Y, Zhang J, Li Q. Autophagy, an accomplice or antagonist of drug resistance in HCC? Cell Death Dis. 2021;12:266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 7. | Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol. 2019;16:411-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 814] [Cited by in RCA: 963] [Article Influence: 160.5] [Reference Citation Analysis (0)] |
| 8. | Han TS, Ban HS, Hur K, Cho HS. The Epigenetic Regulation of HCC Metastasis. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 91] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 9. | Pinyol R, Torrecilla S, Wang H, Montironi C, Piqué-Gili M, Torres-Martin M, Wei-Qiang L, Willoughby CE, Ramadori P, Andreu-Oller C, Taik P, Lee YA, Moeini A, Peix J, Faure-Dupuy S, Riedl T, Schuehle S, Oliveira CP, Alves VA, Boffetta P, Lachenmayer A, Roessler S, Minguez B, Schirmacher P, Dufour JF, Thung SN, Reeves HL, Carrilho FJ, Chang C, Uzilov AV, Heikenwalder M, Sanyal A, Friedman SL, Sia D, Llovet JM. Corrigendum to 'Molecular characterisation of hepatocellular carcinoma in patients with non-alcoholic steatohepatitis' [J Hepatol 75 (2021) 865-878]. J Hepatol. 2021;75:1515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 10. | Al-Shenawy HA. Expression of Beclin-1, an autophagy-related marker, in chronic hepatitis and hepatocellular carcinoma and its relation with apoptotic markers. APMIS. 2016;124:229-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 11. | Allaire M, Rautou PE, Codogno P, Lotersztajn S. Autophagy in liver diseases: Time for translation? J Hepatol. 2019;70:985-998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 288] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 12. | Xie R, Zhang Y. Associations between dietary flavonoid intake with hepatic steatosis and fibrosis quantified by VCTE: Evidence from NHANES and FNDDS. Nutr Metab Cardiovasc Dis. 2023;33:1179-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 41] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 13. | Abdel-Razik A, Mousa N, Shabana W, Refaey M, Elzehery R, Elhelaly R, Zalata K, Abdelsalam M, Eldeeb AA, Awad M, Elgamal A, Attia A, El-Wakeel N, Eldars W. Rifaximin in nonalcoholic fatty liver disease: hit multiple targets with a single shot. Eur J Gastroenterol Hepatol. 2018;30:1237-1246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 14. | Cobbold JFL, Atkinson S, Marchesi JR, Smith A, Wai SN, Stove J, Shojaee-Moradie F, Jackson N, Umpleby AM, Fitzpatrick J, Thomas EL, Bell JD, Holmes E, Taylor-Robinson SD, Goldin RD, Yee MS, Anstee QM, Thursz MR. Rifaximin in non-alcoholic steatohepatitis: An open-label pilot study. Hepatol Res. 2018;48:69-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 15. | Leone P, Mincheva G, Balzano T, Malaguarnera M, Felipo V, Llansola M. Rifaximin Improves Spatial Learning and Memory Impairment in Rats with Liver Damage-Associated Neuroinflammation. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 16. | Gangarapu V, Ince AT, Baysal B, Kayar Y, Kılıç U, Gök Ö, Uysal Ö, Şenturk H. Efficacy of rifaximin on circulating endotoxins and cytokines in patients with nonalcoholic fatty liver disease. Eur J Gastroenterol Hepatol. 2015;27:840-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 17. | de Lima VM, Oliveira CP, Alves VA, Chammas MC, Oliveira EP, Stefano JT, de Mello ES, Cerri GG, Carrilho FJ, Caldwell SH. A rodent model of NASH with cirrhosis, oval cell proliferation and hepatocellular carcinoma. J Hepatol. 2008;49:1055-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 68] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 18. | Longo L, Rampelotto PH, Filippi-Chiela E, de Souza VEG, Salvati F, Cerski CT, da Silveira TR, Oliveira CP, Uribe-Cruz C, Álvares-da-Silva MR. Gut dysbiosis and systemic inflammation promote cardiomyocyte abnormalities in an experimental model of steatohepatitis. World J Hepatol. 2021;13:2052-2070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 19. | Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, Lefkowitch JH, Bower M, Friedman R, Sartor RB, Rabadan R, Schwabe RF. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 854] [Cited by in RCA: 1028] [Article Influence: 79.1] [Reference Citation Analysis (0)] |
| 20. | Liang W, Menke AL, Driessen A, Koek GH, Lindeman JH, Stoop R, Havekes LM, Kleemann R, van den Hoek AM. Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS One. 2014;9:e115922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 260] [Cited by in RCA: 437] [Article Influence: 39.7] [Reference Citation Analysis (1)] |
| 21. | EDMONDSON HA, STEINER PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 22. | Kalambokis GN, Mouzaki A, Rodi M, Tsianos EV. Rifaximin improves thrombocytopenia in patients with alcoholic cirrhosis in association with reduction of endotoxaemia. Liver Int. 2012;32:467-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 23. | Fujinaga Y, Kawaratani H, Kaya D, Tsuji Y, Ozutsumi T, Furukawa M, Kitagawa K, Sato S, Nishimura N, Sawada Y, Takaya H, Kaji K, Shimozato N, Moriya K, Namisaki T, Akahane T, Mitoro A, Yoshiji H. Effective Combination Therapy of Angiotensin-II Receptor Blocker and Rifaximin for Hepatic Fibrosis in Rat Model of Nonalcoholic Steatohepatitis. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 24. | Fujimoto Y, Kaji K, Nishimura N, Enomoto M, Murata K, Takeda S, Takaya H, Kawaratani H, Moriya K, Namisaki T, Akahane T, Yoshiji H. Dual therapy with zinc acetate and rifaximin prevents from ethanol-induced liver fibrosis by maintaining intestinal barrier integrity. World J Gastroenterol. 2021;27:8323-8342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 25. | Li X, Jiang F, Ge Z, Chen B, Yu J, Xin M, Wang J, An L, Wei J, Wu L. Fructose-Bisphosphate Aldolase A Regulates Hypoxic Adaptation in Hepatocellular Carcinoma and Involved with Tumor Malignancy. Dig Dis Sci. 2019;64:3215-3227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 26. | Ashraf TS, Obaid A, Saeed TM, Naz A, Shahid F, Ahmad J, Ali A. Formal model of the interplay between TGFβ1 and MMP-9 and their dynamics in hepatocellular carcinoma. Math Biosci Eng. 2019;16:3285-3310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 27. | Scheau C, Badarau IA, Costache R, Caruntu C, Mihai GL, Didilescu AC, Constantin C, Neagu M. The Role of Matrix Metalloproteinases in the Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma. Anal Cell Pathol (Amst). 2019;2019:9423907. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 234] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 28. | Xie R, Ning Z, Xiao M, Li L, Liu M, Zhang Y. Dietary inflammatory potential and biological aging among US adults: a population-based study. Aging Clin Exp Res. 2023;35:1273-1281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 35] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 29. | Xie R, Xiao M, Li L, Ma N, Liu M, Huang X, Liu Q, Zhang Y. Association between SII and hepatic steatosis and liver fibrosis: A population-based study. Front Immunol. 2022;13:925690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 143] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 30. | Qian H, Chao X, Williams J, Fulte S, Li T, Yang L, Ding WX. Autophagy in liver diseases: A review. Mol Aspects Med. 2021;82:100973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 218] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 31. | Shen S, Wang R, Qiu H, Li C, Wang J, Xue J, Tang Q. Development of an Autophagy-Based and Stemness-Correlated Prognostic Model for Hepatocellular Carcinoma Using Bulk and Single-Cell RNA-Sequencing. Front Cell Dev Biol. 2021;9:743910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 32. | Lomonaco SL, Finniss S, Xiang C, Decarvalho A, Umansky F, Kalkanis SN, Mikkelsen T, Brodie C. The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer. 2009;125:717-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 252] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 33. | Shi YH, Ding ZB, Zhou J, Qiu SJ, Fan J. Prognostic significance of Beclin 1-dependent apoptotic activity in hepatocellular carcinoma. Autophagy. 2009;5:380-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 34. | Li BX, Li CY, Peng RQ, Wu XJ, Wang HY, Wan DS, Zhu XF, Zhang XS. The expression of beclin 1 is associated with favorable prognosis in stage IIIB colon cancers. Autophagy. 2009;5:303-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 124] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 35. | Lin CW, Lin CC, Lee PH, Lo GH, Hsieh PM, Koh KW, Lee CY, Chen YL, Dai CY, Huang JF, Chuang WL, Chen YS, Yu ML. The autophagy marker LC3 strongly predicts immediate mortality after surgical resection for hepatocellular carcinoma. Oncotarget. 2017;8:91902-91913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 36. | Duran A, Hernandez ED, Reina-Campos M, Castilla EA, Subramaniam S, Raghunandan S, Roberts LR, Kisseleva T, Karin M, Diaz-Meco MT, Moscat J. p62/SQSTM1 by Binding to Vitamin D Receptor Inhibits Hepatic Stellate Cell Activity, Fibrosis, and Liver Cancer. Cancer Cell. 2016;30:595-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 185] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 37. | Fei B, Ji F, Chen X, Liu Z, Li S, Mo Z, Fang X. Expression and clinical significance of Beclin-1 in gastric cancer tissues of various clinical stages. Oncol Lett. 2016;11:2271-2277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 38. | Osman NA, Abd El-Rehim DM, Kamal IM. Defective Beclin-1 and elevated hypoxia-inducible factor (HIF)-1α expression are closely linked to tumorigenesis, differentiation, and progression of hepatocellular carcinoma. Tumour Biol. 2015;36:4293-4299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 39. | Lee SJ, Kim HP, Jin Y, Choi AM, Ryter SW. Beclin 1 deficiency is associated with increased hypoxia-induced angiogenesis. Autophagy. 2011;7:829-839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 108] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 40. | Xiao G, Jin LL, Liu CQ, Wang YC, Meng YM, Zhou ZG, Chen J, Yu XJ, Zhang YJ, Xu J, Zheng L. EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J Immunother Cancer. 2019;7:300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 131] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 41. | Baek SH, Kim KI. Epigenetic Control of Autophagy: Nuclear Events Gain More Attention. Mol Cell. 2017;65:781-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 119] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 42. | Wei FZ, Cao Z, Wang X, Wang H, Cai MY, Li T, Hattori N, Wang D, Du Y, Song B, Cao LL, Shen C, Wang L, Yang Y, Xie D, Wang F, Ushijima T, Zhao Y, Zhu WG. Epigenetic regulation of autophagy by the methyltransferase EZH2 through an MTOR-dependent pathway. Autophagy. 2015;11:2309-2322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 138] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 43. | Gumilas NSA, Widodo I, Ratnasari N, Heriyanto DS. Potential relative quantities of miR-122 and miR-150 to differentiate hepatocellular carcinoma from liver cirrhosis. Noncoding RNA Res. 2022;7:34-39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 44. | Chun KH. Molecular Targets and Signaling Pathways of microRNA-122 in Hepatocellular Carcinoma. Pharmaceutics. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 45. | Parizadeh SM, Jafarzadeh-Esfehani R, Ghandehari M, Goldani F, Parizadeh SMR, Hassanian SM, Ghayour-Mobarhan M, Ferns GA, Avan A. MicroRNAs as Potential Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. Curr Drug Targets. 2019;20:1129-1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 46. | Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474:4219-4251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 194] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 47. | Zhang DG, Zheng JN, Pei DS. P53/microRNA-34-induced metabolic regulation: new opportunities in anticancer therapy. Mol Cancer. 2014;13:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 48. | Yin J, Zhao X, Chen X, Shen G. Emodin suppresses hepatocellular carcinoma growth by regulating macrophage polarization via microRNA-26a/transforming growth factor beta 1/protein kinase B. Bioengineered. 2022;13:9548-9563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 49. | Wang Y, Sun B, Sun H, Zhao X, Wang X, Zhao N, Zhang Y, Li Y, Gu Q, Liu F, Shao B, An J. Regulation of proliferation, angiogenesis and apoptosis in hepatocellular carcinoma by miR-26b-5p. Tumour Biol. 2016;37:10965-10979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 50. | Boominathan L. The guardians of the genome (p53, TA-p73, and TA-p63) are regulators of tumor suppressor miRNAs network. Cancer Metastasis Rev. 2010;29:613-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 93] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 51. | Amr KS, Elmawgoud Atia HA, Elazeem Elbnhawy RA, Ezzat WM. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis. 2017;4:215-221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 52. | An F, Wu X, Zhang Y, Chen D, Lin Y, Wu F, Ding J, Xia M, Zhan Q. miR-224 Regulates the Aggressiveness of Hepatoma Cells Through the IL-6/STAT3/SMAD4 Pathway. Turk J Gastroenterol. 2021;32:532-542. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 53. | Wang K, Tan W, Liu X, Deng L, Huang L, Wang X, Gao X. New insight and potential therapy for NAFLD: CYP2E1 and flavonoids. Biomed Pharmacother. 2021;137:111326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 54. | Moreira RO, Valerio CM, Villela-Nogueira CA, Cercato C, Gerchman F, Lottenberg AMP, Godoy-Matos AF, Oliveira RA, Brandão Mello CE, Álvares-da-Silva MR, Leite NC, Cotrim HP, Parisi ER, Silva GF, Miranda PAC, Halpern B, Pinto Oliveira C. Brazilian evidence-based guideline for screening, diagnosis, treatment, and follow-up of metabolic dysfunction-associated steatotic liver disease (MASLD) in adult individuals with overweight or obesity: A joint position statement from the Brazilian Society of Endocrinology and Metabolism (SBEM), Brazilian Society of Hepatology (SBH), and Brazilian Association for the Study of Obesity and Metabolic Syndrome (Abeso). Arch Endocrinol Metab. 2023;67:e230123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 55. | Foerster F, Gairing SJ, Müller L, Galle PR. NAFLD-driven HCC: Safety and efficacy of current and emerging treatment options. J Hepatol. 2022;76:446-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 172] [Article Influence: 57.3] [Reference Citation Analysis (0)] |