Peer-review started: October 26, 2023
First decision: December 1, 2023
Revised: December 4, 2023
Accepted: December 28, 2023
Article in press: December 28, 2023
Published online: January 27, 2024
Processing time: 88 Days and 12.9 Hours
In coronavirus disease 2019 (COVID-19), severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) primarily targets the respiratory system, but evidence suggests extrapulmonary organ involvement, notably in the liver. Viral RNA has been detected in hepatic tissues, and in situ hybridization revealed virions in blood vessels and endothelial cells. Electron microscopy confirmed viral particles in hepatocytes, emphasizing the need for understanding hepatotropism and direct cytopathic effects in COVID-19-related liver injury. Various factors contribute to liver injury, including direct cytotoxicity, vascular changes, inflammatory responses, immune reactions from COVID-19 and vaccinations, and drug-induced liver injury. Although a typical hepatitis presentation is not widely documented, elevated liver biochemical markers are common in hospitalized COVID-19 patients, primarily showing a hepatocellular pattern of elevation. Long-term studies suggest progressive cholestasis may affect 20% of patients with chronic liver disease post-SARS-CoV-2 infection. The molecular mechanisms underlying SARS-CoV-2 infection in the liver and the resulting liver damage are complex. This “Editorial” highlights the expression of the Angiotensin-converting enzyme-2 receptor in liver cells, the role of inflammatory responses, the impact of hypoxia, the involvement of the liver's vascular system, the infection of bile duct epithelial cells, the activation of hepatic stellate cells, and the contribution of monocyte-derived macrophages. It also mentions that pre-existing liver con
Core Tip: The hepatotropism of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a growing concern amid the coronavirus disease 2019 (COVID-19) pandemic. Despite its respiratory focus, the virus significantly affects various organs, notably the liver, leading to complications like inflammation, abnormal function tests, and, in severe cases, organ damage. This complex involvement worsens disease outcomes. Understanding the virus's interplay with the liver, mediated by the Angiotensin-converting enzyme-2 receptor, is crucial for tailored treatments. The liver's pivotal role in the immune response emphasizes the need to comprehend SARS-CoV-2 hepatotropism. Ongoing research is vital for uncovering mechanisms, clinical implications, and effective strategies in managing COVID-19 patients with liver involvement.
- Citation: Quarleri J, Delpino MV. Molecular mechanisms underlying SARS-CoV-2 hepatotropism and liver damage. World J Hepatol 2024; 16(1): 1-11
- URL: https://www.wjgnet.com/1948-5182/full/v16/i1/1.htm
- DOI: https://dx.doi.org/10.4254/wjh.v16.i1.1
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), belonging to the Betacoronavirus genus within the Coronaviridae family, is a positive-sense, single-stranded RNA virus with an enveloped structure. It shares close genetic relatedness with severe acute respiratory syndrome coronavirus-1 (SARS-CoV-1) and Middle East respiratory syndrome CoV. The genome of SARS-CoV-2 is approximately 30000 base pairs long, encoding 16 nonstructural and 4 structural proteins, including spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. The spike protein assumes a critical role in the SARS-CoV-2 life cycle by governing viral attachment, fusion, entry, and transmission. This glycoprotein contains the S1 and S2 domains as functional components able to act as ligands for receptor binding and downstream membrane fusion, respectively. Notably, the receptor binding domain within the S1 unit exhibits significant genetic variability within the coronavirus genome[1,2].
When it comes to infecting the majority of host cells, the SARS-CoV-2 spike engages with its primary receptor, Angiotensin-converting enzyme-2 (ACE2). The process is further facilitated by host transmembrane proteases, such as serine 2 [transmembrane serine protease 2 (TMPRSS2)], which play a crucial role in priming the spike protein for receptor interaction and subsequent entry into the host cell. In the facilitation of viral entry may also act additional host co-factors, such as neuropilin-1, glycosaminoglycans, C-type lectins, and furin. Noteworthy is the spike protein's specific binding to ACE2 and TMPRSS2, which collectively support viral entry. The differential expression of ACE2 and TMPRSS2 in various tissues, including the airways, lungs, nasal/oral mucosa, and intestine, underscores the multifaceted nature of the viral entry process across different cellular environments. The affinity of the spike protein for the ACE2 receptor plays a critical role in determining the replication fitness and severity of SARS-CoV-2 infection[1,2].
In the context of Coronavirus Disease 2019 (COVID-19), produced by the infection with SARS-CoV-2, the most profound pathological modifications are predominantly evident within the respiratory system. Nevertheless, it is of utmost significance to acknowledge that this viral infection imposes deleterious consequences on various other bodily organs. Notably, evidence has been presented of the presence of SARS-CoV-2 viral RNA in extrapulmonary organs, including the liver[3-6]. Building upon the excellent review conducted by Roshanshad et al[6], this editorial seeks to provide supplementary insights into the molecular mechanisms underlying SARS-CoV-2 hepatotropism and liver damage. The specific cellular location of viral replication remains unclear because of the use of whole-tissue homogenization techniques for nucleic acid extraction. Subsequent examinations, employing in situ hybridization analysis, identified the presence of SARS-CoV-2 virions within the lumen of blood vessels and endothelial cells in the portal veins of liver tissues derived from COVID-19 patients[7,8]. Furthermore, electron microscopic assessments of liver specimens from two COVID-19 patients who succumbed to the disease and exhibited elevated liver enzyme levels revealed the presence of intact viral particles within the cytoplasm of hepatocytes[9].
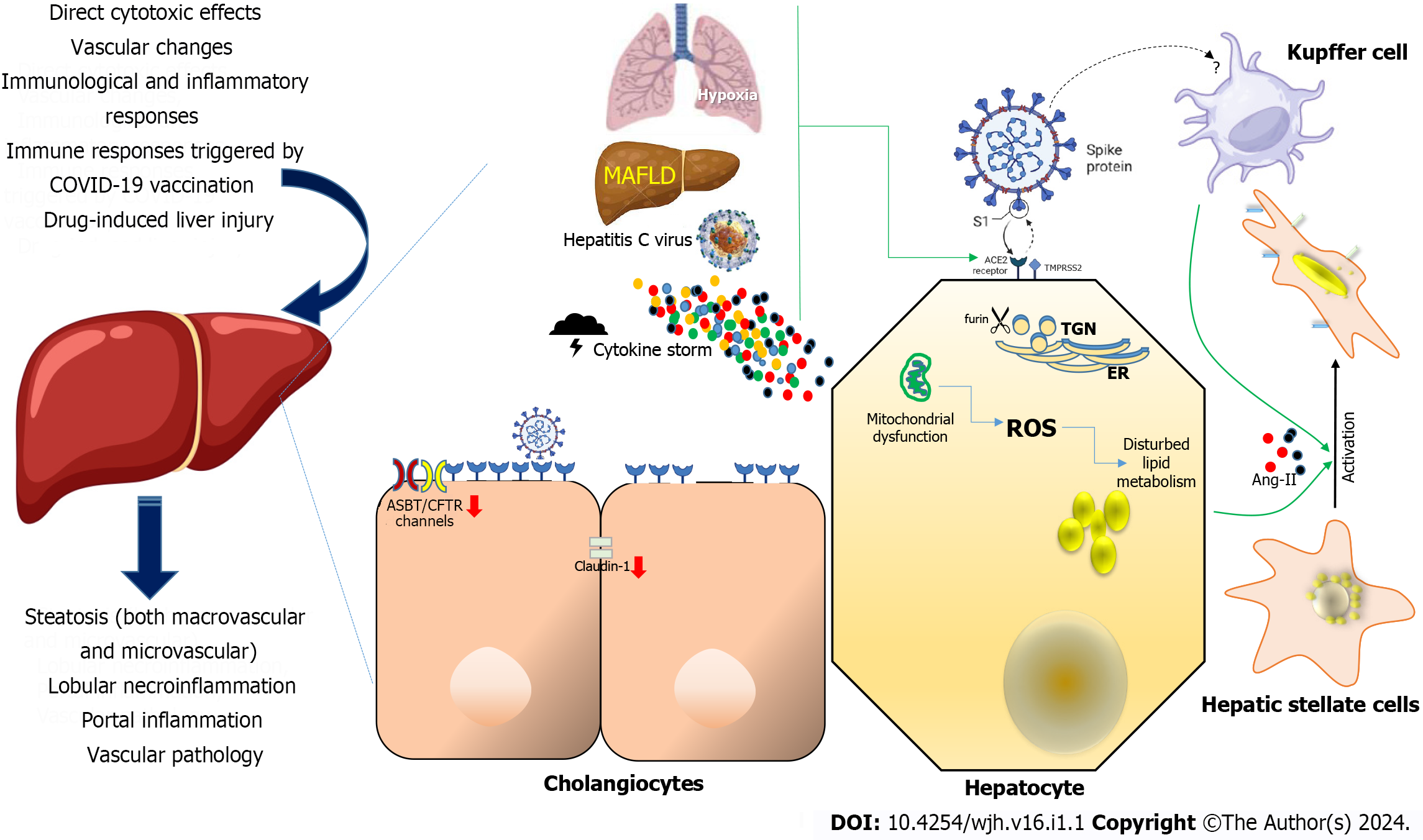
Although the precise etiology of liver injury in the context of COVID-19 remains partially understood, various factors have been postulated to contribute to this phenomenon (Figure 1), including direct cytotoxic effects, vascular changes, immunological and inflammatory responses associated with COVID-19, immune responses triggered by COVID-19 vaccination, and drug-induced liver injury (DILI)[10-12].
The assessment of hepatotropism concerning SARS-CoV-2 and the possible manifestation of direct cytopathic effects are crucial for a comprehensive understanding of the mechanisms underlying liver injury in COVID-19. It is worth noting that a typical hepatitis presentation has not been extensively documented[7,9,13], despite recent albeit limited discoveries.
The prevalence of elevated liver biochemical markers in individuals with COVID-19 varies in different studies but, in hospitalized patients, these abnormalities can be observed in the vast majority of them. These are primarily characterized by a hepatocellular pattern of elevation. The extent of these elevations is typically mild, and the likelihood of encountering substantial increases in alanine aminotransferase or aspartate aminotransferase levels (> 20-fold upper normal limit), liver synthetic dysfunction, or elevated serum bilirubin levels remains relatively uncommon among COVID-19 patients[14-17]. Remarkably, recent extended follow-up investigations have unveiled that after SARS-CoV-2 infection, progressive cholestasis may impact as many as 20% of individuals with chronic liver disease (CLD), demonstrating a proclivity toward increased severity[18].
A comprehensive understanding of tissue reservoirs supporting SARS-CoV-2 replication remains a critical research challenge. This is, in part, attributed to the inherent challenges associated with procuring biopsy samples from individuals presently infected with the virus, coupled with the requisite use of high-level laboratory containment facilities. The well-established understanding includes the interaction of the viral spike protein (S) with ACE2 for cellular entry, emphasizing the crucial roles of TMPRSS2 and furin enzymes in the infection process[1]. Consequently, examination of the expression of these receptors during the early stages of infection provided valuable insights into the potential permissiveness of hepatic cells. Notably, the liver exhibits minimal expression of ACE2 and TMPRSS2 proteins, whereas their highest expression is observed in the intestine and gall bladder. However, it is noteworthy that ACE2 expression appears to be absent in the lungs, where infection unequivocally occurs. Then, studies using single-cell RNA sequencing to analyze samples from healthy human livers revealed that although hepatic ACE2 expression is relatively low but still detectable. The expression level in cholangiocytes, the epithelium lining the bile duct, is similar to that found in lung alveolar cells[12,19]. Interestingly, sinusoidal endothelial cells appear to lack ACE2 expression, which aligns with earlier findings resembling SARS-CoV-1[20]. Recent observations concerning SARS-CoV-2-induced endothelitis in major intrahepatic arteries, coupled with the heightened presence of ACE2 in other endothelial cell types, such as those within the central and portal veins, which are similarly susceptible to infection by the virus, suggest the potential significance of this discovery[7]. TMPRSS2 and furin gene expression are broadly distributed across various liver cell types[21]. Notably, when three distinct single-cell RNA sequencing datasets from healthy liver tissue were collectively analyzed, it was observed that very few hepatocytes co-expressed both ACE2 and TMPRSS2[22].
To investigate the susceptibility of liver cell types to SARS-CoV-2 infection, experimental models involving cellular and organoid cultures have played a pivotal role. Hepatocellular carcinoma-derived cell lines such as Huh-7 and HepG2 have demonstrated the ability to support the entire viral life cycle[23]. A significant expression of ACE2 and TMPRSS2 in liver parenchymal cells was reported using bioinformatic analyses from a single-cell transcriptome database[21]. Permissiveness was demonstrated when pseudotyped lentiviral particles expressing the full-length spike protein of SARS-CoV-2 were inoculated to primary hepatocytes obtained from ACE2-humanized mice[24].
Importantly, research conducted in both murine and human subjects has revealed an increase in hepatic ACE2 expression within hepatocytes in the presence of liver fibrosis or cirrhosis, as already documented[25]. This finding holds significant relevance because pre-existing liver injury may exacerbate the susceptibility of hepatic tissues to the hepatitis C virus, SARS-CoV-2[26]. The impact of liver injury and pre-existing liver conditions on the propensity of SARS-CoV-2 to target the liver is still not well understood, and there is a notable absence of studies that have specifically investigated the histological alterations occurring in individuals with both COVID-19 and CLD. However, it is worth noting that previous investigations conducted before the emergence of COVID-19 have reported a significantly more than 30-fold elevation in ACE2 expression within the livers of patients suffering from cirrhosis related to the hepatitis C virus compared to individuals without underlying liver conditions[25,27] (Figure 1). These findings may be associated with the gene expression patterns observed in metabolic dysfunction-associated fatty liver disease (MAFLD), previously known as non-alcoholic fatty liver disease[28]. The presence of MAFLD within the broader context of metabolic syndrome may contribute to the exacerbation of COVID-19 severity. Molecular investigations have revealed elevated expression levels of crucial viral entry receptors, including ACE2, furin, and TMPRSS2, in individuals diagnosed with MAFLD. Furthermore, the liver mRNA expression of ACE2 and TMPRSS2 was found to be upregulated in individuals without active infection. Moreover, in obese patients with MAFLD, there was an observed upregulation of ACE2 in the liver as well as in subcutaneous and visceral adipose tissues compared with obese individuals lacking MAFLD[17,29,30] (Figure 1).
In addition, it has been established that hypoxia, a characteristic feature of severe cases of COVID-19, serves as a key regulatory factor in the upregulation of ACE2 expression in hepatocytes[17,25,31,32] (Figure 1). This phenomenon may explain the prevalence of extrapulmonary dissemination of SARS-CoV-2 in patients experiencing acute respiratory distress syndrome and other hypoxic conditions. Notably, in a manner analogous to findings in other organ systems, it is conceivable that inflammatory conditions and diseases affecting the liver, as reported[33,34], could elevate the expression of ACE2. Given the potential implication of DILI in the development of liver damage in COVID-19 patients[35,36], it is particularly interesting to investigate whether such conditions or specific pharmaceutical agents may induce excessive ACE2 expression within the hepatic environment. In contrast, while not yet substantiated in human subjects, Brevini et al[37] have recently delineated in a murine model the potential of ursodeoxycholic acid to inhibit ACE2, suggesting its potential as a promising therapeutic and prophylactic strategy against SARS-CoV-2.
In vitro experiments have demonstrated that the spike (S) protein of beta-coronaviruses exhibits a significant increase in its binding affinity for its receptor when it is pre-incubated with trypsin, a process involving proteolytic activation[1]. It's worth noting that liver epithelial cells express trypsin[38] and various other serine proteases, which are continuously involved in extracellular matrix remodeling and liver regeneration[39]. Considering this scenario, there is a plausible suggestion that the expression of ACE2, a pivotal factor for the precise targeting and recognition of SARS-CoV-2 within the liver, might be comparatively diminished in comparison to other tissues where extracellular proteolytic activity is less pronounced[40,41].
In concordance with these findings, recent discoveries have brought attention to the existence of a furin-like proteolytic site within the S protein of SARS-CoV-2, a feature not found in other coronaviruses belonging to the same lineage[1]. It is interesting to note that furin expression is mostly observed in organs that are hypothesized to be susceptible to SARS-CoV-2 infection. These organs include the pancreas, kidney, liver, and salivary glands[21].
While our understanding of the tissue-specific determinants governing SARS-CoV-2 infection remains limited, there is a growing recognition of the involvement of additional accessory receptors in viral entry. Notably, studies have suggested that the high-density lipoprotein scavenger receptor B type 1 (SR-B1) plays a facilitating role in ACE2-dependent coronavirus attachment in vitro, drawing parallels with hepatitis C virus infection. Likewise, therapeutic interventions targeting SR-B1 have shown efficacy in mitigating the lipoprotein-mediated enhancement of SARS-CoV-2 infection. It is important to note, however, that using immunohistochemistry analysis of liver tissue was confirmed only sporadic ACE2 expression within the hepatic tissue[42]. Besides, it is crucial to acknowledge that additional factors, such as ganglioside (GM1)[43], may influence the interaction between the spike (S) protein and ACE2. Consequently, there is an imperative need for more comprehensive research into the S protein-ACE2 interactome to gain a deeper understanding of the molecular mechanisms involved and explore potential therapeutic avenues.
Ou et al[44,45] used pseudovirions carrying the spike (S) protein of SARS-CoV-2 to assess their ability to infect various cell lines. When exposed to viral vectors expressing the SARS-CoV-2 S protein, HuH7 and Calu3 cells (a cell line originating from human lung cancer) were more susceptible to transfection than reference pseudovirus. Additionally, these investigations suggested that the PIKfyve-TCP2 endocytotic pathway, which is expressed at lung-like levels in the liver and gall bladder[15], could be important for the viral entry process[46].
HuH7 cells has been reported as a permissive model to develop a novel and effective functional viromics screening method to forecast the possibility of zoonotic occurrences with known lineage B betacoronaviruses. This model was employed to investigate the binding and recognition processes of both SARS-CoV-1 and SARS-CoV-2[47]. This approach further confirmed the affinity of SARS-CoV-2 for hepatocytes. It is important to note that in their study, HuH7 cells were identified as the third most permissive cell line, following pulmonary (Calu3) and intestinal (CaCo2) cell models, which represent organs with histopathological evidence of SARS-CoV-2 infection[47]. However, it is important to recognize that a cell's ability to attach and internalize viral particles does not always indicate that the particular cell type is also supportive of efficient viral reproduction. In this regard, it has shown that HuH7 cells indeed facilitate SARS-CoV-2 viral multiplication[23,48]. It has been determined that hepatocyte cell lines are robust permissive cell types for infections with SARS-CoV-1 and SARS-CoV-2. Notably, HuH7 cells have recently been used in SARS-CoV-2 immunostaining assays as a positive control[49]. It is essential to underscore that the findings suggesting hepatocytes as potential hosts for SARS-CoV-2 primarily stem from studies conducted with cancer cell lines. To establish the clinical relevance of these observations, it is crucial to conduct a comparison of ACE2 protein expression in HuH7 cells with that observed in primary human hepatocytes.
Post-mortem autopsies have yielded evidence supporting the concept of direct infection of liver cells by SARS-CoV-2. Several studies have recorded the identification of SARS-CoV-2 in a notable portion of post-mortem liver biopsies, employing techniques such as PCR and in situ hybridization. However, the direct invasion of hepatocytes by the virus was not consistently confirmed. Nonetheless, certain researchers managed to demonstrate the presence of distinct coronavirus particles, including spike structures, within the cytoplasm of hepatocytes in individuals with COVID-19. These observations were accompanied by signs of mitochondrial swelling and apoptosis, suggesting a potential link between the virus and cellular damage in the liver[7,9,50]. The diverse spectrum of histological injury patterns observed in individuals infected with SARS-CoV-2, including features such as macrovascular and microvascular steatosis, lobular necroinflammation, portal inflammation, and vascular pathology (Figure 1), likely emphasizes the intricate and multifactorial nature underlying abnormal liver test results in the context of COVID-19-associated liver injury[14-16,51]. Perhaps the most compelling evidence of SARS-CoV-2's ability to infect liver tissue was recently presented by Wanner et al[52]. In their study, the authors presented multiple lines of evidence for SARS-CoV-2 liver tropism, including the direct identification of SARS-CoV-2 genomic material within hepatocytes using in situ hybridization. In our study and theirs, infectious SARS-CoV-2 was isolated from post-mortem liver tissue[53]. Furthermore, Wanner et al[52] delineated activity profiles through transcriptomic and proteomic analyses in hepatic samples, affirming the presence of established SARS-CoV-2 entry receptors and facilitators of infection, encompassing ACE2, TMPRSS2, procathepsin L, and the Ras-related protein Rab-7a. The analyses also unveiled pronounced upregulation in interferon responses, JAK-STAT signaling, and liver-specific metabolic modulation. These findings collectively suggest a viral activity profile bearing notable resemblances to other hepatotropic viral infections, notably hepatitis C virus infection[52]. Moreover, it is imperative to conduct further investigations aimed at unraveling the molecular alterations initiated in hepatocytes subsequent to SARS-CoV-2 infection.
Valuable insights into this matter can be derived from the research conducted by Yang et al[54]. Using organoids created from human hepatocytes generated from pluripotent stem cells and primary adult human hepatocytes, their work confirmed SARS-CoV-2 hepatotropism. Using these organoids, the S-expressing pseudovirus of SARS-CoV-2 demonstrated the ability to infect human hepatocytes, leading to substantial viral replication. Additionally, gene expression analyses indicated that primary hepatocytes infected with SARS-CoV-2 exhibited heightened expression of pro-inflammatory cytokines, coupled with the downregulation of essential metabolic functions, as evidenced by the inhibition of CYP7A1, CYP2A6, CYP1A2, and CYP2D6 expression[54]. Wang et al[9] made a noteworthy advancement when they used electron microscope imaging to examine liver tissues from two deceased COVID-19 patients. They found that the hepatocytes they studied had viral structures that resembled SARS-CoV-2 virions. This data suggests that, even in the absence of a traditional hepatitis pattern, the histological alterations seen in these individuals might be the result of SARS-CoV-2's direct cytopathic effects[55]. It is important, therefore, that more research utilizing more extensive biopsy or autopsy cohorts in conjunction with all-encompassing imaging methods, including immunological electron microscopy, could be necessary to validate these preliminary findings about the existence of SARS-CoV-2 in hepatocytes[56].
Bile duct epithelial cells, also referred to as cholangiocytes, fulfill pivotal functions in both the generation and regulation of bile, while also contributing to immune responses[57]. Single-cell sequencing of long-term liver ductal organoid cultures derived from human tissues revealed the persistence of ACE2 and TMPRSS2 expression[58] (Figure 1). Cholangiocytes were infected with SARS-CoV-2, causing syncytia formation. Twenty-four hours after the infection, there was a notable rise in the amount of SARS-CoV-2 genomic RNA. When the virus was inoculated to adult human cholangiocyte organoids, similar outcomes were seen, thus showing that SARS-CoV-2 infection in vitro may occur in human liver ductal organoids[54], raising the possibility of viral replication within the bile duct epithelium in vivo. Despite the notably elevated expression of ACE2 in cholangiocytes compared to hepatocytes, there are no reports of direct proof of SARS-CoV-2 infection in cholangiocytes in COVID-19 patients. Since hepatocytes and cholangiocytes are the primary producers of bile and because biliary fluids and cholangiocytes' apical membrane interact directly and continuously, the presence of SARS-CoV-2 viral RNA or proteins in bile may be an indirect indicator of cholangiocyte SARS-CoV-2 infection. Currently, there is just one case report that shows SARS-CoV-2 RNA exists in bile, while bile samples from two other small sample series tested negative. Such disparities could be attributed to the circumstance that the bile sample yielding a positive result was obtained during the surgical resolution of bile duct obstruction, whereas the bile sample yielding a negative result was obtained from post-mortem autopsies conducted 48 h after death[59,60].
Tight junctions are essential for cholangiocytes to act as a barrier that protects parenchymal liver cells from potentially hazardous components of bile. Notably, in vitro studies have shown that viral infection with SARS-CoV-2 Leads to a decrease in the mRNA expression of tight junction proteins such as claudin 1 in cholangiocytes, implying a compromised barrier function of these cells[58]. This disruption could result in liver injury, because it may allow toxic bile components to leak into the periductal space and adjacent liver parenchyma. Furthermore, SARS-CoV-2 infection downregulates the expression of hepatobiliary transporters, such as SLC10A2/ASBT and the chloride channel ABCC7/CFTR[58](Figure 1). This downregulation of hepatobiliary transporters could compromise the sensing and signaling of bile acids by cholangiocytes and the secretion of bicarbonate. Consequently, this could contribute to the identified biliary changes in individuals with COVID-19[61]. Additionally, inflammatory pathways were increased in SARS-CoV-2 infected cholangiocytes, indicating the establishment of a reactive phenotype[54]. Prospect investigation are needed to investigate if and how SARS-CoV-2 promoted cytokine production favoring inflammation and fibrosis, potentially playing a role in the development of the "reactive cholangiocyte phenotype". Such alterations have the potential to propagate inflammation and fibrosis[57].
Alveolar macrophages and monocyte-derived macrophages (MDM) are known to express ACE2, and immunohistochemistry has revealed evidence of viral protein infection of alveolar macrophages caused by both SARS-CoV-1 and SARS-CoV-2[62-64]. Nonetheless, during a histopathological assessment of ACE2 tissue distribution, no staining for ACE2 was detected in Kupffer cells and other hepatic immune cells, despite the typical observation of Kupffer cell proliferation in the livers of individuals with COVID-19[9,65] (Figure 1).
Recent investigations in response to the COVID-19 pandemic have involved more comprehensive examinations of ACE2 expression patterns. These investigations included de novo single-cell RNA sequencing analyses and in silico evaluations of RNA sequencing databases. The results of these studies have consistently shown that Kupffer cells do not express ACE2. In contrast, a recent report that differentiates ACE2 expression in tissue macrophages demonstrated a high level of expression even among Kupffer cells[65]. However, it is crucial to emphasize that the evidence and findings reported thus far are based on samples from healthy human livers. Therefore, it may be necessary to quantify ACE2 expression in samples taken from individuals who had either an acute liver injury or underlying chronic liver illness to gain a more comprehensive understanding of different patterns of ACE2 expression in macrophages under such conditions[65,66].
It is worth noting that following liver injury or Kupffer cell depletion, MDM can infiltrate the liver and efficiently replenish the resident hepatic macrophage population[67-69]. While in vitro observations have indicated that MDM may not efficiently support the replication of SARS-CoV-1 (and likely SARS-CoV-2), infected MDM could serve as carriers of the pathogen, facilitating the infection of ACE2-expressing cells in the affected organ[70]. Additionally, Kupffer cell activation and proliferation are commonly observed due to systemic inflammation, and Kupffer cell activation has been reported in liver specimens from deceased COVID-19 patients. Through the propagation of inflammatory signals, monocytic cells may be important in SARS-CoV-2-mediated liver damage, even if ACE2 expression among Kupffer cells is a matter of debate[64].
Pre-existing chronic liver diseases seem to be independent risk factors associated with unfavorable outcomes in COVID-19, with the cirrhosis grade identified as a predictor of mortality in patients infected with SARS-CoV-2[71]. Since hepatic stellate cells are the main source of fibrosis, their activation is a crucial step in the development of chronic liver disease[72,73]. Activation is induced by proinflammatory and profibrotic signals, including angiotensin II, and arises fibrosis through the enzymatic activity of ACE within the profibrotic segment of the renin-angiotensin system[74] (Figure 1). Interestingly, ACE2 acts as an antagonist to ACE, generating the anti-inflammatory and anti-fibrotic peptide angiotensin-(1–7) and lowering the ratio of angiotensin II to angiotensin–(1–7) as a result[74]. Nevertheless, neither fibrogenic nor activated cells nor quiescent hepatic stellate cells have been shown to express ACE2[74-76]. These findings imply that these cells may not serve as highly permissive hosts for SARS-CoV-2. Nevertheless, the pro-inflammatory environment instigated by direct or indirect injury to hepatocytes and cholangiocytes in the context of COVID-19 may establish conditions conducive to the activation of hepatic stellate cells, thereby initiating the process of fibrosis (Figure 1). This scenario may be particularly pertinent for individuals who have already underlying chronic liver diseases, such as MAFLD as a condition characterized by steatosis in > 5% of the liver parenchyma. While available data indicate that liver injury caused by COVID-19 is typically mild and temporary, long-term surveillance studies are essential to fully assess the possibility of hepatic fibrosis developing as a long-term effect of COVID-19, especially in patients with pre-existing liver diseases. In the context of MAFLD, inflamed hepatocytes, along with other somatic cells, may manifest mito
The understanding of the interaction of SARS-CoV-2 with the liver is still evolving, and more research is needed to fully elucidate the molecular mechanisms involved in liver tropism and damage in COVID-19. The complexity of these mechanisms underscores the importance of monitoring and managing liver function in patients with COVID-19, particularly those with underlying liver conditions.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Argentina
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Manautou JE, United States; Mukhopadhyay A, India; Papadopoulos K, Thailand S-Editor: Lin C L-Editor: A P-Editor: Zhao S
| 1. | Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022;23:3-20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1831] [Cited by in RCA: 1911] [Article Influence: 637.0] [Reference Citation Analysis (0)] |
| 2. | V'kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155-170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2164] [Cited by in RCA: 2013] [Article Influence: 503.3] [Reference Citation Analysis (0)] |
| 3. | Delorey TM, Ziegler CGK, Heimberg G, Normand R, Yang Y, Segerstolpe Å, Abbondanza D, Fleming SJ, Subramanian A, Montoro DT, Jagadeesh KA, Dey KK, Sen P, Slyper M, Pita-Juárez YH, Phillips D, Biermann J, Bloom-Ackermann Z, Barkas N, Ganna A, Gomez J, Melms JC, Katsyv I, Normandin E, Naderi P, Popov YV, Raju SS, Niezen S, Tsai LT, Siddle KJ, Sud M, Tran VM, Vellarikkal SK, Wang Y, Amir-Zilberstein L, Atri DS, Beechem J, Brook OR, Chen J, Divakar P, Dorceus P, Engreitz JM, Essene A, Fitzgerald DM, Fropf R, Gazal S, Gould J, Grzyb J, Harvey T, Hecht J, Hether T, Jané-Valbuena J, Leney-Greene M, Ma H, McCabe C, McLoughlin DE, Miller EM, Muus C, Niemi M, Padera R, Pan L, Pant D, Pe'er C, Pfiffner-Borges J, Pinto CJ, Plaisted J, Reeves J, Ross M, Rudy M, Rueckert EH, Siciliano M, Sturm A, Todres E, Waghray A, Warren S, Zhang S, Zollinger DR, Cosimi L, Gupta RM, Hacohen N, Hibshoosh H, Hide W, Price AL, Rajagopal J, Tata PR, Riedel S, Szabo G, Tickle TL, Ellinor PT, Hung D, Sabeti PC, Novak R, Rogers R, Ingber DE, Jiang ZG, Juric D, Babadi M, Farhi SL, Izar B, Stone JR, Vlachos IS, Solomon IH, Ashenberg O, Porter CBM, Li B, Shalek AK, Villani AC, Rozenblatt-Rosen O, Regev A. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595:107-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 590] [Cited by in RCA: 555] [Article Influence: 138.8] [Reference Citation Analysis (0)] |
| 4. | Kariyawasam JC, Jayarajah U, Abeysuriya V, Riza R, Seneviratne SL. Involvement of the Liver in COVID-19: A Systematic Review. Am J Trop Med Hyg. 2022;106:1026-1041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 5. | Yadav DK, Singh A, Zhang Q, Bai X, Zhang W, Yadav RK, Zhiwei L, Adhikari VP, Liang T. Involvement of liver in COVID-19: systematic review and meta-analysis. Gut. 2021;70:807-809. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 6. | Roshanshad R, Roshanshad A, Fereidooni R, Hosseini-Bensenjan M. COVID-19 and liver injury: Pathophysiology, risk factors, outcome and management in special populations. World J Hepatol. 2023;15:441-459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Sonzogni A, Previtali G, Seghezzi M, Grazia Alessio M, Gianatti A, Licini L, Morotti D, Zerbi P, Carsana L, Rossi R, Lauri E, Pellegrinelli A, Nebuloni M. Liver histopathology in severe COVID 19 respiratory failure is suggestive of vascular alterations. Liver Int. 2020;40:2110-2116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 218] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 8. | D'Ardes D, Boccatonda A, Cocco G, Fabiani S, Rossi I, Bucci M, Guagnano MT, Schiavone C, Cipollone F. Impaired coagulation, liver dysfunction and COVID-19: Discovering an intriguing relationship. World J Gastroenterol. 2022;28:1102-1112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 41] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 9. | Wang Y, Liu S, Liu H, Li W, Lin F, Jiang L, Li X, Xu P, Zhang L, Zhao L, Cao Y, Kang J, Yang J, Li L, Liu X, Li Y, Nie R, Mu J, Lu F, Zhao S, Lu J, Zhao J. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J Hepatol. 2020;73:807-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 457] [Article Influence: 91.4] [Reference Citation Analysis (0)] |
| 10. | Saviano A, Wrensch F, Ghany MG, Baumert TF. Liver Disease and Coronavirus Disease 2019: From Pathogenesis to Clinical Care. Hepatology. 2021;74:1088-1100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 11. | Romano C, Cozzolino D, Nevola R, Abitabile M, Carusone C, Cinone F, Cuomo G, Nappo F, Sellitto A, Umano GR, Adinolfi LE, Marrone A, Rinaldi L. Liver Involvement during SARS-CoV-2 Infection Is Associated with a Worse Respiratory Outcome in COVID-19 Patients. Viruses. 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 12. | Luo M, Ballester MP, Soffientini U, Jalan R, Mehta G. SARS-CoV-2 infection and liver involvement. Hepatol Int. 2022;16:755-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 13. | Piano S, Dalbeni A, Vettore E, Benfaremo D, Mattioli M, Gambino CG, Framba V, Cerruti L, Mantovani A, Martini A, Luchetti MM, Serra R, Cattelan A, Vettor R, Angeli P; COVID-LIVER study group. Abnormal liver function tests predict transfer to intensive care unit and death in COVID-19. Liver Int. 2020;40:2394-2406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 92] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 14. | Li P, Liu Y, Cheng Z, Yu X, Li Y. COVID-19-associated liver injury: Clinical characteristics, pathophysiological mechanisms and treatment management. Biomed Pharmacother. 2022;154:113568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 15. | Nardo AD, Schneeweiss-Gleixner M, Bakail M, Dixon ED, Lax SF, Trauner M. Pathophysiological mechanisms of liver injury in COVID-19. Liver Int. 2021;41:20-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 281] [Cited by in RCA: 271] [Article Influence: 67.8] [Reference Citation Analysis (2)] |
| 16. | Marjot T, Webb GJ, Barritt AS 4th, Moon AM, Stamataki Z, Wong VW, Barnes E. COVID-19 and liver disease: mechanistic and clinical perspectives. Nat Rev Gastroenterol Hepatol. 2021;18:348-364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 278] [Cited by in RCA: 272] [Article Influence: 68.0] [Reference Citation Analysis (2)] |
| 17. | Ali FEM, Abd El-Aziz MK, Ali MM, Ghogar OM, Bakr AG. COVID-19 and hepatic injury: cellular and molecular mechanisms in diverse liver cells. World J Gastroenterol. 2023;29:425-449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Hartl L, Haslinger K, Angerer M, Semmler G, Schneeweiss-Gleixner M, Jachs M, Simbrunner B, Bauer DJM, Eigenbauer E, Strassl R, Breuer M, Kimberger O, Laxar D, Lampichler K, Halilbasic E, Stättermayer AF, Ba-Ssalamah A, Mandorfer M, Scheiner B, Reiberger T, Trauner M. Progressive cholestasis and associated sclerosing cholangitis are frequent complications of COVID-19 in patients with chronic liver disease. Hepatology. 2022;76:1563-1575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 56] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 19. | Bucurica S, Ionita Radu F, Bucurica A, Socol C, Prodan I, Tudor I, Sirbu CA, Plesa FC, Jinga M. Risk of New-Onset Liver Injuries Due to COVID-19 in Preexisting Hepatic Conditions-Review of the Literature. Medicina (Kaunas). 2022;59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 20. | Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631-637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3643] [Cited by in RCA: 4149] [Article Influence: 197.6] [Reference Citation Analysis (0)] |
| 21. | Pirola CJ, Sookoian S. SARS-CoV-2 virus and liver expression of host receptors: Putative mechanisms of liver involvement in COVID-19. Liver Int. 2020;40:2038-2040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 22. | De Smet V, Verhulst S, van Grunsven LA. Single cell RNA sequencing analysis did not predict hepatocyte infection by SARS-CoV-2. J Hepatol. 2020;73:993-995. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 23. | Chu H, Chan JF, Yuen TT, Shuai H, Yuan S, Wang Y, Hu B, Yip CC, Tsang JO, Huang X, Chai Y, Yang D, Hou Y, Chik KK, Zhang X, Fung AY, Tsoi HW, Cai JP, Chan WM, Ip JD, Chu AW, Zhou J, Lung DC, Kok KH, To KK, Tsang OT, Chan KH, Yuen KY. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14-e23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 507] [Cited by in RCA: 600] [Article Influence: 120.0] [Reference Citation Analysis (0)] |
| 24. | Mercado-Gómez M, Prieto-Fernández E, Goikoetxea-Usandizaga N, Vila-Vecilla L, Azkargorta M, Bravo M, Serrano-Maciá M, Egia-Mendikute L, Rodríguez-Agudo R, Lachiondo-Ortega S, Lee SY, Eguileor Giné A, Gil-Pitarch C, González-Recio I, Simón J, Petrov P, Jover R, Martínez-Cruz LA, Ereño-Orbea J, Delgado TC, Elortza F, Jiménez-Barbero J, Nogueiras R, Prevot V, Palazon A, Martínez-Chantar ML. The spike of SARS-CoV-2 promotes metabolic rewiring in hepatocytes. Commun Biol. 2022;5:827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 25. | Paizis G, Tikellis C, Cooper ME, Schembri JM, Lew RA, Smith AI, Shaw T, Warner FJ, Zuilli A, Burrell LM, Angus PW. Chronic liver injury in rats and humans upregulates the novel enzyme angiotensin converting enzyme 2. Gut. 2005;54:1790-1796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 281] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 26. | Domovitz T, Ayoub S, Werbner M, Alter J, Izhaki Tavor L, Yahalom-Ronen Y, Tikhonov E, Meirson T, Maman Y, Paran N, Israely T, Dessau M, Gal-Tanamy M. HCV Infection Increases the Expression of ACE2 Receptor, Leading to Enhanced Entry of Both HCV and SARS-CoV-2 into Hepatocytes and a Coinfection State. Microbiol Spectr. 2022;10:e0115022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 27. | Zhang W, Miao J, Li P, Wang Y, Zhang Y. Up-regulation of components of the renin-angiotensin system in liver fibrosis in the rat induced by CCL₄. Res Vet Sci. 2013;95:54-58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 28. | Méndez-Sánchez N, Bugianesi E, Gish RG, Lammert F, Tilg H, Nguyen MH, Sarin SK, Fabrellas N, Zelber-Sagi S, Fan JG, Shiha G, Targher G, Zheng MH, Chan WK, Vinker S, Kawaguchi T, Castera L, Yilmaz Y, Korenjak M, Spearman CW, Ungan M, Palmer M, El-Shabrawi M, Gruss HJ, Dufour JF, Dhawan A, Wedemeyer H, George J, Valenti L, Fouad Y, Romero-Gomez M, Eslam M; Global multi-stakeholder consensus on the redefinition of fatty liver disease. Global multi-stakeholder endorsement of the MAFLD definition. Lancet Gastroenterol Hepatol. 2022;7:388-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 188] [Article Influence: 62.7] [Reference Citation Analysis (0)] |
| 29. | Fondevila MF, Mercado-Gómez M, Rodríguez A, Gonzalez-Rellan MJ, Iruzubieta P, Valentí V, Escalada J, Schwaninger M, Prevot V, Dieguez C, Crespo J, Frühbeck G, Martinez-Chantar ML, Nogueiras R. Obese patients with NASH have increased hepatic expression of SARS-CoV-2 critical entry points. J Hepatol. 2021;74:469-471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 30. | Meijnikman AS, Bruin S, Groen AK, Nieuwdorp M, Herrema H. Increased expression of key SARS-CoV-2 entry points in multiple tissues in individuals with NAFLD. J Hepatol. 2021;74:748-749. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 31. | Xia CY, Li L, Liu HM, Cong WM. High expression of angiotensin-converting enzyme and angiotensin-converting enzyme 2 in preservation injury after liver transplantation in rats. Hepatol Res. 2009;39:1118-1124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 32. | Herath CB, Warner FJ, Lubel JS, Dean RG, Jia Z, Lew RA, Smith AI, Burrell LM, Angus PW. Upregulation of hepatic angiotensin-converting enzyme 2 (ACE2) and angiotensin-(1-7) levels in experimental biliary fibrosis. J Hepatol. 2007;47:387-395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 127] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 33. | Suárez-Fariñas M, Tokuyama M, Wei G, Huang R, Livanos A, Jha D, Levescot A, Irizar H, Kosoy R, Cording S, Wang W, Losic B, Ungaro RC, Di'Narzo A, Martinez-Delgado G, Suprun M, Corley MJ, Stojmirovic A, Houten SM, Peters L, Curran M, Brodmerkel C, Perrigoue J, Friedman JR, Hao K, Schadt EE, Zhu J, Ko HM, Cho J, Dubinsky MC, Sands BE, Ndhlovu L, Cerf-Bensusan N, Kasarskis A, Colombel JF, Harpaz N, Argmann C, Mehandru S. Intestinal Inflammation Modulates the Expression of ACE2 and TMPRSS2 and Potentially Overlaps With the Pathogenesis of SARS-CoV-2-related Disease. Gastroenterology. 2021;160:287-301.e20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 97] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 34. | Penninger JM, Grant MB, Sung JJY. The Role of Angiotensin Converting Enzyme 2 in Modulating Gut Microbiota, Intestinal Inflammation, and Coronavirus Infection. Gastroenterology. 2021;160:39-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 103] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 35. | Muhović D, Bojović J, Bulatović A, Vukčević B, Ratković M, Lazović R, Smolović B. First case of drug-induced liver injury associated with the use of tocilizumab in a patient with COVID-19. Liver Int. 2020;40:1901-1905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 36. | Delgado A, Stewart S, Urroz M, Rodríguez A, Borobia AM, Akatbach-Bousaid I, González-Muñoz M, Ramírez E. Characterisation of Drug-Induced Liver Injury in Patients with COVID-19 Detected by a Proactive Pharmacovigilance Program from Laboratory Signals. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 37. | Brevini T, Maes M, Webb GJ, John BV, Fuchs CD, Buescher G, Wang L, Griffiths C, Brown ML, Scott WE 3rd, Pereyra-Gerber P, Gelson WTH, Brown S, Dillon S, Muraro D, Sharp J, Neary M, Box H, Tatham L, Stewart J, Curley P, Pertinez H, Forrest S, Mlcochova P, Varankar SS, Darvish-Damavandi M, Mulcahy VL, Kuc RE, Williams TL, Heslop JA, Rossetti D, Tysoe OC, Galanakis V, Vila-Gonzalez M, Crozier TWM, Bargehr J, Sinha S, Upponi SS, Fear C, Swift L, Saeb-Parsy K, Davies SE, Wester A, Hagström H, Melum E, Clements D, Humphreys P, Herriott J, Kijak E, Cox H, Bramwell C, Valentijn A, Illingworth CJR; UK-PBC Consortium, Dahman B, Bastaich DR, Ferreira RD, Marjot T, Barnes E, Moon AM, Barritt AS 4th, Gupta RK, Baker S, Davenport AP, Corbett G, Gorgoulis VG, Buczacki SJA, Lee JH, Matheson NJ, Trauner M, Fisher AJ, Gibbs P, Butler AJ, Watson CJE, Mells GF, Dougan G, Owen A, Lohse AW, Vallier L, Sampaziotis F. FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2. Nature. 2023;615:134-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 195] [Article Influence: 97.5] [Reference Citation Analysis (0)] |
| 38. | Koshikawa N, Hasegawa S, Nagashima Y, Mitsuhashi K, Tsubota Y, Miyata S, Miyagi Y, Yasumitsu H, Miyazaki K. Expression of trypsin by epithelial cells of various tissues, leukocytes, and neurons in human and mouse. Am J Pathol. 1998;153:937-944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 166] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 39. | Huang W, Han N, Du L, Wang M, Chen L, Tang H. A narrative review of liver regeneration-from models to molecular basis. Ann Transl Med. 2021;9:1705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 40. | Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pöhlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88:1293-1307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 602] [Cited by in RCA: 669] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 41. | Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271-280.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11946] [Cited by in RCA: 14253] [Article Influence: 2850.6] [Reference Citation Analysis (0)] |
| 42. | Wei C, Wan L, Yan Q, Wang X, Zhang J, Yang X, Zhang Y, Fan C, Li D, Deng Y, Sun J, Gong J, Wang Y, Li J, Yang H, Li H, Zhang Z, Wang R, Du P, Zong Y, Yin F, Zhang W, Wang N, Peng Y, Lin H, Feng J, Qin C, Chen W, Gao Q, Zhang R, Cao Y, Zhong H. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat Metab. 2020;2:1391-1400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 210] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 43. | Fantini J, Di Scala C, Chahinian H, Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents. 2020;55:105960. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 354] [Cited by in RCA: 400] [Article Influence: 80.0] [Reference Citation Analysis (0)] |
| 44. | Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. Author Correction: Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2021;12:2144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 45. | Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2003] [Cited by in RCA: 2289] [Article Influence: 457.8] [Reference Citation Analysis (0)] |
| 46. | Aganovic A. pH-dependent endocytosis mechanisms for influenza A and SARS-coronavirus. Front Microbiol. 2023;14:1190463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 47. | Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562-569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1933] [Cited by in RCA: 2212] [Article Influence: 442.4] [Reference Citation Analysis (0)] |
| 48. | Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, Queen K, Tao Y, Paden CR, Zhang J, Li Y, Uehara A, Wang H, Goldsmith C, Bullock HA, Wang L, Whitaker B, Lynch B, Gautam R, Schindewolf C, Lokugamage KG, Scharton D, Plante JA, Mirchandani D, Widen SG, Narayanan K, Makino S, Ksiazek TG, Plante KS, Weaver SC, Lindstrom S, Tong S, Menachery VD, Thornburg NJ. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States. Emerg Infect Dis. 2020;26:1266-1273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 384] [Cited by in RCA: 462] [Article Influence: 92.4] [Reference Citation Analysis (0)] |
| 49. | Zhang H, Zhou P, Wei Y, Yue H, Wang Y, Hu M, Zhang S, Cao T, Yang C, Li M, Guo G, Chen X, Chen Y, Lei M, Liu H, Zhao J, Peng P, Wang CY, Du R. Histopathologic Changes and SARS-CoV-2 Immunostaining in the Lung of a Patient With COVID-19. Ann Intern Med. 2020;172:629-632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 308] [Cited by in RCA: 376] [Article Influence: 75.2] [Reference Citation Analysis (0)] |
| 50. | Lagana SM, Kudose S, Iuga AC, Lee MJ, Fazlollahi L, Remotti HE, Del Portillo A, De Michele S, de Gonzalez AK, Saqi A, Khairallah P, Chong AM, Park H, Uhlemann AC, Lefkowitch JH, Verna EC. Hepatic pathology in patients dying of COVID-19: a series of 40 cases including clinical, histologic, and virologic data. Mod Pathol. 2020;33:2147-2155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 181] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 51. | Díaz LA, Idalsoaga F, Cannistra M, Candia R, Cabrera D, Barrera F, Soza A, Graham R, Riquelme A, Arrese M, Leise MD, Arab JP. High prevalence of hepatic steatosis and vascular thrombosis in COVID-19: A systematic review and meta-analysis of autopsy data. World J Gastroenterol. 2020;26:7693-7706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 61] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (1)] |
| 52. | Wanner N, Andrieux G, Badia-I-Mompel P, Edler C, Pfefferle S, Lindenmeyer MT, Schmidt-Lauber C, Czogalla J, Wong MN, Okabayashi Y, Braun F, Lütgehetmann M, Meister E, Lu S, Noriega MLM, Günther T, Grundhoff A, Fischer N, Bräuninger H, Lindner D, Westermann D, Haas F, Roedl K, Kluge S, Addo MM, Huber S, Lohse AW, Reiser J, Ondruschka B, Sperhake JP, Saez-Rodriguez J, Boerries M, Hayek SS, Aepfelbacher M, Scaturro P, Puelles VG, Huber TB. Molecular consequences of SARS-CoV-2 liver tropism. Nat Metab. 2022;4:310-319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 122] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 53. | Maffia-Bizzozero S, Cevallos C, Lenicov FR, Freiberger RN, Lopez CAM, Guano Toaquiza A, Sviercz F, Jarmoluk P, Bustos C, D'Addario AC, Quarleri J, Delpino MV. Viable SARS-CoV-2 Omicron sub-variants isolated from autopsy tissues. Front Microbiol. 2023;14:1192832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 54. | Yang L, Han Y, Nilsson-Payant BE, Gupta V, Wang P, Duan X, Tang X, Zhu J, Zhao Z, Jaffré F, Zhang T, Kim TW, Harschnitz O, Redmond D, Houghton S, Liu C, Naji A, Ciceri G, Guttikonda S, Bram Y, Nguyen DT, Cioffi M, Chandar V, Hoagland DA, Huang Y, Xiang J, Wang H, Lyden D, Borczuk A, Chen HJ, Studer L, Pan FC, Ho DD, tenOever BR, Evans T, Schwartz RE, Chen S. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell. 2020;27:125-136.e7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 529] [Cited by in RCA: 511] [Article Influence: 102.2] [Reference Citation Analysis (0)] |
| 55. | Stein SR, Ramelli SC, Grazioli A, Chung JY, Singh M, Yinda CK, Winkler CW, Sun J, Dickey JM, Ylaya K, Ko SH, Platt AP, Burbelo PD, Quezado M, Pittaluga S, Purcell M, Munster VJ, Belinky F, Ramos-Benitez MJ, Boritz EA, Lach IA, Herr DL, Rabin J, Saharia KK, Madathil RJ, Tabatabai A, Soherwardi S, McCurdy MT; NIH COVID-19 Autopsy Consortium, Peterson KE, Cohen JI, de Wit E, Vannella KM, Hewitt SM, Kleiner DE, Chertow DS. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature. 2022;612:758-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 526] [Article Influence: 175.3] [Reference Citation Analysis (0)] |
| 56. | Heinrich F, Mertz KD, Glatzel M, Beer M, Krasemann S. Using autopsies to dissect COVID-19 pathogenesis. Nat Microbiol. 2023;8:1986-1994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 57. | Banales JM, Huebert RC, Karlsen T, Strazzabosco M, LaRusso NF, Gores GJ. Cholangiocyte pathobiology. Nat Rev Gastroenterol Hepatol. 2019;16:269-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 341] [Article Influence: 56.8] [Reference Citation Analysis (1)] |
| 58. | Zhao B, Ni C, Gao R, Wang Y, Yang L, Wei J, Lv T, Liang J, Zhang Q, Xu W, Xie Y, Wang X, Yuan Z, Zhang R, Lin X. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell. 2020;11:771-775. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 268] [Cited by in RCA: 311] [Article Influence: 62.2] [Reference Citation Analysis (0)] |
| 59. | Skok K, Stelzl E, Trauner M, Kessler HH, Lax SF. Post-mortem viral dynamics and tropism in COVID-19 patients in correlation with organ damage. Virchows Arch. 2021;478:343-353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 71] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 60. | Han D, Fang Q, Wang X. SARS-CoV-2 was found in the bile juice from a patient with severe COVID-19. J Med Virol. 2021;93:102-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 61. | Lax SF, Skok K, Zechner P, Kessler HH, Kaufmann N, Koelblinger C, Vander K, Bargfrieder U, Trauner M. Pulmonary Arterial Thrombosis in COVID-19 With Fatal Outcome : Results From a Prospective, Single-Center, Clinicopathologic Case Series. Ann Intern Med. 2020;173:350-361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 600] [Cited by in RCA: 618] [Article Influence: 123.6] [Reference Citation Analysis (0)] |
| 62. | Schaefer IM, Padera RF, Solomon IH, Kanjilal S, Hammer MM, Hornick JL, Sholl LM. In situ detection of SARS-CoV-2 in lungs and airways of patients with COVID-19. Mod Pathol. 2020;33:2104-2114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 235] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 63. | Shieh WJ, Hsiao CH, Paddock CD, Guarner J, Goldsmith CS, Tatti K, Packard M, Mueller L, Wu MZ, Rollin P, Su IJ, Zaki SR. Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum Pathol. 2005;36:303-309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 122] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 64. | Labzin LI, Chew KY, Eschke K, Wang X, Esposito T, Stocks CJ, Rae J, Patrick R, Mostafavi H, Hill B, Yordanov TE, Holley CL, Emming S, Fritzlar S, Mordant FL, Steinfort DP, Subbarao K, Nefzger CM, Lagendijk AK, Gordon EJ, Parton RG, Short KR, Londrigan SL, Schroder K. Macrophage ACE2 is necessary for SARS-CoV-2 replication and subsequent cytokine responses that restrict continued virion release. Sci Signal. 2023;16:eabq1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 65. | Song X, Hu W, Yu H, Zhao L, Zhao Y, Zhao X, Xue HH. Little to no expression of angiotensin-converting enzyme-2 on most human peripheral blood immune cells but highly expressed on tissue macrophages. Cytometry A. 2023;103:136-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 83] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 66. | Qi F, Qian S, Zhang S, Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 2020;526:135-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 523] [Cited by in RCA: 741] [Article Influence: 148.2] [Reference Citation Analysis (0)] |
| 67. | Scott CL, Zheng F, De Baetselier P, Martens L, Saeys Y, De Prijck S, Lippens S, Abels C, Schoonooghe S, Raes G, Devoogdt N, Lambrecht BN, Beschin A, Guilliams M. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun. 2016;7:10321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 614] [Cited by in RCA: 651] [Article Influence: 72.3] [Reference Citation Analysis (1)] |
| 68. | Bonnardel J, T'Jonck W, Gaublomme D, Browaeys R, Scott CL, Martens L, Vanneste B, De Prijck S, Nedospasov SA, Kremer A, Van Hamme E, Borghgraef P, Toussaint W, De Bleser P, Mannaerts I, Beschin A, van Grunsven LA, Lambrecht BN, Taghon T, Lippens S, Elewaut D, Saeys Y, Guilliams M. Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity. 2019;51:638-654.e9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 336] [Cited by in RCA: 417] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 69. | Musrati MA, De Baetselier P, Movahedi K, Van Ginderachter JA. Ontogeny, functions and reprogramming of Kupffer cells upon infectious disease. Front Immunol. 2023;14:1238452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 70. | Abassi Z, Knaney Y, Karram T, Heyman SN. The Lung Macrophage in SARS-CoV-2 Infection: A Friend or a Foe? Front Immunol. 2020;11:1312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 138] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 71. | Moon AM, Webb GJ, Aloman C, Armstrong MJ, Cargill T, Dhanasekaran R, Genescà J, Gill US, James TW, Jones PD, Marshall A, Mells G, Perumalswami PV, Qi X, Su F, Ufere NN, Barnes E, Barritt AS, Marjot T. High mortality rates for SARS-CoV-2 infection in patients with pre-existing chronic liver disease and cirrhosis: Preliminary results from an international registry. J Hepatol. 2020;73:705-708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 203] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 72. | Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, Pradere JP, Schwabe RF. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 750] [Cited by in RCA: 1052] [Article Influence: 95.6] [Reference Citation Analysis (0)] |
| 73. | Cogliati B, Yashaswini CN, Wang S, Sia D, Friedman SL. Friend or foe? The elusive role of hepatic stellate cells in liver cancer. Nat Rev Gastroenterol Hepatol. 2023;20:647-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 67] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 74. | Zhang HF, Gao X, Wang X, Chen X, Huang Y, Wang L, Xu ZW. The mechanisms of renin-angiotensin system in hepatocellular carcinoma: From the perspective of liver fibrosis, HCC cell proliferation, metastasis and angiogenesis, and corresponding protection measures. Biomed Pharmacother. 2021;141:111868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 75. | Marcher AB, Bendixen SM, Terkelsen MK, Hohmann SS, Hansen MH, Larsen BD, Mandrup S, Dimke H, Detlefsen S, Ravnskjaer K. Transcriptional regulation of Hepatic Stellate Cell activation in NASH. Sci Rep. 2019;9:2324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 76. | Akil A, Endsley M, Shanmugam S, Saldarriaga O, Somasunderam A, Spratt H, Stevenson HL, Utay NS, Ferguson M, Yi M. Fibrogenic Gene Expression in Hepatic Stellate Cells Induced by HCV and HIV Replication in a Three Cell Co-Culture Model System. Sci Rep. 2019;9:568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 77. | Longo M, Meroni M, Paolini E, Macchi C, Dongiovanni P. Mitochondrial dynamics and nonalcoholic fatty liver disease (NAFLD): new perspectives for a fairy-tale ending? Metabolism. 2021;117:154708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 76] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 78. | Yu L, Zhang X, Ye S, Lian H, Wang H, Ye J. Obesity and COVID-19: Mechanistic Insights From Adipose Tissue. J Clin Endocrinol Metab. 2022;107:1799-1811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 79. | Shang C, Liu Z, Zhu Y, Lu J, Ge C, Zhang C, Li N, Jin N, Li Y, Tian M, Li X. SARS-CoV-2 Causes Mitochondrial Dysfunction and Mitophagy Impairment. Front Microbiol. 2021;12:780768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 91] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 80. | Sánchez A, García-Pardo G, Gómez-Bertomeu F, López-Dupla M, Foguet-Romero E, Buzón MJ, Almirante B, Olona M, Fernández-Veledo S, Vidal F, Chafino S, Rull A, Peraire J; COVIDOMICS Study Group. Mitochondrial dysfunction, lipids metabolism, and amino acid biosynthesis are key pathways for COVID-19 recovery. iScience. 2023;26:107948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 81. | Santana MF, Guerra MT, Hundt MA, Ciarleglio MM, Pinto RAA, Dutra BG, Xavier MS, Lacerda MVG, Ferreira AJ, Wanderley DC, Borges do Nascimento IJ, Araújo RFA, Pinheiro SVB, Araújo SA, Leite MF, Ferreira LCL, Nathanson MH, Vieira Teixeira Vidigal P. Correlation Between Clinical and Pathological Findings of Liver Injury in 27 Patients With Lethal COVID-19 Infections in Brazil. Hepatol Commun. 2022;6:270-280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 82. | Lim JK, Njei B. Clinical and Histopathological Discoveries in Patients with Hepatic Injury and Cholangiopathy Who Have Died of COVID-19: Insights and Opportunities for Intervention. Hepat Med. 2023;15:151-164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 83. | Dietrich CG, Geier A, Merle U. Non-alcoholic fatty liver disease and COVID-19: Harmless companions or disease intensifier? World J Gastroenterol. 2023;29:367-377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 84. | Miranda C, Garlatti E, Da Porto A, Rinaldo E, Grazioli S, Zanette G, Tonizzo M. Liver injury in COVID-19 patients with non-alcoholic fatty liver disease: an update. Arch Med Sci Atheroscler Dis. 2023;8:e1-e10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |