Published online Jan 27, 2024. doi: 10.4254/wjh.v16.i1.65
Peer-review started: September 30, 2023
First decision: October 23, 2023
Revised: November 3, 2023
Accepted: November 28, 2023
Article in press: November 28, 2023
Published online: January 27, 2024
Processing time: 114 Days and 23.7 Hours
The function of prohibitin 1 (Phb1) during liver regeneration (LR) remains relatively unexplored. Our previous research identified downregulation of Phb1 in rat liver mitochondria 24 h after 70% partial hepatectomy (PHx), as determined by subcellular proteomic analysis.
To investigate the potential role of Phb1 during LR.
We examined changes in Phb1 mRNA and protein levels, subcellular distribution, and abundance in rat liver during LR following 70% PHx. We also evaluated mitochondrial changes and apoptosis using electron microscopy and flow cytometry. RNA-interference-mediated knockdown of Phb1 (PHBi) was performed in BRL-3A cells.
Compared with sham-operation control groups, Phb1 mRNA and protein levels in 70% PHx test groups were downregulated at 24 h, then upregulated at 72 and 168 h. Phb1 was mainly located in mitochondria, showed a reduced abundance at 24 h, significantly increased at 72 h, and almost recovered to normal at 168 h. Phb1 was also present in nuclei, with continuous increase in abundance observed 72 and 168 h after 70% PHx. The altered ultrastructure and reduced mass of mitochondria during LR had almost completely recovered to normal at 168 h. PHBi in BRL-3A cells resulted in increased S-phase entry, a higher number of apoptotic cells, and disruption of mitochondrial membrane potential.
Phb1 may contribute to maintaining mitochondrial stability and could play a role in regulating cell proliferation and apoptosis of rat liver cells during LR.
Core tip: Using subcellular proteomic analysis, we previously found that prohibitin 1 (Phb1) was downregulated in rat liver mitochondria at 24 h after 70% partial hepatectomy (PHx). Phb1 has various functions, but little is known about its role during liver regeneration (LR). To explore the function of Phb1 in mitochondria during LR, we investigated the changes of Phb1 expression, the alterations of mitochondrial mass and ultrastructure, and the subcellular distribution of Phb1 at 24, 72 and 168 h in rat liver after 70% PHx. Using RNA-interference-mediated knockdown of Phb1, the potential functions of Phb1 were analyzed. Phb1 was differentially expressed during LR.
- Citation: Sun QJ, Liu T. Subcellular distribution of prohibitin 1 in rat liver during liver regeneration and its cellular implication. World J Hepatol 2024; 16(1): 65-74
- URL: https://www.wjgnet.com/1948-5182/full/v16/i1/65.htm
- DOI: https://dx.doi.org/10.4254/wjh.v16.i1.65
It is well known that the liver has the capacity to regenerate and restore its original size and function after 70% partial hepatectomy (PHx), or injury[1,2]. It would be important clinically to develop therapeutic strategies to enhance liver regeneration (LR) or support the liver in its attempt to restore its functional integrity under pathophysiological circumstances[3,4]. However, the complexity of the regulatory mechanisms of LR, together with our limited understanding of the functional priorities of the hepatocytes have rendered difficult the identification of targets for therapeutic interventions.
As the hub of energy metabolism, mitochondria have been investigated due to their direct involvement in the process of LR[5]. In an attempt to identify mitochondrial proteins that are correlated with the early phase of LR, using subcellular proteomic analysis in our recent study, our recent study revealed that Prohibitin 1 (Phb1), a potential tumor suppressor protein, was downregulated in rat liver mitochondria at 24 h after 70% PHx[6].
Phb1 is a ubiquitously expressed highly conserved protein among eukaryotes. Previous research has proposed that Phb1 is involved in many cellular processes, such as cell cycle regulation, senescence, transcription regulation, tumor suppression and apoptosis[7-11]. Phb1 is reported to mainly localize in mitochondria, with its expression upregulated by mitochondrial stress and downregulated during cellular senescence[12]. Therefore, Phb1 is thought to have a crucial role in mitochondria function. One study identified a novel function of Phb1 in the maintenance of mitochondrial DNA (mtDNA). In Phb1-knockdown cells, the status of mtDNA is altered in several ways[13]. Despite such information, our understanding of the overall functions of Phb1 in mitochondria remains incomplete and its potential role during LR is largely unexplored. LR is a complicated biological procedure involving various signal transduction pathways and molecular events[14,15]. Thus, we hypothesized that Phb1 could play a crucial role during LR. This study aimed to investigate the function of Phb1 in mitochondria during changes in Phb1 expression, mitochondrial mass and ultrastructure, and the subcellular distribution of Phb1 at 24, 72 and 168 h post 70% PHx in rat liver. Using RNA-interference-mediated knockdown of Phb1 (PHBi), we also analyzed the potential functions of Phb1. Our results revealed differential expression of Phb1 during LR, with its primary localization in mitochondria, where its altered expression may be associated with the recovery of mitochondrial mass and ultrastructure. Phb1 was also present in the nuclei, with increased abundance during LR. PHBi in BRL-3A cells, a widely used cell line in liver research, led to increased S-phase entry and apoptotic cell count. We also observed disruption of mitochondrial membrane potential following Phb1 knockdown in BRL-3A cells, mirroring our previous findings. Collectively, these results suggest that Phb1 may contribute to maintaining mitochondrial stability and regulating the cell cycle and apoptosis during LR.
Adult male Sprague–Dawley rats (220–250 g) were obtained from the Experimental Animal House at Second Military Medical University (Shanghai, China). The rats were randomly divided into two groups: Five served as the sham-operation control group and the other five comprised the 70% PHx test group. PHx (~70%) was performed according to the method of Higgins et al[16]. The experimental rats were anesthetized by intraperitoneal injection of 2% pentobarbital (40 mg/kg). In the test group, the median and left lateral lobes were removed without injuring the remaining liver tissue. The control group underwent a sham operation identical to the test group procedure, but without liver removal. After surgery, the rats were kept on a standard diet until they were killed by cervical dislocation under anesthesia.
Liver specimens were fixed with 4% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 4 h at 4C. After fixation, they were washed overnight in sodium cacodylate buffer at 4C. The specimens were then postfixed with 1% osmium tetroxide in sodium cacodylate buffer for 1 h at 4C, dehydrated in alcohol, embedded in araldite resin, and semithin sections were removed for optical microscopy. Ultrathin sections were mounted on copper mesh grids and stained with uranyl acetate and lead citrate as described[17] before examination with a Hitachi H-800 electron microscope.
Rat livers were removed and treated as previously reported[18] for isolation of nuclei, cytosol and mitochondria. Livers were collected and homogenized. Subsequent centrifugation at increasingly higher speeds at 4C yielded the following fractions: Nuclear fraction at 1000 g for 10 min; mitochondrial fraction at 15 000 g for 15 min; and microsomes at 144 000 g for 90 min. The final supernatant was the cytosolic fraction. Purification of mitochondria was performed by Nycodenz density gradient purification[19]. The mitochondrial pellets obtained from differential centrifugation were suspended in 12 mL 25% Nycodenz and placed onto a discontinuous Nycodenz gradient consisting of 5 mL 34% Nycodenz and 8 mL 30% Nycodenz, followed by 8 mL 23% Nycodenz, and finally, 3 mL 20% Nycodenz. The sealed tubes were centrifuged for 90 min at 52 000 g at 4C. The mitochondria were in the band at the 25%/30% interface which was collected and diluted with the same volume of homogenization buffer and then centrifuged twice at 15 000 g for 20 min. The preparation of each subcellular fraction protein of rat livers was performed as previously described[19]. Protein concentration of each fraction was determined with a Quick Start Bradford Assay Kit (Bio-Rad).
Protein extracts of each sample were separated on 12% SDS-PAGE and transferred to nitrocellulose membranes (Millipore). The blots were probed by anti-Phb1 antibody (Neomarker) and proteins were normalized with anti--actin antibody (Neomarker) or anti-COX IV antibody (Cell Signaling) or anti-histone H3 antibody (Cell Signaling) and were visualized by Amersham ECL system. The digital image was obtained by scanning the membrane, and then subjected to gray value analysis. For a better understanding of western blotting results and derived ratio changes, a detailed methodology introduction can be found in the subsequent figures and legends.
The normal rat liver cell line BRL-3A was obtained from the Shanghai Institute of Biochemistry and Cell Biology. The BRL-3A cells were maintained as a monolayer in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum, 100 U/mL penicillin, and 100 mg /mL streptomycin. The cells were maintained at 37C in an atmosphere with 5% CO2.
Duplex siRNA was obtained from GeneChem (Shanghai, China). The siRNA sequence targeting rat Phb1 was 5’-GCCAGAUUUGUGGUGGAAAtt-3’ (sense) and 5’-UUUCCACCACAAAUCUGGCtt-3’ (antisense). A nonsense duplex was used as the control (mock). BRL-3A cells were plated on six-well plates with antibiotic-free DMEM overnight and transfected with siRNA by Lipofectamine2000 (Invitrogen). The final concentration of siRNA duplex was 100 nM. Six hours after transfection, the medium was switched to DMEM supplemented with antibiotics.
Total RNA of each sample was isolated by TRIzol reagent (Invitrogen). After treatment with DNase I, each RNA sample was reverse-transcribed with random primers (dN6) (MBI Fermantas, Vilnius, Lithuania). The single-stranded cDNA was used in quantitative real-time PCR to evaluate the relative expression levels of Phb1 (5’-GCGGTGGAAGCCAAACAG-3’ and 5’-TTCTTCTGCTGCTCAGCCTTT-3’), compared to -actin (5’-ATGGTGGGTATGGGTCAGAAG-3’ and 5’- TGGCTGGGGTGTTGAAGGTC-3’) used as an internal control for determining cell number and metabolic status. Quantitative real-time PCR (ABI7300, Applied Biosystems) was done with the SYBR Green I reagents (TOYOBO) and the primers were designed according to the ABI manufacturer’s protocol. Forty cycles of PCR were performed with cycling conditions of 15 s at 95C and 60 s at 60C. The real-time PCR signals were analyzed with LightCycler 3.5 software (Roche Diagnostics).
Cells were stained with propidium iodide (PI; BD Clontech) as previously described[20]. A suspension of 104 cells was analyzed for each DNA histogram, and from the analysis of DNA histograms, the percentages of cells in different phases of cell cycle were evaluated. Flow cytometry was performed on a FACSCalibur and analyzed using CellQuest software (BD Bioscience). The Annexin V/PI method was used to quantify numbers of apoptotic cells. Cells were washed twice with phosphate-buffered saline and stained with Annexin V and PI for 20 min at room temperature. The level of apoptosis was determined by measuring the fluorescence of the cells by flow cytometry analysis.
The data presented are the means ± SD of three independent experiments. Statistical significance was estimated with Student’s t-test for unpaired observations. P < 0.05 was considered significant.
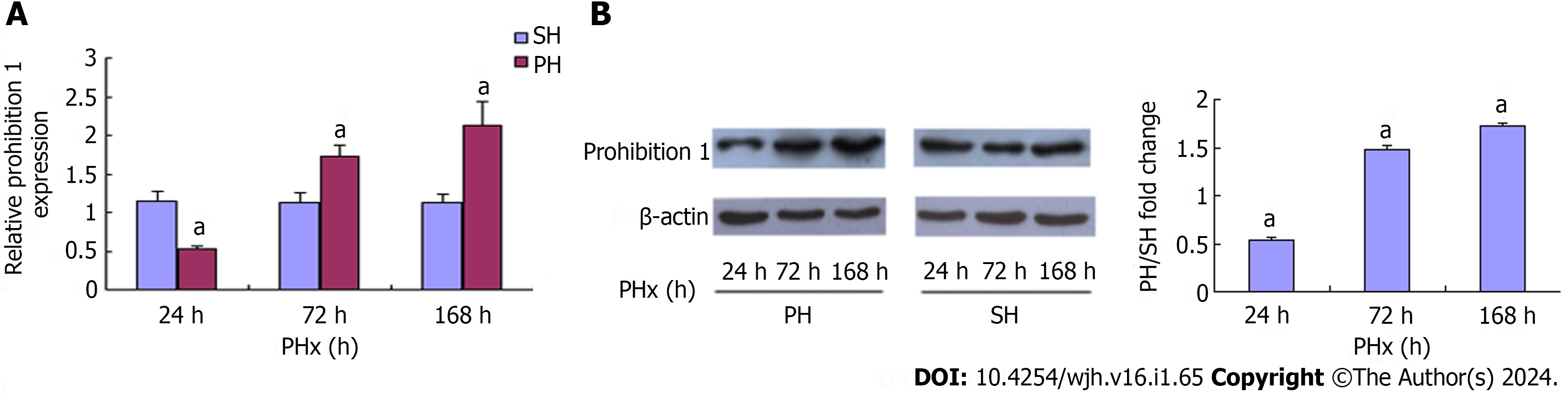
Phb1 mRNA expression was examined by real-time PCR. Compared with the control group, expression of Phb1 mRNA in the 70% PHx test group was 0.46-fold decreased at 24 h, and 1.54-fold and 1.89-fold increased at 72 and 168 h, respectively (Figure 1A). Western blotting showed that Phb1 protein expression during LR was 0.54-fold decreased at 24 h, and 1.48-fold and 1.73-fold increased at 72 and 168 h, respectively, after 70% PHx (Figure 1B), which was consistent with the expression of Phb1 mRNA.
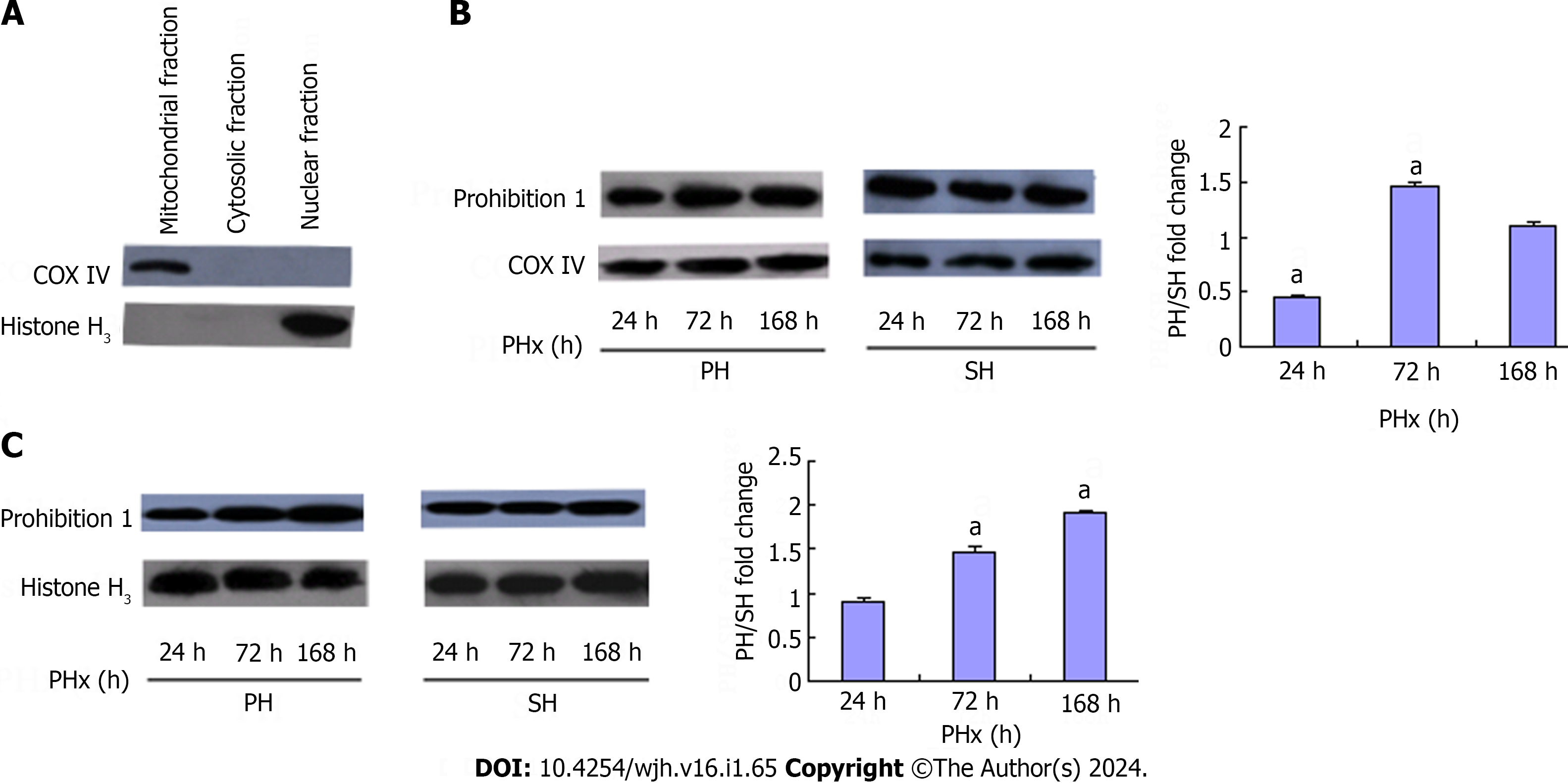
Previous observations suggest that examining the subcellular distribution of Phb1 might yield important information about physiological or pathological processes that are taking place in cells. To verify the cellular distribution pattern of Phb1 during LR, we fractionated cytosolic, mitochondrial and nuclear fractions of rat liver cells and performed western blotting analysis. The purity of subcellular fractionation was controlled by several marker proteins (Figure 2A). Phb1 was mainly located in mitochondria and its abundance was reduced 0.47-fold at 24 h, and induced 1.47-fold at 72 h and almost recovered to normal at 168 h after 70% PHx (Figure 2B). Phb1 was also located in nuclei and its abundance was increased during LR after 70% PHx (Figure 2C). No Phb1 was found in the cytosol.
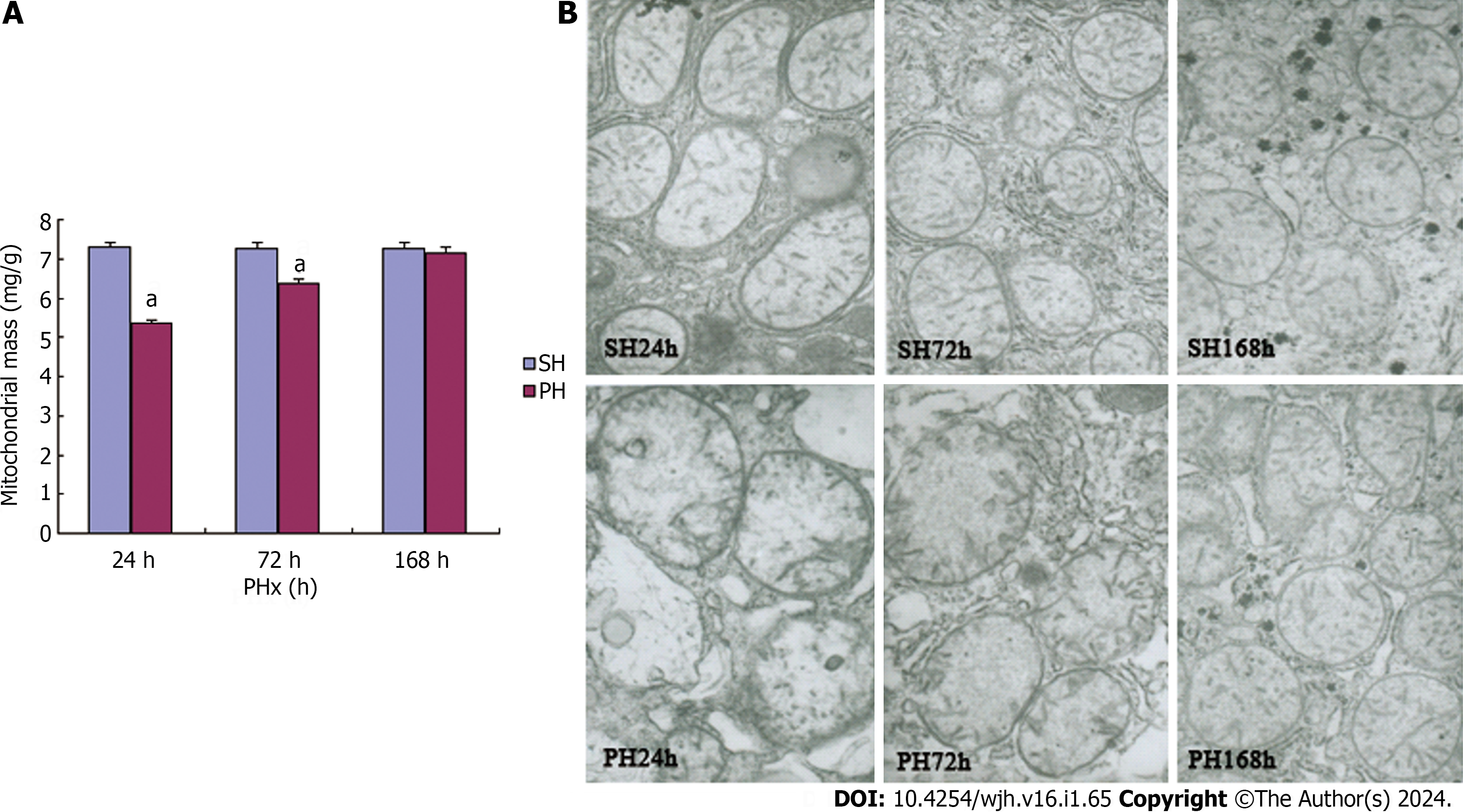
Mitochondrial mass and ultrastructural alterations during LR were observed to determine whether Phb1 changes were associated with mitochondrial stabilization or biogenesis. The mitochondrial mass was quantified by examining the protein contents of mitochondrial fractions[21] extracted from equivalent weights (1 g) of liver tissues from each experimental group. The results indicated an increase in mitochondrial protein contents during LR with 5.37 ± 0.08, 6.38 ± 0.10 and 7.16 ± 0.16 mg at 24 , 72 and 168 h respectively, after 70% PHx. The mitochondrial protein contents at 168 h in the 70% PHx test group closely mirrored that of the control group (Figure 3A).
Mitochondrial ultrastructural alterations during LR were observed by electron microscopy. The mitochondrial morphologies of control livers (Figure 3B upper panel) (SH 24 h, SH 72 h, SH 168 h), were characterized by a consistent basic architecture featuring a folded internal membrane and a dense matrix. The alterations in mitochondrial ultrast
To downregulate Phb1 cellular expression, PHBi was performed in BRL-3A cells. PHBi resulted in a dramatic reduction in both Phb1 mRNA and protein level compared with that of the control group (mock). Detailed results are available in our previous publication[4].
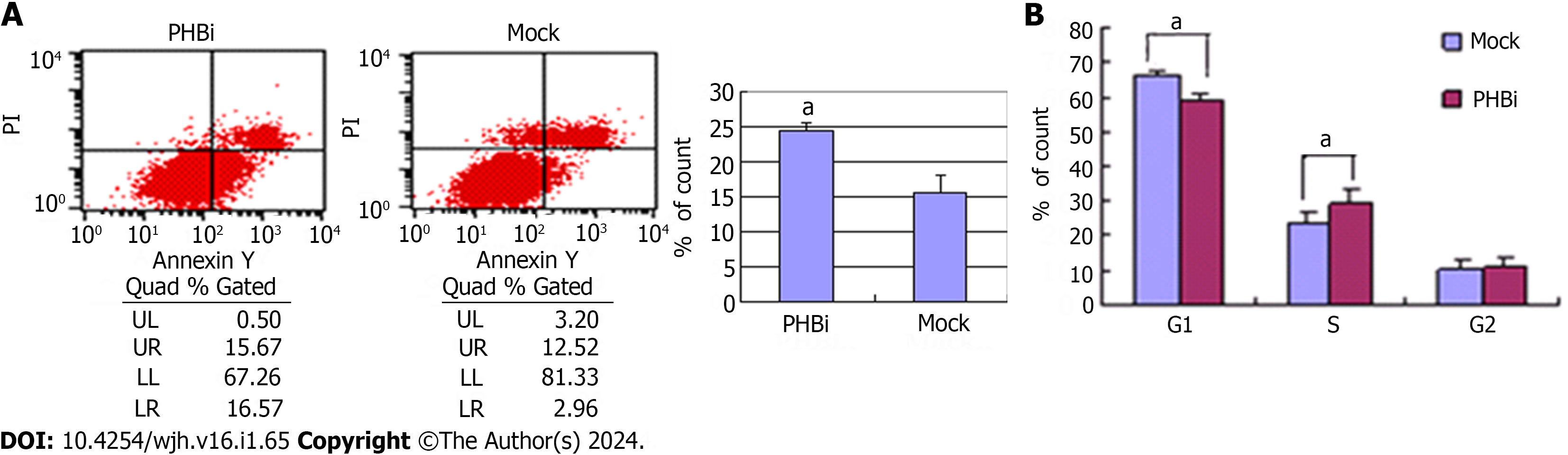
Previous reports have suggested that Phb1 could serve an antiapoptotic role in undifferentiated granulosa cells[22]. In this study, to evaluate whether Phb1 was involved in modulating apoptosis in rat liver cells, flow cytometry was used to evaluate percentage of apoptotic cells by Annexin V/PI staining. Phb1 knockdown cells displayed a 1.56-fold increase in the percentage of apoptotic cells compared with controls (Figure 4A).
We investigated whether the decrease of Phb1 by PHBi had any effect on cell growth and proliferation. The cell cycle distribution in Phb1 knockdown cells showed a 1.26-fold increase in the S-phase compared to control cells (Figure 4B). Although the increase in the S-phase was not dramatic, the difference was significant. Nuell et al[23] also previously reported a cell cycle modulatory role of Phb1, indicating that Phb1 could function as a negative cell cycle regulator.
Phb1, a potential tumor suppressor protein, was initially cloned due to its ability to induce G1/S phase arrest. Phb1 is proposed to be involved in numerous cellular processes. However, most studies to date have focused on the role of Phb1 in various types of tumors, with its role during LR remaining largely unexplored. In recent years, some studies have explored the role of Phb1 in liver injury and liver cancer[10,24-27]. However, the role of Phb1 in LR remained unstudied.
In this study, Phb1 mRNA and protein expression underwent concordant changes during LR after 70% PHx. Compared to sham-operation control groups, 70% PHx test groups showed downregulation of Phb1 mRNA and protein expression at 24 h, and upregulation at 72 and 168 h (Figure 1). A previous study found that the gene encoding Phb1 might have additional antiproliferative effects that do not require translation[11]. Manjeshwar et al[28] reported that the 3’ untranslated region of the Phb1 gene encoded a functional RNA that arrested cell-cycle proliferation between the G1 and S phases. In light of previous reports, we propose that Phb1 might regulate cell proliferation during LR in a complex manner, potentially involving mechanisms mediated by both Phb1 mRNA and protein.
The well-characterized function of Phb1 is as a chaperone involved in the stabilization of mitochondrial proteins. Mitochondrial-localized Phb1 is confirmed as a high-molecular-weight hetero-complex (ring-shaped structure) by single-particle structures[7]. The interaction of no assembled respiratory chain subunits with the Phb1 complex has led to the proposal of a chaperone activity of Phb1 during the biogenesis of the respiratory chain[29]. Recently, Phb1 was reported to be essential for normal mitochondrial development, and Phb1 deficiency was showed to be associated with deficient mitochondrial biogenesis[30]. PHBi showed enhanced sensitivity to anthralin-induced cell death due to enhanced loss of mitochondrial membrane potential in psoriatic lesions[31]. Mitochondria are the center of energy metabolism and play a crucial role in regulating cell life. Various stimuli can induce dysfunction and structural injury in mitochondria, which triggers a series of cellular events ultimately leading to apoptosis or necrosis. We found that Phb1 was mainly located in the mitochondria in rat liver, and its abundance underwent a 0.47-fold reduction at 24 h, a 1.47-fold induction at 72 h, and nearly recovered to normal level at 168 h after 70% PHx (Figure 2B). Mitochondria showed significant changes in the ultrastructure at 24 h, and nearly recovered to normal at 168 h after 70% PHx (Figure 3B). The reduced mitochondrial mass also nearly recovered to normal at 168 h after 70% PHx. Mitochondrial membrane potential is an important parameter of mitochondrial function. In our previous study, we found that knockdown of Phb1 in BRL-3A cells resulted in disruption of mitochondrial membrane potential, implying a potential role of Phb1 in maintaining mitochondrial integrity[6]. Ross et al[32] also reported that siRNA-mediated knockdown of Phb1 in Kit225 cells resulted in disruption of mitochondrial membrane potential and Phb1 proteins were novel phosphoproteins upregulated during T-cell activation that function to maintain mitochondrial integrity. In this study, using PHBi, we also observed that Phb1 knockdown cells exhibited a 1.56-fold increase in the number of apoptotic cells (Figure 4A). Although these results provide evidence for a functional role of Phb1 in suppressing apoptosis in rat liver cells, the involved molecular mechanisms remain unknown. It is likely that the mechanism by which knockdown of Phb1 results in apoptosis targets the mitochondria in agreement with previous findings[22]. All these results suggest that Phb1 has a role in regulating stabilization of mitochondria during LR, which might affect mitochondrial function.
Although it has been reported that Phb1 is primarily located in mitochondria[12,30,33,34], other studies have reported that Phb1 is also located in the nuclei[35,36]. We found that Phb1 was located in nuclei as well as mitochondria in rat liver and its abundance increased during LR (Figure 2B). Previous studies reported that Phb1 was present in the nuclei and interacted with transcription factors important in cell-cycle progression[35,36]. In this study, using PHBi, we observed that Phb1 knockdown cells showed an increase in S-phase entry (Figure 4B). The involvement of Phb1 in the cell cycle was also observed in a prostate cancer cell line, in which downregulation of Phb1 led to an increase in cell-cycle entry from G1 to S phase[30]. Although most data suggest that Phb1 has an antiproliferative effect by interacting with the p53 and pRb pathways in the nuclei[9,37], it appears that Phb1 can also have antiapoptotic effects. In osteosarcoma cells, Phb1 was identified as a gene with downregulated expression in response to cytotoxic drugs, and the transient overexpression of the Phb1 coding sequence significantly reduced cytotoxic drug-induced apoptosis in these cells[38]. In this study, we also observed that Phb1 knockdown cells showed an increase in the number of apoptotic cells (Figure 4A). It has been reported that the subcellular localization of Phb1 may depend on the cell type examined and its physiological status, and Phb1 might have distinct but overlapping functions in each of these cellular compartments[39]. Although there is controversy concerning the function of nuclear-localized Phb1, in combination with previous reports, we suggest that the upregulated Phb1 in the nuclei in rat liver cells might have a function, at least in part, in regulating cell-cycle progression of rat liver cells. It might regulate the balance between proliferation and apoptosis during LR after 70% PHx, but this needs further investigation.
In summary, our results demonstrate that Phb1 plays two roles in the LR process: one is to regulate cell cycle and apoptosis, and the other is to regulate and maintain mitochondrial stability. Whether the two effects are directly linked or show two different effects remains unclear. Further in-depth studies will aid in us better understanding the complexities and roles of Phb1 in the LR process.
It is clinically important to develop therapeutic strategies to enhance liver regeneration (LR) or support the liver in its attempt to restore its functional integrity under pathophysiological circumstances. However, the complexity of the regulatory mechanisms of LR, together with our limited understanding of the functional priorities of the hepatocytes have rendered difficult the identification of targets for therapeutic interventions.
Prohibitin 1 (Phb1) is a ubiquitously expressed highly conserved protein among eukaryotes. Previous research has proposed that Phb1 was involved in many cellular processes. Phb1 was reported to mainly localize in mitochondria, with its expression upregulated by mitochondrial stress and downregulated during cellular senescence. Therefore, Phb1 is thought to have a crucial role in mitochondrial function. One study identified a novel function of Phb1 in the maintenance of mitochondrial DNA (mtDNA). In Phb1-knockdown cells, the status of mtDNA is altered in several ways. Despite such information, our understanding of the overall functions of Phb1 in mitochondria remains incomplete and its potential role during LR is largely unexplored. LR is a very complicated biological procedure involving various signal transduction pathways and molecular events. Thus, we hypothesized that Phb1 could play a crucial role during LR.
This study aimed to further investigate the function of Phb1 in mitochondria during changes in Phb1 expression, mitochondrial mass and ultrastructure, and the subcellular distribution of Phb1 at 24, 72 and 168 h post 70% partial hepatectomy (PHx) in rat liver. Using RNA-interference-mediated knockdown of Phb1 (PHBi), we also analyzed the potential functions of Phb1.
We examined changes in Phb1 mRNA and protein levels, subcellular distribution, and abundance in rat liver during LR following 70% PHx. We also evaluated mitochondrial changes and apoptosis levels using electron microscopy and flow cytometry. PHBi was performed in BRL-3A cells.
Compared with sham-operation control groups, Phb1 mRNA and protein levels in 70% PHx test groups were downregulated at 24 h, then upregulated at 72 and 168 h. Phb1 was mainly located in mitochondria, showed a reduced abundance at 24 h, significantly increased at 72 h, and almost recovered to normal at 168 h. Phb1 was also present in nuclei, with continuous increase in abundance observed at 72 and 168 h after 70% PHx. The altered ultrastructure and reduced mass of mitochondria during LR had almost completely recovered to normal at 168 h. PHBi in BRL-3A cells resulted in increased S-phase entry, a higher number of apoptotic cells, and disruption of mitochondrial membrane potential.
In summary, our results demonstrate that Phb1 plays two roles in the LR process: one is to regulate cell cycle and apoptosis, and the other is to regulate and maintain mitochondrial stability.
Whether the two effects are directly linked or show two different effects remains unclear. Further in-depth studies will aid in us better understanding the complexities and roles of Phb1 in the LR process.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zharikov YO, Russia S-Editor: Liu JH L-Editor: Kerr C P-Editor: Cai YX
| 1. | Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2649] [Cited by in RCA: 2468] [Article Influence: 88.1] [Reference Citation Analysis (0)] |
| 2. | Shu W, Yang M, Yang J, Lin S, Wei X, Xu X. Cellular crosstalk during liver regeneration: unity in diversity. Cell Commun Signal. 2022;20:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 3. | Kiseleva YV, Antonyan SZ, Zharikova TS, Tupikin KA, Kalinin DV, Zharikov YO. Molecular pathways of liver regeneration: A comprehensive review. World J Hepatol. 2021;13:270-290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (3)] |
| 4. | Hadjittofi C, Feretis M, Martin J, Harper S, Huguet E. Liver regeneration biology: Implications for liver tumour therapies. World J Clin Oncol. 2021;12:1101-1156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 5. | Guerrieri F, Muolo L, Cocco T, Capozza G, Turturro N, Cantatore P, Papa S. Correlation between rat liver regeneration and mitochondrial energy metabolism. Biochim Biophys Acta. 1995;1272:95-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Sun Q, Miao M, Jia X, Guo W, Wang L, Yao Z, Liu C, Jiao B. Subproteomic analysis of the mitochondrial proteins in rats 24 h after partial hepatectomy. J Cell Biochem. 2008;105:176-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 7. | Della-Flora Nunes G, Wilson ER, Marziali LN, Hurley E, Silvestri N, He B, O'Malley BW, Beirowski B, Poitelon Y, Wrabetz L, Feltri ML. Prohibitin 1 is essential to preserve mitochondria and myelin integrity in Schwann cells. Nat Commun. 2021;12:3285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 8. | Jackson DN, Panopoulos M, Neumann WL, Turner K, Cantarel BL, Thompson-Snipes L, Dassopoulos T, Feagins LA, Souza RF, Mills JC, Blumberg RS, Venuprasad K, Thompson WE, Theiss AL. Mitochondrial dysfunction during loss of prohibitin 1 triggers Paneth cell defects and ileitis. Gut. 2020;69:1928-1938. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 9. | Wang S, Nath N, Adlam M, Chellappan S. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene. 1999;18:3501-3510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 184] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 10. | Barbier-Torres L, Lu SC. Prohibitin 1 in liver injury and cancer. Exp Biol Med (Maywood). 2020;245:385-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 11. | Roskams AJ, Friedman V, Wood CM, Walker L, Owens GA, Stewart DA, Altus MS, Danner DB, Liu XT, McClung JK. Cell cycle activity and expression of prohibitin mRNA. J Cell Physiol. 1993;157:289-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 12. | Coates PJ, Nenutil R, McGregor A, Picksley SM, Crouch DH, Hall PA, Wright EG. Mammalian prohibitin proteins respond to mitochondrial stress and decrease during cellular senescence. Exp Cell Res. 2001;265:262-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 160] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 13. | Kasashima K, Sumitani M, Satoh M, Endo H. Human prohibitin 1 maintains the organization and stability of the mitochondrial nucleoids. Exp Cell Res. 2008;314:988-996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 112] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 14. | Verma AK, Sharma A, Subramaniyam N, Gandhi CR. Augmenter of liver regeneration: Mitochondrial function and steatohepatitis. J Hepatol. 2022;77:1410-1421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 15. | Li W, Li L, Hui L. Cell Plasticity in Liver Regeneration. Trends Cell Biol. 2020;30:329-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 99] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 16. | Higgins GM A, RM. Experimental pathology of the liver I: Restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186-202. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 17. | Guerrieri F, Pellecchia G, Lopriore B, Papa S, Esterina Liquori G, Ferri D, Moro L, Marra E, Greco M. Changes in ultrastructure and the occurrence of permeability transition in mitochondria during rat liver regeneration. Eur J Biochem. 2002;269:3304-3312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 18. | Okado-Matsumoto A, Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J Biol Chem. 2001;276:38388-38393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 732] [Cited by in RCA: 734] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 19. | Jiang XS, Zhou H, Zhang L, Sheng QH, Li SJ, Li L, Hao P, Li YX, Xia QC, Wu JR, Zeng R. A high-throughput approach for subcellular proteome: identification of rat liver proteins using subcellular fractionation coupled with two-dimensional liquid chromatography tandem mass spectrometry and bioinformatic analysis. Mol Cell Proteomics. 2004;3:441-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 62] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Kim HJ, Kim HJ, Lim SC, Kim SH, Kim TY. Induction of apoptosis and expression of cell cycle regulatory proteins in response to a phytosphingosine derivative in HaCaT human keratinocyte cells. Mol Cells. 2003;16:331-337. [PubMed] |
| 21. | Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci USA. 1994;91:10771-10778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1464] [Cited by in RCA: 1458] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 22. | Chowdhury I, Xu W, Stiles JK, Zeleznik A, Yao X, Matthews R, Thomas K, Thompson WE. Apoptosis of rat granulosa cells after staurosporine and serum withdrawal is suppressed by adenovirus-directed overexpression of prohibitin. Endocrinology. 2007;148:206-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 54] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Nuell MJ, Stewart DA, Walker L, Friedman V, Wood CM, Owens GA, Smith JR, Schneider EL, Dell' Orco R, Lumpkin CK. Prohibitin, an evolutionarily conserved intracellular protein that blocks DNA synthesis in normal fibroblasts and HeLa cells. Mol Cell Biol. 1991;11:1372-1381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 71] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 24. | Xia L, Liu Y, Zhang S, Yang Y, Zhou Z, Tu J. Can Prohibitin 1 be a Safeguard against liver disease? Ann Hepatol. 2019;18:790-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 25. | Mavila N, Tang Y, Berlind J, Ramani K, Wang J, Mato JM, Lu SC. Prohibitin 1 Acts As a Negative Regulator of Wingless/Integrated-Beta-Catenin Signaling in Murine Liver and Human Liver Cancer Cells. Hepatol Commun. 2018;2:1583-1600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 26. | Fan W, Yang H, Liu T, Wang J, Li TW, Mavila N, Tang Y, Yang J, Peng H, Tu J, Annamalai A, Noureddin M, Krishnan A, Gores GJ, Martínez-Chantar ML, Mato JM, Lu SC. Prohibitin 1 suppresses liver cancer tumorigenesis in mice and human hepatocellular and cholangiocarcinoma cells. Hepatology. 2017;65:1249-1266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 27. | Sánchez-Quiles V, Segura V, Bigaud E, He B, O'Malley BW, Santamaría E, Prieto J, Corrales FJ. Prohibitin-1 deficiency promotes inflammation and increases sensitivity to liver injury. J Proteomics. 2012;75:5783-5792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 28. | Manjeshwar S, Branam DE, Lerner MR, Brackett DJ, Jupe ER. Tumor suppression by the prohibitin gene 3'untranslated region RNA in human breast cancer. Cancer Res. 2003;63:5251-5256. [PubMed] |
| 29. | Nijtmans LG, de Jong L, Artal Sanz M, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000;19:2444-2451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 438] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 30. | Artal-Sanz M, Tsang WY, Willems EM, Grivell LA, Lemire BD, van der Spek H, Nijtmans LG. The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J Biol Chem. 2003;278:32091-32099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 165] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 31. | Kim SY, Kim Y, Hwang HY, Kim TY. Altered expression of prohibitin in psoriatic lesions and its cellular implication. Biochem Biophys Res Commun. 2007;360:653-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 32. | Ross JA, Nagy ZS, Kirken RA. The PHB1/2 phosphocomplex is required for mitochondrial homeostasis and survival of human T cells. J Biol Chem. 2008;283:4699-4713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 33. | Yan Y, Tang J, Yuan Q, Liu C, Chen X, Liu H, Huang J, Bao C, Hsiang T, Zheng L. Mitochondrial prohibitin complex regulates fungal virulence via ATG24-assisted mitophagy. Commun Biol. 2022;5:698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 34. | Thompson WE, Ramalho-Santos J, Sutovsky P. Ubiquitination of prohibitin in mammalian sperm mitochondria: possible roles in the regulation of mitochondrial inheritance and sperm quality control. Biol Reprod. 2003;69:254-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 128] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 35. | Wang S, Fusaro G, Padmanabhan J, Chellappan SP. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene. 2002;21:8388-8396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 175] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 36. | Fusaro G, Wang S, Chellappan S. Differential regulation of Rb family proteins and prohibitin during camptothecin-induced apoptosis. Oncogene. 2002;21:4539-4548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 83] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 37. | Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem. 2003;278:47853-47861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 270] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 38. | Fellenberg J, Dechant MJ, Ewerbeck V, Mau H. Identification of drug-regulated genes in osteosarcoma cells. Int J Cancer. 2003;105:636-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 39. | Mishra S, Murphy LC, Nyomba BL, Murphy LJ. Prohibitin: a potential target for new therapeutics. Trends Mol Med. 2005;11:192-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 188] [Article Influence: 9.4] [Reference Citation Analysis (0)] |