Published online Jul 27, 2022. doi: 10.4254/wjh.v14.i7.1307
Peer-review started: January 17, 2022
First decision: March 8, 2022
Revised: April 20, 2022
Accepted: July 6, 2022
Article in press: July 6, 2022
Published online: July 27, 2022
Processing time: 191 Days and 1.2 Hours
Extracellular vesicles (EVs), especially small EVs (sEVs) derived from liver cells, have been the focus of much attention in the normal physiology and pathogenesis of various diseases affecting the liver. sEVs are approximately 100 nm in size, enclosed within lipid bilayers, and are very stable. The lipids, proteins, and nucleic acids, including miRNAs, contained within these vesicles are known to play important roles in intercellular communication. This mini-review summarizes the application of sEVs. First, liver diseases and the related diagnostic markers are described, and the current active status of miRNA research in diagnosis of hepatocellular carcinoma (HCC) is reported. Second, the biodistribution and pharmacokinetics of sEVs are described, and the liver is highlighted as the organ with the highest accumulation of sEVs. Third, the relationship between sEVs and the pathogenesis of liver disorders is described with emphesis on the current active status of miRNA research in HCC recurrence and survival. Finally, the possibility of future therapy using sEVs from mesenchymal stem (stromal) cells for cirrhosis and other diseases is described.
Core Tip: Extracellular vesicles (EVs), especially small EVs (sEVs) derived from liver cells, have been the focus of much attention in the normal physiology and pathogenesis of various diseases affecting the liver. sEVs are approximately 100 nm in size, enclosed within lipid bilayers, and are very stable. The proteins and nucleic acids, including miRNAs, contained within these vesicles are known to play important roles in intercellular communication. This mini-review summarizes the application of sEVs in the diagnosis of liver diseases, along with their distribution post administration, their role in pathogenesis, and their potential therapeutic effects in hepatic disorders.
- Citation: Tsuchiya A, Natsui K, Ishii Y, Koseki Y, Takeda N, Tomiyoshi K, Yamazaki F, Yoshida Y, Terai S. Small extracellular vesicles and liver diseases: From diagnosis to therapy. World J Hepatol 2022; 14(7): 1307-1318
- URL: https://www.wjgnet.com/1948-5182/full/v14/i7/1307.htm
- DOI: https://dx.doi.org/10.4254/wjh.v14.i7.1307
The study of extracellular vesicles (EVs) is an active area of research. Recent evidence has established that these EVs are released not only by human cells, but also by plant, bacterial, and yeast cells[1]. EVs are sub-organellar entities that act as "cargo" carriers that transmit information between cells thus exerting a variety of effects on biological activities1. Since the vesicles released are unique to the cells that release them, they have been widely studied in diseases of various organs and systems, including diseases of the liver in the context of diagnosis, pathogenesis, and therapeutic applications[2-5]. In particular, small extracellular vesicles (sEVs; referred to as exosomes), with a particle size of approximately 100 nm, have garnered much attention in recent years[6-11].
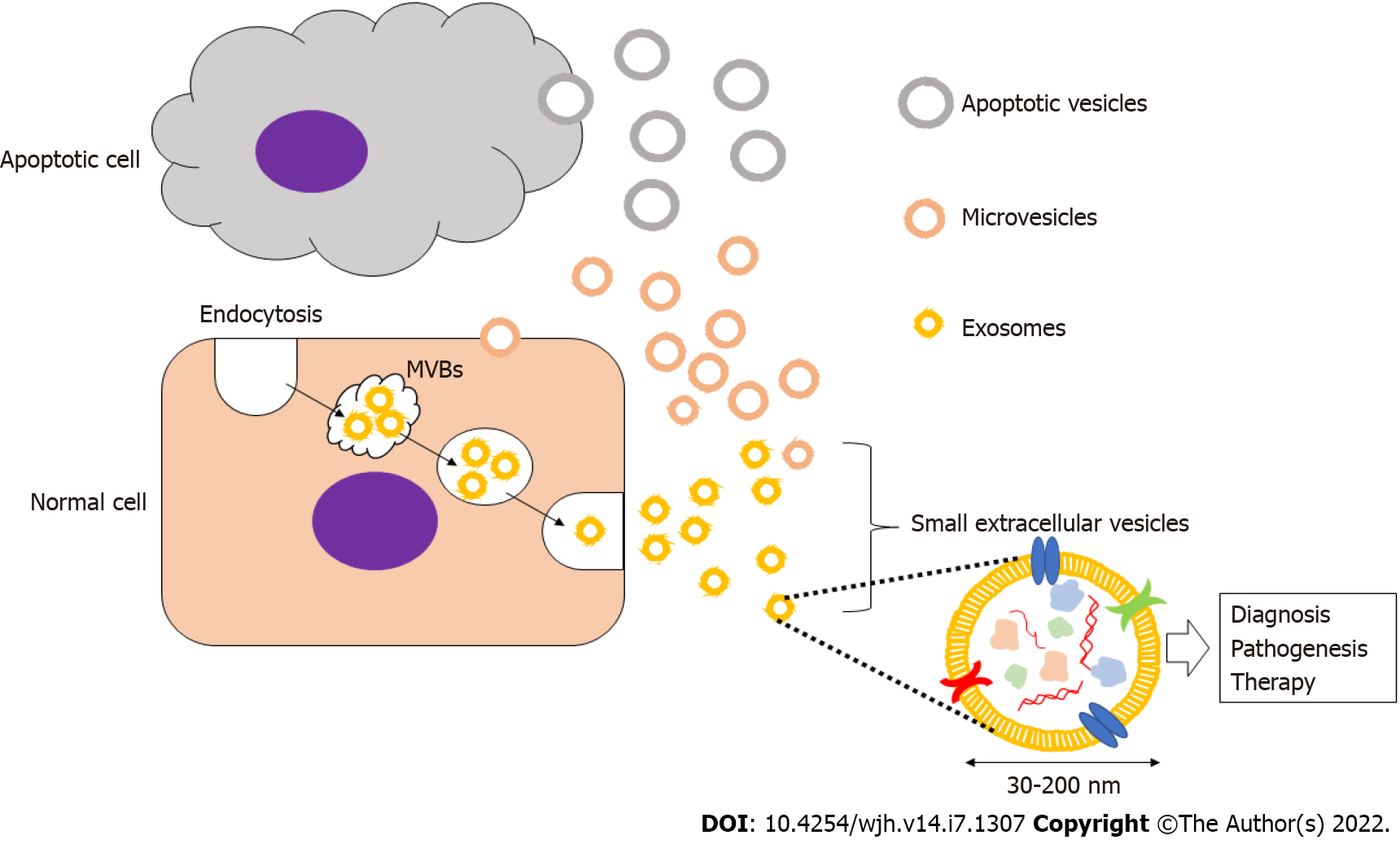
Secretory vesicles were first described in the 1980s, and they have been referred to by a number of different names based on their size and cellular origin such as exosomes, ectosomes, microvesicles, shedding vesicles, apoptotic bodies, oncosomes, and prostasomes. The International Society for Extracellular Vesicles recommends the usage of extracellular vesicles as a general term for these entities. Small EVs (sEVs), or exosomes, are formed from early endosomes that are generated by endocytosis and subsequently mature into late endosomes[12,13].
The late endosomes expand to form intraluminal membrane vesicles, also referred to as multivesicular bodies (MVBs), which then fuse with the plasma membrane and are released into the extracellular space. Secreted vesicles with a diameter of 30–200 nm are called sEVs or exosomes, and they are known to encapsulate a content of proteins, mRNAs, and miRNAs within a membrane composed of cholesterol, sphingomyelin, ceramide, and lipid rafts[13]. Although these vesicles vary between cells, there are common markers that characterize most exosomes including membrane transport and fusion proteins (GTPases, annexins, flotillin, etc.), heat shock proteins (HSP60, HSP70, HSP90, etc.), tetraspanins (CD9, CD63, CD81, etc.), MVB formation and transport proteins (TSG101, ALIX, Annexins, etc.), and cytoskeletal proteins (actin, tubulin, etc.)[11-13]. sEVs can be have important applications in the diagnosis and treamtent of various diseases and malignancies, and their study can also contribute to the ellucidation of the pathogenesis of these disease. For example, the stable inclusion of drugs within the lipid bilayers of sEVs creates novel therapeutic drug delivery systems that can be implied in the treatment of different diseases[13] (Figure 1).
Investigations on EVs are rapidly moving beyond basic research to clinical trials, and the global market of the diagnostic and treatment strategies that use sEVs, although still in its infancy, is expected to progress rapidly. This paper reviews the role of sEVs in the context of diagnosis, pathogenesis, and treatment of liver diseases.
Although different types of EVs were studied in the context of liver disorders, in this report we focus mainly on the role sEVs in the pathogenesis, diagnosis, and treatment of liver diseases. sEVs in particular have been analyzed in various chronic liver diseases such as non-alcoholic steatohepatitis (NASH), alcoholic liver disease (ALD), viral hepatitis caused by hepatitis B virus (HBV) and hepatitis C virus (HCV), cirrhosis, acute liver disease, and hepatocellular carcinoma (HCC); and in various specimens including blood, urine, bile, and ascitic fluid5. There are various techniques that were employed in the collection ofsEVs including ultracentrifugation, size exclusion chromatography, and methods utilizing precipitation kits and bead kits[14]. Details of sEV collection have been described in the minimal information for studies of extracellular vesicles guidelines 2018 (MISEV2018)[13]. After their collection, sEVs have been evaluated by western blotting, ELISA, flow cytometry, and nano tracking analysis to study the expression of common sEVs proteins, such as tetraspanins, and to identify markers including lipids, proteins, and nucleic acids, such as miRNAs and lincRNAs. Although the aforementioned studies are in the pre-clinical stage, they are expected to yield specific markers that can aid the processes of early and definitive diagnosis, treatment, and follow-up of liver diseases, in addition to helping in the ellucidation of the pathophysiology governing many of these liver disorders. Table 1[15-71] summarizes the various liver diseases and their related sEVs diagnostic markers. The bulk of the studies reported on miRNAs as sEVs diagnostic biomarkers of liver diseases which may be due to the ease of evaluating them using qRT-PCR. Markers of HCC have been the most frequently analyzed, and diseases such as NASH and ALD have received the most attention in recent years. Extracting sEVs produced by target cells and using them as markers of disease can contribute greatly to the field of diagnosis and treatment of liver disorders. However, we believe that there are some limitations and challenges to be acknowledged and addressed in the future, such as the efficient collection of target sEVs, recognition of target molecules (e.g., protein miRNA), cost, and high reproducibility.
| Diseases | Types of molecules | Markers | Refs. |
| HCC | Protein | Aminopeptidase N, Galectin-3-binding protein, SMAD, ANGPT2, 14-3-3 ζ, β-catenin, P120-catenin, EPCAM | [15–21] |
| RNA | miR-21, miR-21, -96, miR-122, miR-18a, -221, -222, -224, miR-10b-5p, -215-5p, miR-101, -106b, 12, -195, miR-519d, -595, -939, miR-19b,-92, miR-125, miR-9-3, miR-122, 148a, -1246, miR-122, miR-93, miR 144-3p, -21-5p, miR210, miR-638, miR-665, miR-774, miR-1262, miR-320d, miR-23a/b, miR-45-1a, miR-224, miR-21, -10b, miR-122, -125b, -145, -192, -194, 29a, 17-5p, -106a, miR-26a, -29c, -21, lncRNA Jpx, lncRNA FAL1, lncRNA-RP11-513I15.6, mRNA RAB11A, miR-1262, lncRNA HEIH, lncRNA LINC00161, lnc RNA HULC, AFP mRNA | [22–50] | |
| HBV | miRNA | miR-21-5p | [33] |
| HCV | RNA | HCV-RNA, miR-122-5p, -222-3p, -146-5p, -150-5p, -30c-5p, -378a-3p, -20a-5p, | [51–53] |
| NAFLD/NASH | Protein | ITGβ1, CD68 | [54,55] |
| RNA | miR-192-5p | [56] | |
| Lipid | ceramides and sphingosin 1-phosphate | [57] | |
| ALD | Protein | ASGR2 and CYP2E1, CD163, 206, ASGPR, CD40L, CK18, Glutathione synthetase | [55,58–62] |
| RNA | miR-122, -155, miR-Let-7f, 29a, -340, miR-122, let7f, -21, -29a, -146a, miR-192-5p, miR-192, -30a | [56,63–66] | |
| Lipid | Sphingosin 1-phosphate | [67] | |
| Cirrhosis | Protein | CD163, 206, PDGFRβ, urinary maltase and glucoamylase (for AKI during cirrhosis) | [59,68–70] |
| RNA | miR-19a, -19b, -92, 17a, -20a | [27] | |
| ALI | Protein | Apolipoprotein A-1, Argininosuccinate synthase-1 | [62] |
| RNA | Gnb21 mRNA, | [71] |
Numerous studies have demonstrated the importance of the liver in the biodistribution and pharmacokinetics of sEVs. This has been accomplished by employing techniques such as lipophilic fluorescent and luminescent, radio-labeling, and magnetic resonance imaging. Studies have conclusively shown that post systemic administration of sEVs, these vesicles are cleared from the bloodstream within a few minutes of their half-life via phagocytosis by macrophages and neutrophils[72]. While they disappear from the blood, they have been reported to persist longer within organs, with the largest accumulation occurring in the liver. This accumulation peaks in the liver and kidneys approximately 1 h post administration, which is earlier than that in the lungs where maximal accumulation is achieved 2–12 h post administration. It has been shown that high concentrations of sEVs can be maintained in the liver for about 12-24 h, although there have been contradictory reports about this[72,73]. Some studies suggest that the macrophages primarily take up scaffold in the liver, while others report that hepatocytes and other cells also do the same. The abundant expression of scavenger receptors in macrophages is thought to play a crucial role in this process[73]. Additionally, phosphatidyl serine (PS) has been found to easily accumulate in the liver unlike the phosphatidylcholine-rich lipids[73]. Hoshino et al[74] have demonstrated the importance of integrins by showing that integrin αvβ5 in sEVs is essential for its accumulation in macrophages. However, results from these studies must be interpreted with caution since most of them employed the technique of labeling lipid bilayers, which may have resulted in the visualization of cells that ingested phospholipids rather than the sEVs. Given their miniscule size, sEVs by themselves have never been directly visualized in isolation. Furthermore, the majority of these reports have made observations under conditions of normal physiology, so it is possible that the biodistribution and pharmacokinetics of sEVs in pathological conditions may be significantly different.
Many reports described the implication of sEVs in various aspects of the pathogenisis of liver diseases. These entities are highly stable in vivo and play an important role in the communication between both neighboring and distant cells. Table 2[75-99] summarizes the different sEV markers that have been linked to certain processes of liver pathogenesis.
| Pathophysiology | Types of molecules | Markers | Refs. |
| NASH inflammation and fibrosis | miRNA | miR-122, -192 | [75] |
| NASH fibrosis | miRNA | miR-122 | [76] |
| ALD outcome | protein | ASGR2 and CYP2E1 | [58] |
| HBV fibrosis | miRNA | miR-150, -192, -200b, -91a | [77] |
| HCV treatment response | miRNA | miR-122, -199a, miR-122, miR-122-5p, -222-3p, -146-5p, -150-5p, -30c-5p, -378a-3p, -20a-5p | [53,78,79] |
| HCV fibrosis | protein | CD81 | [80] |
| miRNA | Let-7s, miR-122, -150, -192, 200b, 92a, miR-19a | [77,81] | |
| HCC recurrence | protein | SMAD3, CASC9 | [16,82] |
| RNA | miR-718, miR-125, miR-21, miR-103, miR1247-3p, miR-92b, miR-21 and lncRNA-ATB, miR-21, -10b, miR-215-5p, miR-155, mRNA RAB11A, miR-211-3p, -6826-3p, -1236-3p, 4448 | [25,26,36,42,83–90] | |
| HCC survival | RNA | miR-125, miR-21, miR-103, miR-22a-3p, miR-335, miR-25-5p, miR-320a-PBX3, miR-718, miR-210, miR-122, miR-93, miR-21, -96, -122, miR-1247-3p, miR-638, miR-665, miR-21 and lncRNA-ATB, miR-30d, -140, miR-106a, miR-224, miR-320d, long non-coding RNA (ENSG00000258332.1 and LINC00635), hnRNPH1, circPTGR1, circRNA-100, -338, circ DB | [18,23,31,32,34,36,38,41,43,82–86,88,91–99] |
sEVs exert their effect on inter-cellular communication between neighboring cells via the peri-sinusoidal space. For instance, it has been reported that sEVs secreted by HCV-infected hepatocytes exert an effect on hepatic stellate cells (HSCs) driving hepatic fibrosis[100]. Additionally, as shown in Table 2, sEVs produced by hepatocellular carcinoma (HCC) have a profound effect on the surrounding environment. This effect is mediated by the modulation of the immune system by sEVs that have an inhibitory effect on macrophages, monocytes, NK cells, B cells, and T cells[101]. These vesicles can also promote HSC and the transformation of fibroblasts to cancer-associated fibroblasts (CAFs), promote migration of hepatocellular carcinoma cells in the vicinity, act on vascular endothelial cells to promote angiogenesis, and induce drug resistance in surrounding cancer cells[101]. Additionally, sEVs released from hepatocytes are believed to function as drivers of inflammation and state formation in inflammatory cells such as macrophages in NASH[102].
It has been also reported that these vesicles drive the pathogenesis of disease through an effect on distant cells. This is exemplified by the crosstalk between the sEVs produced by adipocytes and those produced by hepatocellular carcinoma cells, which contributes to cell proliferation, angiogenesis, invasion, epithelial-mesenchymal transition, and the creation of a favorable environment for metastasis[103]. Recent reports have also demonstrated that microbiota-derived EVs in the intestine affect other organs and tissues in the body, including the liver, heart, brain, kidney, lung, and adipose tissue[104]. Though there have been some studies describing the effects of sEVs on various cells in the body, it is still unclear how these sEVs that are produced by specific cells selectively reach their target cells. Consequently, further analysis from a broader perspective is essential to describe the specificities and dynamics of this interaction between sEVs and their target cells.
To date, there have been no reports or ongoing trials on the application of sEVs in the treatment of liver diseases. This scarcity might be attributed to the fact that there are still many unknowns regarding the effects of sEVs on liver disease, but further mechanistic analysis in the future may lead to the development of new therapies. However, the potential of sEVs as anti-fibrotic and anti-cancer therapeutic agents needs to be explored. In the case of anti-fibrotic therapy, sEVs may be the most convenient therapeutic agents that target macrophages on account of their massive accumulation in these cells within the liver[5,105]. Furthermore, Mesenchymal stem (stromal) cells (MSCs) have been investigated for their anti-fibrotic properties due to their ability to suppress fibrogenesis by reducing the inflammatory responses of inflammatory cells and by inducing fibrolysis via their effect on macrophages and matrix metalloproteinases[106].
Basic research has recently revealed that sEVs secreted by MSCs transmit information to macrophages. Additionally, the potency of these sEVs has been enhanced by pre-conditioning the MSCs with IFN-γ to augment their therapeutic effects in a mouse model of liver cirrhosis[105]. In view of this, it might be possible to create and evaluate a therapeutic strategy that employs sEVs obtained from pre-conditioned or modified MSCs to transmit information to macrophages and exert anti-fibrotic effects suppressing fibrogenesis. Although such attempts have been made, the production of sEVs from preconditioned/modified MSCs has not yet been successful due to regulatory concerns, lack of appropriate quality control, and difficulties associated with mass purification.
Warnecke et al[107] reported a first-in-human case study that utilized MSC-EVs derived from umbilical cord tissues to reduce inflammation during cochlear implantation. Briefly, the authors obtained 1.03 × 1011 particles/mL of EVs with a diameter range of 110–130 nm, as measured by nanoparticle tracking analysis, via a combination of tangential flow filtration (TFF) and diafiltration techniques post culture. This showed that MSCs-derived exosomes may find successful application in clinical use. Furthermore, improvements in the methods that can reduce the quantity of exosomes needed for efficient treatment will further expedite their use in clinical scenarios[107].
The development of cancer therapies that employ sEVs are also theoretically possible, such as those that aim to suppress sEVs derived from cancer cells. Mendt et al[108] have developed a therapeutic strategy for pancreatic cancer using sEVs that is currently under clinical trials. Their study involved the optimization of iExosomes to enable them to deliver higher concentrations of sEVs to pancreatic cancers via a two-pronged strategy. This includes the selection of CD47 that protects exosomes from phagocytosis by macrophages and engineering exosomes to carry siRNA or shRNA specifically targeted against the oncogenic KRASG12D, the key driver of pancreatic cancer. The study also reported the feasibility of large-scale production of clinical grade iExosomes by a bioreactor-based methodology[108,109]. Therefore, sEVs have a promising potential in anti-fibrotic and anti-cancer therapy, and their applications may be expanded by developing techniques that efficiently load therapy enhancing substances, aid their incorporation into target cells, and improve high-throughput collection methodologies.
In summary, small extracellular vesicles (sEVs) have a promising potential in the diagnosis and treatment of liver diseases. The challenge in the therapeutic uses of sEVs is that it is not easy to harvest large amout of sEVs for human systemic therapy. However, the collection of sEVs can be greatly enhanced by pre-conditioning or modifying the source cells, thereby greatly expanding their possible applications. Furthermore, the potential for using EVs in therapy may be enhanced by utilizing larger EVs in addition to sEVs. These vesicles can potentially be harvested from cell sources other than mesenchymal stem cells (MSCs), such as induced pluripotent stem cells (iPS cells). In addition, with using these larger EVs there are fewer risk of embolization especially to the lungs after the administration. Consequently, in spite of issues with future regulatory trends and establishment of manufacturing processes, sEVs remain a promising therapeutic option for liver ailments.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Naserian S, France; Ullah K, Pakistan S-Editor: Ma YJ L-Editor: A P-Editor: Ma YJ
| 1. | Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6920] [Cited by in RCA: 6566] [Article Influence: 1313.2] [Reference Citation Analysis (0)] |
| 2. | Othman N, Jamal R, Abu N. Cancer-Derived Exosomes as Effectors of Key Inflammation-Related Players. Front Immunol. 2019;10:2103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 3. | An T, Qin S, Xu Y, Tang Y, Huang Y, Situ B, Inal JM, Zheng L. Exosomes serve as tumour markers for personalized diagnostics owing to their important role in cancer metastasis. J Extracell Vesicles. 2015;4:27522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 223] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 4. | Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5:31292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 294] [Cited by in RCA: 306] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 5. | Watanabe T, Tsuchiya A, Takeuchi S, Nojiri S, Yoshida T, Ogawa M, Itoh M, Takamura M, Suganami T, Ogawa Y, Terai S. Development of a non-alcoholic steatohepatitis model with rapid accumulation of fibrosis, and its treatment using mesenchymal stem cells and their small extracellular vesicles. Regen Ther. 2020;14:252-261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 6. | Allan D, Tieu A, Lalu M, Burger D. Mesenchymal stromal cell-derived extracellular vesicles for regenerative therapy and immune modulation: Progress and challenges toward clinical application. Stem Cells Transl Med. 2020;9:39-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 7. | He C, Zheng S, Luo Y, Wang B. Exosome Theranostics: Biology and Translational Medicine. Theranostics. 2018;8:237-255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 700] [Cited by in RCA: 916] [Article Influence: 130.9] [Reference Citation Analysis (0)] |
| 8. | Reiner AT, Witwer KW, van Balkom BWM, de Beer J, Brodie C, Corteling RL, Gabrielsson S, Gimona M, Ibrahim AG, de Kleijn D, Lai CP, Lötvall J, Del Portillo HA, Reischl IG, Riazifar M, Salomon C, Tahara H, Toh WS, Wauben MHM, Yang VK, Yang Y, Yeo RWY, Yin H, Giebel B, Rohde E, Lim SK. Concise Review: Developing Best-Practice Models for the Therapeutic Use of Extracellular Vesicles. Stem Cells Transl Med. 2017;6:1730-1739. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 236] [Cited by in RCA: 253] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 9. | Rohde E, Pachler K, Gimona M. Manufacturing and characterization of extracellular vesicles from umbilical cord-derived mesenchymal stromal cells for clinical testing. Cytotherapy. 2019;21:581-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 152] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 10. | Russell AE, Sneider A, Witwer KW, Bergese P, Bhattacharyya SN, Cocks A, Cocucci E, Erdbrügger U, Falcon-Perez JM, Freeman DW, Gallagher TM, Hu S, Huang Y, Jay SM, Kano SI, Lavieu G, Leszczynska A, Llorente AM, Lu Q, Mahairaki V, Muth DC, Noren Hooten N, Ostrowski M, Prada I, Sahoo S, Schøyen TH, Sheng L, Tesch D, Van Niel G, Vandenbroucke RE, Verweij FJ, Villar AV, Wauben M, Wehman AM, Yin H, Carter DRF, Vader P. Biological membranes in EV biogenesis, stability, uptake, and cargo transfer: an ISEV position paper arising from the ISEV membranes and EVs workshop. J Extracell Vesicles. 2019;8:1684862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 183] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 11. | Witwer KW, Van Balkom BWM, Bruno S, Choo A, Dominici M, Gimona M, Hill AF, De Kleijn D, Koh M, Lai RC, Mitsialis SA, Ortiz LA, Rohde E, Asada T, Toh WS, Weiss DJ, Zheng L, Giebel B, Lim SK. Defining mesenchymal stromal cell (MSC)-derived small extracellular vesicles for therapeutic applications. J Extracell Vesicles. 2019;8:1609206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 340] [Cited by in RCA: 443] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 12. | Lener T, Gimona M, Aigner L, Börger V, Buzas E, Camussi G, Chaput N, Chatterjee D, Court FA, Del Portillo HA, O'Driscoll L, Fais S, Falcon-Perez JM, Felderhoff-Mueser U, Fraile L, Gho YS, Görgens A, Gupta RC, Hendrix A, Hermann DM, Hill AF, Hochberg F, Horn PA, de Kleijn D, Kordelas L, Kramer BW, Krämer-Albers EM, Laner-Plamberger S, Laitinen S, Leonardi T, Lorenowicz MJ, Lim SK, Lötvall J, Maguire CA, Marcilla A, Nazarenko I, Ochiya T, Patel T, Pedersen S, Pocsfalvi G, Pluchino S, Quesenberry P, Reischl IG, Rivera FJ, Sanzenbacher R, Schallmoser K, Slaper-Cortenbach I, Strunk D, Tonn T, Vader P, van Balkom BW, Wauben M, Andaloussi SE, Théry C, Rohde E, Giebel B. Applying extracellular vesicles based therapeutics in clinical trials - an ISEV position paper. J Extracell Vesicles. 2015;4:30087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 950] [Cited by in RCA: 1078] [Article Influence: 107.8] [Reference Citation Analysis (0)] |
| 13. | Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borràs FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MÁ, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Górecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzás EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D'Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekström K, El Andaloussi S, Elie-Caille C, Erdbrügger U, Falcón-Pérez JM, Fatima F, Fish JE, Flores-Bellver M, Försönits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gámez-Valero A, Gardiner C, Gärtner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Görgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanović MM, Kovács ÁF, Krämer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lässer C, Laurent LC, Lavieu G, Lázaro-Ibáñez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Linē A, Linnemannstöns K, Llorente A, Lombard CA, Lorenowicz MJ, Lörincz ÁM, Lötvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr, Meehan KL, Mertens I, Minciacchi VR, Möller A, Møller Jørgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-'t Hoen EN, Noren Hooten N, O'Driscoll L, O'Grady T, O'Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Østergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saá P, Sahoo S, Salas-Huenuleo E, Sánchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schøyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sódar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr, Veit TD, Vella LJ, Velot É, Verweij FJ, Vestad B, Viñas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yáñez-Mó M, Yin H, Yuana Y, Zappulli V, Zarubova J, Žėkas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6453] [Cited by in RCA: 7698] [Article Influence: 1099.7] [Reference Citation Analysis (1)] |
| 14. | Thietart S, Rautou PE. Extracellular vesicles as biomarkers in liver diseases: A clinician's point of view. J Hepatol. 2020;73:1507-1525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 15. | Arbelaiz A, Azkargorta M, Krawczyk M, Santos-Laso A, Lapitz A, Perugorria MJ, Erice O, Gonzalez E, Jimenez-Agüero R, Lacasta A, Ibarra C, Sanchez-Campos A, Jimeno JP, Lammert F, Milkiewicz P, Marzioni M, Macias RIR, Marin JJG, Patel T, Gores GJ, Martinez I, Elortza F, Falcon-Perez JM, Bujanda L, Banales JM. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology. 2017;66:1125-1143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 228] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 16. | Fu Q, Zhang Q, Lou Y, Yang J, Nie G, Chen Q, Chen Y, Zhang J, Wang J, Wei T, Qin H, Dang X, Bai X, Liang T. Primary tumor-derived exosomes facilitate metastasis by regulating adhesion of circulating tumor cells via SMAD3 in liver cancer. Oncogene. 2018;37:6105-6118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 117] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 17. | Xie JY, Wei JX, Lv LH, Han QF, Yang WB, Li GL, Wang PX, Wu SB, Duan JX, Zhuo WF, Liu PQ, Min J. Angiopoietin-2 induces angiogenesis via exosomes in human hepatocellular carcinoma. Cell Commun Signal. 2020;18:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 18. | Wang X, Shen H, Zhangyuan G, Huang R, Zhang W, He Q, Jin K, Zhuo H, Zhang Z, Wang J, Sun B, Lu X. 14-3-3ζ delivered by hepatocellular carcinoma-derived exosomes impaired anti-tumor function of tumor-infiltrating T lymphocytes. Cell Death Dis. 2018;9:159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 19. | Han Q, Lv L, Wei J, Lei X, Lin H, Li G, Cao J, Xie J, Yang W, Wu S, You J, Lu J, Liu P, Min J. Vps4A mediates the localization and exosome release of β-catenin to inhibit epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 2019;457:47-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 20. | Cheng Z, Lei Z, Yang P, Si A, Xiang D, Tang X, Guo G, Zhou J, Hüser N. Exosome-transmitted p120-catenin suppresses hepatocellular carcinoma progression via STAT3 pathways. Mol Carcinog. 2019;58:1389-1399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 21. | Julich-Haertel H, Urban SK, Krawczyk M, Willms A, Jankowski K, Patkowski W, Kruk B, Krasnodębski M, Ligocka J, Schwab R, Richardsen I, Schaaf S, Klein A, Gehlert S, Sänger H, Casper M, Banales JM, Schuppan D, Milkiewicz P, Lammert F, Lukacs-Kornek V, Kornek M. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J Hepatol. 2017;67:282-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 131] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 22. | Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int. 2014;2014:864894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 181] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 23. | Wang S, Yang Y, Sun L, Qiao G, Song Y, Liu B. Exosomal MicroRNAs as Liquid Biopsy Biomarkers in Hepatocellular Carcinoma. Onco Targets Ther. 2020;13:2021-2030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 24. | Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, Shim SG, Paik YH. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47:e184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 259] [Cited by in RCA: 343] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 25. | Cho HJ, Eun JW, Baek GO, Seo CW, Ahn HR, Kim SS, Cho SW, Cheong JY. Serum Exosomal MicroRNA, miR-10b-5p, as a Potential Diagnostic Biomarker for Early-Stage Hepatocellular Carcinoma. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 26. | Fornari F, Ferracin M, Trerè D, Milazzo M, Marinelli S, Galassi M, Venerandi L, Pollutri D, Patrizi C, Borghi A, Foschi FG, Stefanini GF, Negrini M, Bolondi L, Gramantieri L. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, Identify Cirrhotic Patients with HCC. PLoS One. 2015;10:e0141448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 116] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 27. | Brandon-Warner E, Feilen NA, Culberson CR, Field CO, deLemos AS, Russo MW, Schrum LW. Processing of miR17-92 Cluster in Hepatic Stellate Cells Promotes Hepatic Fibrogenesis During Alcohol-Induced Injury. Alcohol Clin Exp Res. 2016;40:1430-1442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 28. | Zuwała-Jagiełło J, Simon KA, Pazgan-Simon M. Elevated circulating endothelial cell-derived microparticle levels in patients with liver cirrhosis: a preliminary report. Clin Exp Hepatol. 2015;1:105-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 29. | Tang J, Li Y, Liu K, Zhu Q, Yang WH, Xiong LK, Guo DL. Exosomal miR-9-3p suppresses HBGF-5 expression and is a functional biomarker in hepatocellular carcinoma. Minerva Med. 2018;109:15-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 30. | Wang Y, Zhang C, Zhang P, Guo G, Jiang T, Zhao X, Jiang J, Huang X, Tong H, Tian Y. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018;7:1670-1679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 31. | Suehiro T, Miyaaki H, Kanda Y, Shibata H, Honda T, Ozawa E, Miuma S, Taura N, Nakao K. Serum exosomal microRNA-122 and microRNA-21 as predictive biomarkers in transarterial chemoembolization-treated hepatocellular carcinoma patients. Oncol Lett. 2018;16:3267-3273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 32. | Xue X, Wang X, Zhao Y, Hu R, Qin L. Exosomal miR-93 promotes proliferation and invasion in hepatocellular carcinoma by directly inhibiting TIMP2/TP53INP1/CDKN1A. Biochem Biophys Res Commun. 2018;502:515-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 33. | Pu C, Huang H, Wang Z, Zou W, Lv Y, Zhou Z, Zhang Q, Qiao L, Wu F, Shao S. Extracellular Vesicle-Associated mir-21 and mir-144 Are Markedly Elevated in Serum of Patients With Hepatocellular Carcinoma. Front Physiol. 2018;9:930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 34. | Lin XJ, Fang JH, Yang XJ, Zhang C, Yuan Y, Zheng L, Zhuang SM. Hepatocellular Carcinoma Cell-Secreted Exosomal MicroRNA-210 Promotes Angiogenesis In Vitro and In Vivo. Mol Ther Nucleic Acids. 2018;11:243-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 186] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 35. | Shi M, Jiang Y, Yang L, Yan S, Wang YG, Lu XJ. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem. 2018;119:4711-4716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 136] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 36. | Abd El Gwad A, Matboli M, El-Tawdi A, Habib EK, Shehata H, Ibrahim D, Tash F. Role of exosomal competing endogenous RNA in patients with hepatocellular carcinoma. J Cell Biochem. 2018;119:8600-8610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 37. | Wang G, Zhao W, Wang H, Qiu G, Jiang Z, Wei G, Li X. Exosomal MiR-744 Inhibits Proliferation and Sorafenib Chemoresistance in Hepatocellular Carcinoma by Targeting PAX2. Med Sci Monit. 2019;25:7209-7217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 38. | Li W, Ding X, Wang S, Xu L, Yin T, Han S, Geng J, Sun W. Downregulation of serum exosomal miR-320d predicts poor prognosis in hepatocellular carcinoma. J Clin Lab Anal. 2020;34:e23239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 39. | Liu Y, Tan J, Ou S, Chen J, Chen L. Adipose-derived exosomes deliver miR-23a/b to regulate tumor growth in hepatocellular cancer by targeting the VHL/HIF axis. J Physiol Biochem. 2019;75:391-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 40. | Zhao S, Li J, Zhang G, Wang Q, Wu C, Zhang Q, Wang H, Sun P, Xiang R, Yang S. Exosomal miR-451a Functions as a Tumor Suppressor in Hepatocellular Carcinoma by Targeting LPIN1. Cell Physiol Biochem. 2019;53:19-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 41. | Cui Y, Xu HF, Liu MY, Xu YJ, He JC, Zhou Y, Cang SD. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J Gastroenterol. 2019;25:1890-1898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 60] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (1)] |
| 42. | Tian XP, Wang CY, Jin XH, Li M, Wang FW, Huang WJ, Yun JP, Xu RH, Cai QQ, Xie D. Acidic Microenvironment Up-Regulates Exosomal miR-21 and miR-10b in Early-Stage Hepatocellular Carcinoma to Promote Cancer Cell Proliferation and Metastasis. Theranostics. 2019;9:1965-1979. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 185] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 43. | Xue X, Zhao Y, Wang X, Qin L, Hu R. Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120:135-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 44. | Lin H, Zhang Z. Diagnostic value of a microRNA signature panel in exosomes for patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2019;12:1478-1487. [PubMed] |
| 45. | Ma X, Yuan T, Yang C, Wang Z, Zang Y, Wu L, Zhuang L. X-inactive-specific transcript of peripheral blood cells is regulated by exosomal Jpx and acts as a biomarker for female patients with hepatocellular carcinoma. Ther Adv Med Oncol. 2017;9:665-677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 46. | Li B, Mao R, Liu C, Zhang W, Tang Y, Guo Z. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 177] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 47. | Zhang C, Yang X, Qi Q, Gao Y, Wei Q, Han S. lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular carcinoma. Cancer Biomark. 2018;21:651-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 115] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 48. | Sun L, Su Y, Liu X, Xu M, Chen X, Zhu Y, Guo Z, Bai T, Dong L, Wei C, Cai X, He B, Pan Y, Sun H, Wang S. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J Cancer. 2018;9:2631-2639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 105] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 49. | Cao SQ, Zheng H, Sun BC, Wang ZL, Liu T, Guo DH, Shen ZY. Long non-coding RNA highly up-regulated in liver cancer promotes exosome secretion. World J Gastroenterol. 2019;25:5283-5299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 45] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 50. | Wang X, Kwak KJ, Yang Z, Zhang A, Zhang X, Sullivan R, Lin D, Lee RL, Castro C, Ghoshal K, Schmidt C, Lee LJ. Extracellular mRNA detected by molecular beacons in tethered lipoplex nanoparticles for diagnosis of human hepatocellular carcinoma. PLoS One. 2018;13:e0198552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 51. | Liu Z, Zhang X, Yu Q, He JJ. Exosome-associated hepatitis C virus in cell cultures and patient plasma. Biochem Biophys Res Commun. 2014;455:218-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 84] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 52. | Bukong TN, Momen-Heravi F, Kodys K, Bala S, Szabo G. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog. 2014;10:e1004424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 277] [Cited by in RCA: 342] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 53. | Santangelo L, Bordoni V, Montaldo C, Cimini E, Zingoni A, Battistelli C, D'Offizi G, Capobianchi MR, Santoni A, Tripodi M, Agrati C. Hepatitis C virus direct-acting antivirals therapy impacts on extracellular vesicles microRNAs content and on their immunomodulating properties. Liver Int. 2018;38:1741-1750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 54. | Guo Q, Furuta K, Lucien F, Gutierrez Sanchez LH, Hirsova P, Krishnan A, Kabashima A, Pavelko KD, Madden B, Alhuwaish H, Gao Y, Revzin A, Ibrahim SH. Integrin β1-enriched extracellular vesicles mediate monocyte adhesion and promote liver inflammation in murine NASH. J Hepatol. 2019;71:1193-1205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 143] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 55. | Sukriti S, Maras JS, Bihari C, Das S, Vyas AK, Sharma S, Hussain S, Shasthry S, Choudhary A, Premkumar M, Kumar D, Kumar G, Mukhopadhyay C, Kumar A, Trehanpati N, Rautou PE, Moreau R, Sarin SK. Microvesicles in hepatic and peripheral vein can predict nonresponse to corticosteroid therapy in severe alcoholic hepatitis. Aliment Pharmacol Ther. 2018;47:1151-1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 56. | Liu XL, Pan Q, Cao HX, Xin FZ, Zhao ZH, Yang RX, Zeng J, Zhou H, Fan JG. Lipotoxic Hepatocyte-Derived Exosomal MicroRNA 192-5p Activates Macrophages Through Rictor/Akt/Forkhead Box Transcription Factor O1 Signaling in Nonalcoholic Fatty Liver Disease. Hepatology. 2020;72:454-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 218] [Article Influence: 43.6] [Reference Citation Analysis (1)] |
| 57. | Kakazu E, Mauer AS, Yin M, Malhi H. Hepatocytes release ceramide-enriched pro-inflammatory extracellular vesicles in an IRE1α-dependent manner. J Lipid Res. 2016;57:233-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 233] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 58. | Sehrawat TS, Arab JP, Liu M, Amrollahi P, Wan M, Fan J, Nakao Y, Pose E, Navarro-Corcuera A, Dasgupta D, Liao CY, He L, Mauer AS, Avitabile E, Ventura-Cots M, Bataller RA, Sanyal AJ, Chalasani NP, Heimbach JK, Watt KD, Gores GJ, Gines P, Kamath PS, Simonetto DA, Hu TY, Shah VH, Malhi H. Circulating Extracellular Vesicles Carrying Sphingolipid Cargo for the Diagnosis and Dynamic Risk Profiling of Alcoholic Hepatitis. Hepatology. 2021;73:571-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 64] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 59. | Nielsen MC, Andersen MN, Grønbæk H, Damgaard Sandahl T, Møller HJ. Extracellular vesicle-associated soluble CD163 and CD206 in patients with acute and chronic inflammatory liver disease. Scand J Gastroenterol. 2020;55:588-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 60. | Verma VK, Li H, Wang R, Hirsova P, Mushref M, Liu Y, Cao S, Contreras PC, Malhi H, Kamath PS, Gores GJ, Shah VH. Alcohol stimulates macrophage activation through caspase-dependent hepatocyte derived release of CD40L containing extracellular vesicles. J Hepatol. 2016;64:651-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 185] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 61. | Bissonnette J, Altamirano J, Devue C, Roux O, Payancé A, Lebrec D, Bedossa P, Valla D, Durand F, Ait-Oufella H, Sancho-Bru P, Caballeria J, Ginès P, Boulanger CM, Bataller R, Rautou PE. A prospective study of the utility of plasma biomarkers to diagnose alcoholic hepatitis. Hepatology. 2017;66:555-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 62. | Cho YE, Im EJ, Moon PG, Mezey E, Song BJ, Baek MC. Increased liver-specific proteins in circulating extracellular vesicles as potential biomarkers for drug- and alcohol-induced liver injury. PLoS One. 2017;12:e0172463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 63. | Momen-Heravi F, Bala S, Kodys K, Szabo G. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci Rep. 2015;5:9991. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 280] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 64. | Eguchi A, Lazaro RG, Wang J, Kim J, Povero D, Willliams B, Ho SB, Stärkel P, Schnabl B, Ohno-Machado L, Tsukamoto H, Feldstein AE. Extracellular vesicles released by hepatocytes from gastric infusion model of alcoholic liver disease contain a MicroRNA barcode that can be detected in blood. Hepatology. 2017;65:475-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 92] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 65. | Eguchi A, Franz N, Kobayashi Y, Iwasa M, Wagner N, Hildebrand F, Takei Y, Marzi I, Relja B. Circulating Extracellular Vesicles and Their miR "Barcode" Differentiate Alcohol Drinkers With Liver Injury and Those Without Liver Injury in Severe Trauma Patients. Front Med (Lausanne). 2019;6:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 66. | Momen-Heravi F, Saha B, Kodys K, Catalano D, Satishchandran A, Szabo G. Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J Transl Med. 2015;13:261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 252] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 67. | Wang R, Ding Q, Yaqoob U, de Assuncao TM, Verma VK, Hirsova P, Cao S, Mukhopadhyay D, Huebert RC, Shah VH. Exosome Adherence and Internalization by Hepatic Stellate Cells Triggers Sphingosine 1-Phosphate-dependent Migration. J Biol Chem. 2015;290:30684-30696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 185] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 68. | Kostallari E, Hirsova P, Prasnicka A, Verma VK, Yaqoob U, Wongjarupong N, Roberts LR, Shah VH. Hepatic stellate cell-derived platelet-derived growth factor receptor-alpha-enriched extracellular vesicles promote liver fibrosis in mice through SHP2. Hepatology. 2018;68:333-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 69. | Lambrecht J, Verhulst S, Mannaerts I, Sowa JP, Best J, Canbay A, Reynaert H, van Grunsven LA. A PDGFRβ-based score predicts significant liver fibrosis in patients with chronic alcohol abuse, NAFLD and viral liver disease. EBioMedicine. 2019;43:501-512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 70. | Awdishu L, Tsunoda S, Pearlman M, Kokoy-Mondragon C, Ghassemian M, Naviaux RK, Patton HM, Mehta RL, Vijay B, RamachandraRao SP. Identification of Maltase Glucoamylase as a Biomarker of Acute Kidney Injury in Patients with Cirrhosis. Crit Care Res Pract. 2019;2019:5912804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 71. | Royo F, Schlangen K, Palomo L, Gonzalez E, Conde-Vancells J, Berisa A, Aransay AM, Falcon-Perez JM. Transcriptome of extracellular vesicles released by hepatocytes. PLoS One. 2013;8:e68693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (1)] |
| 72. | Choi H, Choi Y, Yim HY, Mirzaaghasi A, Yoo JK, Choi C. Biodistribution of Exosomes and Engineering Strategies for Targeted Delivery of Therapeutic Exosomes. Tissue Eng Regen Med. 2021;18:499-511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 146] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 73. | Kang M, Jordan V, Blenkiron C, Chamley LW. Biodistribution of extracellular vesicles following administration into animals: A systematic review. J Extracell Vesicles. 2021;10:e12085. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 244] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 74. | Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329-335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2697] [Cited by in RCA: 3761] [Article Influence: 376.1] [Reference Citation Analysis (0)] |
| 75. | Lee YS, Kim SY, Ko E, Lee JH, Yi HS, Yoo YJ, Je J, Suh SJ, Jung YK, Kim JH, Seo YS, Yim HJ, Jeong WI, Yeon JE, Um SH, Byun KS. Exosomes derived from palmitic acid-treated hepatocytes induce fibrotic activation of hepatic stellate cells. Sci Rep. 2017;7:3710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 184] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 76. | Akuta N, Kawamura Y, Watanabe C, Nishimura A, Okubo M, Mori Y, Fujiyama S, Sezaki H, Hosaka T, Kobayashi M, Saitoh S, Suzuki F, Suzuki Y, Arase Y, Ikeda K, Kumada H. Impact of sodium glucose cotransporter 2 inhibitor on histological features and glucose metabolism of non-alcoholic fatty liver disease complicated by diabetes mellitus. Hepatol Res. 2019;49:531-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 77. | Lambrecht J, Jan Poortmans P, Verhulst S, Reynaert H, Mannaerts I, van Grunsven LA. Circulating ECV-Associated miRNAs as Potential Clinical Biomarkers in Early Stage HBV and HCV Induced Liver Fibrosis. Front Pharmacol. 2017;8:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 78. | Jiao X, Fan Z, Chen H, He P, Li Y, Zhang Q, Ke C. Serum and exosomal miR-122 and miR-199a as a biomarker to predict therapeutic efficacy of hepatitis C patients. J Med Virol. 2017;89:1597-1605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 79. | Fan Z, Zhang Q, Chen H, He P, Li Y, Si M, Jiao X. Circulating microRNAs as a biomarker to predict therapy efficacy in hepatitis C patients with different genotypes. Microb Pathog. 2017;112:320-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 80. | Welker MW, Reichert D, Susser S, Sarrazin C, Martinez Y, Herrmann E, Zeuzem S, Piiper A, Kronenberger B. Soluble serum CD81 is elevated in patients with chronic hepatitis C and correlates with alanine aminotransferase serum activity. PLoS One. 2012;7:e30796. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 81. | Devhare PB, Sasaki R, Shrivastava S, Di Bisceglie AM, Ray R, Ray RB. Exosome-Mediated Intercellular Communication between Hepatitis C Virus-Infected Hepatocytes and Hepatic Stellate Cells. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 132] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 82. | Gramantieri L, Baglioni M, Fornari F, Laginestra MA, Ferracin M, Indio V, Ravaioli M, Cescon M, De Pace V, Leoni S, Coadă CA, Negrini M, Bolondi L, Giovannini C. LncRNAs as novel players in hepatocellular carcinoma recurrence. Oncotarget. 2018;9:35085-35099. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 83. | Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, Shinden Y, Iguchi T, Eguchi H, Shirabe K, Ochiya T, Maehara Y, Mimori K. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112:532-538. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 243] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 84. | Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, Shi X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res. 2018;37:324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 329] [Cited by in RCA: 356] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 85. | Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin YF, Yuan Y, Zhuang SM. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. 2018;68:1459-1475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 264] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 86. | Fang T, Lv H, Lv G, Li T, Wang C, Han Q, Yu L, Su B, Guo L, Huang S, Cao D, Tang L, Tang S, Wu M, Yang W, Wang H. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 749] [Article Influence: 107.0] [Reference Citation Analysis (0)] |
| 87. | Nakano T, Chen IH, Wang CC, Chen PJ, Tseng HP, Huang KT, Hu TH, Li LC, Goto S, Cheng YF, Lin CC, Chen CL. Circulating exosomal miR-92b: Its role for cancer immunoediting and clinical value for prediction of posttransplant hepatocellular carcinoma recurrence. Am J Transplant. 2019;19:3250-3262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 88. | Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, Park JG, Lee HW, Han YS, Chun JM, Park SY, Hur K. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer. 2019;144:1444-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 202] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 89. | Matsuura Y, Wada H, Eguchi H, Gotoh K, Kobayashi S, Kinoshita M, Kubo M, Hayashi K, Iwagami Y, Yamada D, Asaoka T, Noda T, Kawamoto K, Takeda Y, Tanemura M, Umeshita K, Doki Y, Mori M. Exosomal miR-155 Derived from Hepatocellular Carcinoma Cells Under Hypoxia Promotes Angiogenesis in Endothelial Cells. Dig Dis Sci. 2019;64:792-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 90. | Itami-Matsumoto S, Hayakawa M, Uchida-Kobayashi S, Enomoto M, Tamori A, Mizuno K, Toyoda H, Tamura T, Akutsu T, Ochiya T, Kawada N, Murakami Y. Circulating Exosomal miRNA Profiles Predict the Occurrence and Recurrence of Hepatocellular Carcinoma in Patients with Direct-Acting Antiviral-Induced Sustained Viral Response. Biomedicines. 2019;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 91. | Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, Dai Z, Cao Y, Fan J, Zhou J. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther. 2017;10:3843-3851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 124] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 92. | Liu J, Fan L, Yu H, Zhang J, He Y, Feng D, Wang F, Li X, Liu Q, Li Y, Guo Z, Gao B, Wei W, Wang H, Sun G. Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-regulate Programmed Death Ligand 1 Expression in Macrophages. Hepatology. 2019;70:241-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 314] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 93. | Wang F, Li L, Piontek K, Sakaguchi M, Selaru FM. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology. 2018;67:940-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 214] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 94. | Qu Z, Wu J, Ji A, Qiang G, Jiang Y, Jiang C, Ding Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:80666-80678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 95. | Yu LX, Zhang BL, Yang Y, Wang MC, Lei GL, Gao Y, Liu H, Xiao CH, Xu JJ, Qin H, Xu XY, Chen ZS, Zhang DD, Li FG, Zhang SG, Liu R. Exosomal microRNAs as potential biomarkers for cancer cell migration and prognosis in hepatocellular carcinoma patient-derived cell models. Oncol Rep. 2019;41:257-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 96. | Xu H, Chen Y, Dong X, Wang X. Serum Exosomal Long Noncoding RNAs ENSG00000258332.1 and LINC00635 for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Cancer Epidemiol Biomarkers Prev. 2018;27:710-716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 97. | Xu H, Dong X, Chen Y, Wang X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin Chem Lab Med. 2018;56:479-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 98. | Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, Zhang Y, Li H, Zhang Q, Yang Y, Chen G. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432-445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 194] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 99. | Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, Zhou J, Tang ZY. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 299] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 100. | Khatun M, Ray RB. Mechanisms Underlying Hepatitis C Virus-Associated Hepatic Fibrosis. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 101. | He R, Wang Z, Shi W, Yu L, Xia H, Huang Z, Liu S, Zhao X, Xu Y, Yam JWP, Cui Y. Exosomes in hepatocellular carcinoma microenvironment and their potential clinical application value. Biomed Pharmacother. 2021;138:111529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 102. | Srinivas AN, Suresh D, Santhekadur PK, Suvarna D, Kumar DP. Extracellular Vesicles as Inflammatory Drivers in NAFLD. Front Immunol. 2020;11:627424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 103. | Rios-Colon L, Arthur E, Niture S, Qi Q, Moore JT, Kumar D. The Role of Exosomes in the Crosstalk between Adipocytes and Liver Cancer Cells. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 104. | Díaz-Garrido N, Badia J, Baldomà L. Microbiota-derived extracellular vesicles in interkingdom communication in the gut. J Extracell Vesicles. 2021;10:e12161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 171] [Article Influence: 42.8] [Reference Citation Analysis (0)] |
| 105. | Takeuchi S, Tsuchiya A, Iwasawa T, Nojiri S, Watanabe T, Ogawa M, Yoshida T, Fujiki K, Koui Y, Kido T, Yoshioka Y, Fujita M, Kikuta J, Itoh T, Takamura M, Shirahige K, Ishii M, Ochiya T, Miyajima A, Terai S. Small extracellular vesicles derived from interferon-γ pre-conditioned mesenchymal stromal cells effectively treat liver fibrosis. NPJ Regen Med. 2021;6:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 64] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 106. | Watanabe Y, Tsuchiya A, Seino S, Kawata Y, Kojima Y, Ikarashi S, Starkey Lewis PJ, Lu WY, Kikuta J, Kawai H, Yamagiwa S, Forbes SJ, Ishii M, Terai S. Mesenchymal Stem Cells and Induced Bone Marrow-Derived Macrophages Synergistically Improve Liver Fibrosis in Mice. Stem Cells Transl Med. 2019;8:271-284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 112] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 107. | Warnecke A, Prenzler N, Harre J, Köhl U, Gärtner L, Lenarz T, Laner-Plamberger S, Wietzorrek G, Staecker H, Lassacher T, Hollerweger J, Gimona M, Rohde E. First-in-human intracochlear application of human stromal cell-derived extracellular vesicles. J Extracell Vesicles. 2021;10:e12094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 108. | Mendt M, Kamerkar S, Sugimoto H, McAndrews KM, Wu CC, Gagea M, Yang S, Blanko EVR, Peng Q, Ma X, Marszalek JR, Maitra A, Yee C, Rezvani K, Shpall E, LeBleu VS, Kalluri R. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. 2018;3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 578] [Article Influence: 82.6] [Reference Citation Analysis (0)] |
| 109. | Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498-503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1167] [Cited by in RCA: 1905] [Article Influence: 238.1] [Reference Citation Analysis (1)] |