Published online Jun 27, 2022. doi: 10.4254/wjh.v14.i6.1131
Peer-review started: December 3, 2021
First decision: February 8, 2022
Revised: February 22, 2022
Accepted: May 16, 2022
Article in press: May 16, 2022
Published online: June 27, 2022
Processing time: 202 Days and 1.9 Hours
Coronavirus disease 19 (COVID-19) has not only been shown to affect the respiratory system, but has also demonstrated variable clinical presentations including gastrointestinal tract disorders. In addition, abnormalities in liver enzymes have been reported indicating hepatic injury. It is known that severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) might infect cells via the viral receptor angiotensin-converting enzyme 2 (ACE2) which is expressed in several organs including the liver. The viral Spike glycoprotein binds to ACE2 and must be cleaved by Furin and Type 2 Serine Protease to enter the cells. After that, the Akt/mTOR signaling pathway is activated and several COVID-19 changes are triggered.
To analyze liver and gastrointestinal symptoms and cell signaling pathways triggered by SARS-CoV-2 infection due to virus-liver interactions in silico.
In this in silico study, the three-dimensional structures of the Akt, mTORC1 and Furin (receptors) were selected from the Protein Data Bank (PDB) and the structures of inhibitors (ligands) MK-2206, CC-223 and Naphthofluorescein were selected from PubChem and ZINC databases. Ligand files were downloaded as 2D structures and converted to optimized 3D structures using ViewerLite 4.2 software. Marvin Sketch® software was used to calculate prediction of the protonated form of inhibitors in a physiological environment (pH 7.4). AutoDock Tools (ADT) software was used to calculate and delimit the Grid box used in the molecular docking of each structure selected in the PDB. In addition, protonated ligands were prepared for molecular docking using ADT software. Molecular docking was performed using ADT software tools connected to Vina software. Analysis of the amino acid residues involved in ligand interactions, as well as ligand twists, the atoms involved in interactions, bond type and strength of interactions were performed using PyMol® and Discovery Studio® (BIOVIA) software.
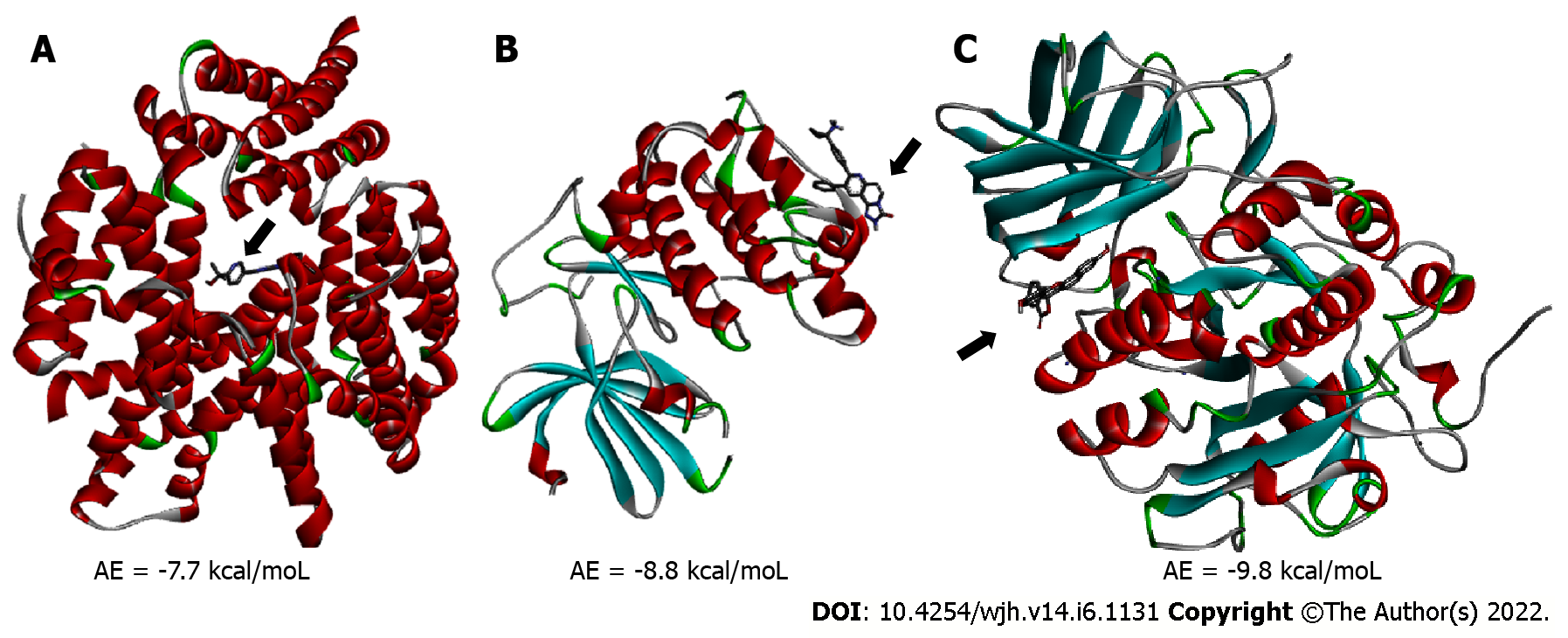
Molecular docking analysis showed that the mTORC1/CC-223 complex had affinity energy between the receptor and ligand of -7.7 kcal/moL with interactions ranging from 2.7 to 4.99 Å. There were four significant chemical bonds which involved two of five polypeptide chains that formed the FKBP12–Rapamycin-Binding (FRB) domain. The strongest was a hydrogen bond, the only polar interaction, and Van der Waals interactions shown to be present in 12 residues of mTORC1’s FRB domain. With regard to the Akt/MK-2206 complex there were three Van der Waals interactions and 12 chemical bonds in which seven residues of Akt were involved with all five rings of the MK-2206 structure. In this way, both ASP 388 and GLN 391 bind to the same MK-2206 ring, the smaller one. However, LYS 386 had four chemical bonds with the inhibitor, one with each structure ring, while LYS 387 binds two distinct rings. One of the MK-2206 inhibitor's rings which binds to LYS 387 also binds simultaneously to ILE 367 and LEU 385 residues, and the fifth ring of the structure was involved in a bond with the ALA 382 residue. The hydrogen bonds were the shortest bonds in the complex (2.61 and 3.08 Å) and all interactions had an affinity energy of -8.8 kcal/moL. The affinity energy in the Furin/Naphhofluorescein complex was -9.8 kcal/moL and involved six interactions ranging from 2.57 to 4.98 Å. Among them, two were polar and the others were non-polar, in addition to twelve more Van der Waals interactions. Two distinct hydrogen bonds were formed between Furin and its inhibitor involving GLN 388 and ALA 532 residues. ALA 532 also binds to two distinct rings of Naphthofluorescein, while TRP 531 residue has two simultaneous bonds with the inhibitor.
Liver infection and signaling pathways altered by SARS-CoV-2 can be modulated by inhibitors that demonstrate significant interaction affinity with human proteins, which could prevent the development of infection and symptoms.
Core Tip: The classification of coronavirus disease 19 (COVID-19) as a respiratory disease caused the focus of studies to be directed in this direction. Therefore, mild clinical symptoms, such as gastrointestinal symptoms, have been little studied. Following the knowledge that COVID-19 is a systemic disease, studies on liver damage have become important. This study analyzed liver molecules targeted by severe acute respiratory syndrome coronavirus-2 infection using bioinformatics. Although these molecules are present in various organs due to the liver's central role in systemic metabolism, trying to understand metabolic changes in this organ will help understand systemic changes induced by the virus.
- Citation: Peiter GC, de Souza CBT, de Oliveira LM, Pagliarin LG, dos Anjos VNF, da Silva FAF, de Melo FF, Teixeira KN. COVID-19 liver and gastroenterology findings: An in silico analysis of SARS-CoV-2 interactions with liver molecules. World J Hepatol 2022; 14(6): 1131-1141
- URL: https://www.wjgnet.com/1948-5182/full/v14/i6/1131.htm
- DOI: https://dx.doi.org/10.4254/wjh.v14.i6.1131
At the end of 2019, in Wuhan (China), the emergence of a new coronavirus [severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)] was reported, which is classified as belonging to the Beta-coronavirus genus and possessing as genetic material a single strip positive RNA, being capable of resulting in disease [coronavirus disease 19 (COVID-19)] that can evolve to severe acute respiratory syndrome. Although it is known that this disease mainly affects the respiratory system, it was identified that it can manifest clinic signs and symptoms related to other organs, such as nausea, vomiting, abdominal pain and diarrhea. With regard to the liver, alterations in lesion markers including alanine aminotransferase (ALT), aspartate aminotransferase (AST) and total serum bilirubin were reported, indicating hepatic injury[1,2]. Between 14%-53% of patients with COVID-19 present with abnormalities in transaminase levels[3,4] which is associated with severity of the disease, and abnormal transaminase levels can indicate a higher chance of poor prognosis and Intensive Care Unit requirement[5-7]. The associated causes of liver injury, beyond direct viral impact, include the use of drugs during treatment, hypoxia due to pulmonary symptoms, previous hepatic lesions and co-morbidities[4]. Some of the drugs used can cause hepatotoxicity, liver injury and dysregulation, as shown for hydroxychloroquine, Azithromycin and Remdesivir, respectively[8,9]. One of the SARS-CoV-2 presentations on this system can be acute non-icteric hepatitis before the most common symptoms (fever and respiratory symptoms)[10] and those already related to the liver, nausea, diarrhea and abdominal pain. The viral cellular entrance and its dissemination is on a vast spectrum of the organism’s systems, for example the liver-related, is a consequence of the expression of a common cellular receptor [angiotensin-converting enzyme 2 (ACE2)]. However, for effective cellular intrusion the participation of other human proteins such as Type 2 Serine Protease (TMPRSS2) and Furin (Convertase Proprotein of the Subtilisin Type)[1,2,11-14]. Therefore, concerning the entrance mechanisms, it has been identified that the viral Spike glycoprotein possesses affinity and binds to the ACE2 (responsible for the adhesion stage), as it is structurally divided into two subunits: S1 (N-terminus, that connects to the receptor) and S2 (C-terminus, that takes part in the penetration process)[2,12,13]. Between both units, S1 and S2, there is a cleavage site for Furin, that triggers the activation and conformational change of viral Spike glycoprotein after it completes this action[2,13,14]. Following the initial activation process, further cleavage between the S1/S2 and S2’ sites is essential for viral entrance. The TMPRSS2 protein performs this activity on the Spike glycoprotein of SARS-CoV-2, which allows fusion between the viral and cellular membranes, entry of the viral genetic material and the development of infection[1,11,12]. Following viral entrance, during the course of infection, modulation of the signaling pathway Akt/mTOR, that regulates apoptosis, cell survival, transcription and translation, occurs which also occurs during infection by other viruses[15-17]. This signaling possibly increases factors of viral translation while blocking mechanisms of cellular death, generating greater pathogenicity. Based on this, recent studies have indicated the possibility of using already existing drugs that interfere with this pathway for the treatment of COVID-19, including MK-2206, an Akt inhibitor[18].
Despite these facts, studies directly investigating the interaction between SARS-CoV-2 and liver cells, specifically the entrance mechanisms, the biochemical cascades and methods for possible infection inhibition still require further investigation. Therefore, the present study aims to associate the hepatic alterations triggered by SARS-CoV-2 with the activation and/or inhibition of transduction pathways of cellular signals in the viral infection process. In addition, possible intervention in the signaling pathways with inhibitors was analyzed to suggest potential treatments for SARS-CoV-2 infection.
Akt, mTORC1 and Furin enzymes are inhibited by MK-2206, CC-223 and Naphthofluorescein, respectively. The three-dimensional structures of proteins/enzymes (receptors) Akt, mTORC1 and Furin were selected from the Protein Data Bank (PDB) (http://www.rcsb.org/pdb/home/home.do) and their files were obtained in the extension.pdb (input file). The structures of the inhibitors (ligands) MK-2206, CC-223 and Naphthofluorescein were selected from PubChem (https://pubchem.ncbi.nlm.nih.gov/) and ZINC databases (https://zinc12.docking.org/). The ligand files were downloaded as 2D structures in the extension.sdf and converted into.pdb (input file) by optimizing the 3D structure using ViewerLite 4.2 software (Accelrys Inc.). The selected receptor's three-dimensional structure was prepared for molecular docking simulations using AutoDock Tools (ADT) software (MGL, The Scripps Research Institute); possible ligands (ions, peptides) were eliminated, water molecules were deleted and hydrogen atoms were added. The location and dimensions of the Grid box (virtual box that delimits the region where the ligand will perform possible interactions with the receptor) were then determined using ADT software. The Grid box data and coordinates were used in molecular docking. The ligands interact with their receptors in a physiological environment at pH 7.4; thus, to reliably simulate their interaction, calculations for the prediction of their protonation state were carried out using Marvin Sketch® software (ChemAxon®). Protonated ligands were prepared for molecular docking using ADT software. This software detects the torsion points of the inhibitors and calculates their angle of torsion.
Molecular docking procedures for a rigid receptor and a flexible ligand were used. A grid of points in x, y, and z directions was built with a grid spacing of 1.0 Å using the AutoGrid component of the software. Molecular docking simulations were performed using ADT software connected to Vina software[19]. The software used associates two components: A search algorithm and a score function. The algorithm is responsible for the search of possible combinations in the bonds, exploring the rotational, translational and conformational degrees of freedom of the ligand, as well as of the proteins. The score function is then used to choose the best binding modes. These functions are obtained according to the force fields of molecular mechanics and empirical parameters from free energy calculations, thus an affinity energy is calculated. The cut-off for stable interactions is considered an affinity energy < −6.0 kcal/moL[20]. The results are based on the first docking conformer of the ligands with reference Root-Mean-Square Deviation of atomic positions (RMSD) of 0[21]. The analyses of amino acid residues of receptors involved in the interactions with ligands, as well as the twists of the ligand, atoms involved in the interactions, type, strength and length of the interactions were performed using PyMol® software (pymol.org/2) and Discovery Studio® software from BIOVIA.
The receptors’ 3D structures selected were the 5WBH-FKBP12–Rapamycin-Binding (FRB) domain of mTOR; 1GZN-PKB kinase domain and 5JXI-Furin convertase. All structures are human and present less than 2.5 Å of resolution. The structure of inhibitor MK-2206 was selected from ZINC databank-ZINC36382821 and the structures of inhibitors CC-223 (CID58298316), and Naphthofluorescin (CID3124834) were selected from the PubChem databank.
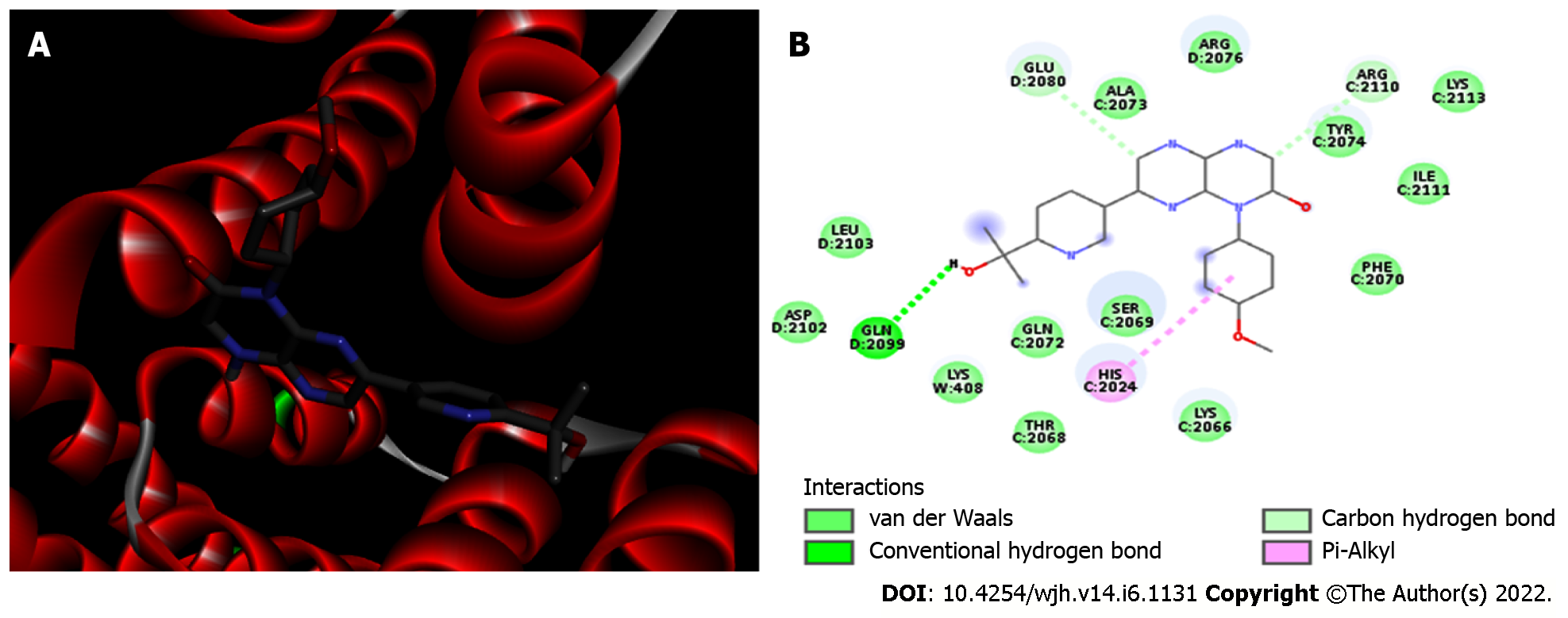
mTORC1/CC-223 complex: Molecular docking was used to analyze if there was an interaction between mTORC1 enzyme and CC-223 in its protonated form. In this way, the most stable mTORC1/CC-223 complex showed an affinity energy of -7.7 kcal/moL (Figure 1). Four significant chemical bonds were observed between the receptor and the ligand involving four distinct residues and bond size ranging from 2.7 to 4.99 Å (Table 1). Of these bonds only one was polar (hydrogen bond) and according to their size, this was the strongest (2.7 Å); the other bonds had a non-polar character. In addition, 12 other residues were involved in Van der Waals interactions (Figure 2). The four significant bonds in the mTORC1/CC-223 complex involved two (C and D) of five polypeptide chains (A-E) that formed the FRB domain.
| Protein | Residues | Type | Bond size (Å) |
| mTORC1 | HIS 2024 | Pi-Alkyl | 4.99 |
| GLU 2080 | Carbon hydrogen bond | 3.45 | |
| GLN 2099 | Hydrogen bond | 2.70 | |
| ARG 2110 | Carbon hydrogen bond | 3.44 | |
| 12 residues | Van der Waals | - | |
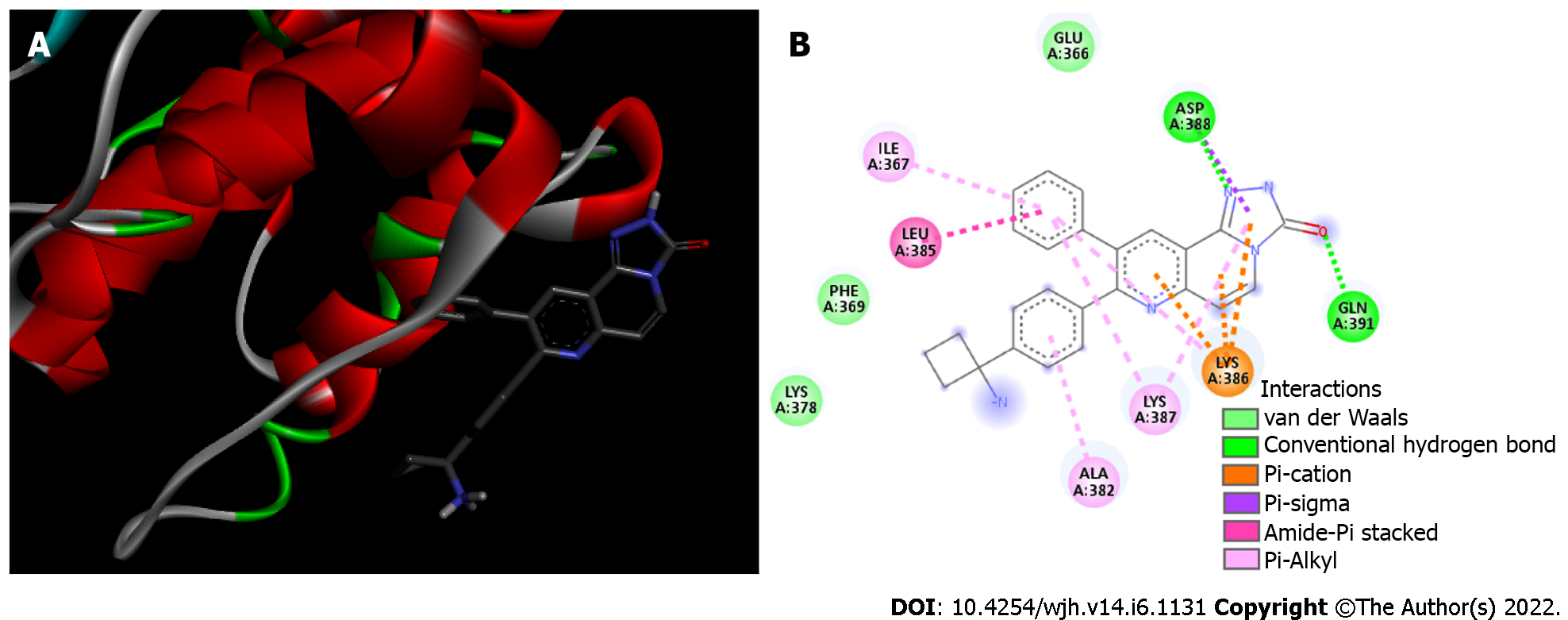
| Akt | ILE 367 | Pi-Alkyl | 5.04 |
| ALA 382 | Pi-Alkyl | 5.02 | |
| LEU 385 | Amide-Pi | 3.90 | |
| LYS 386 | Pi-Alkyl | 5.25 | |
| LYS 386 | Pi-Cation | 3.08 | |
| LYS 386 | Pi-Cation | 4.19 | |
| LYS 386 | Pi-Cation | 4.94 | |
| LYS 387 | Pi-Alkyl | 5.22 | |
| LYS 387 | Pi-Alkyl | 5.19 | |
| ASP 388 | Hydrogen bond | 3.08 | |
| ASP 388 | Pi-Sigma | 3.67 | |
| GLN 391 | Hydrogen bond | 2.61 | |
| 3 residues | Van der Waals | - | |
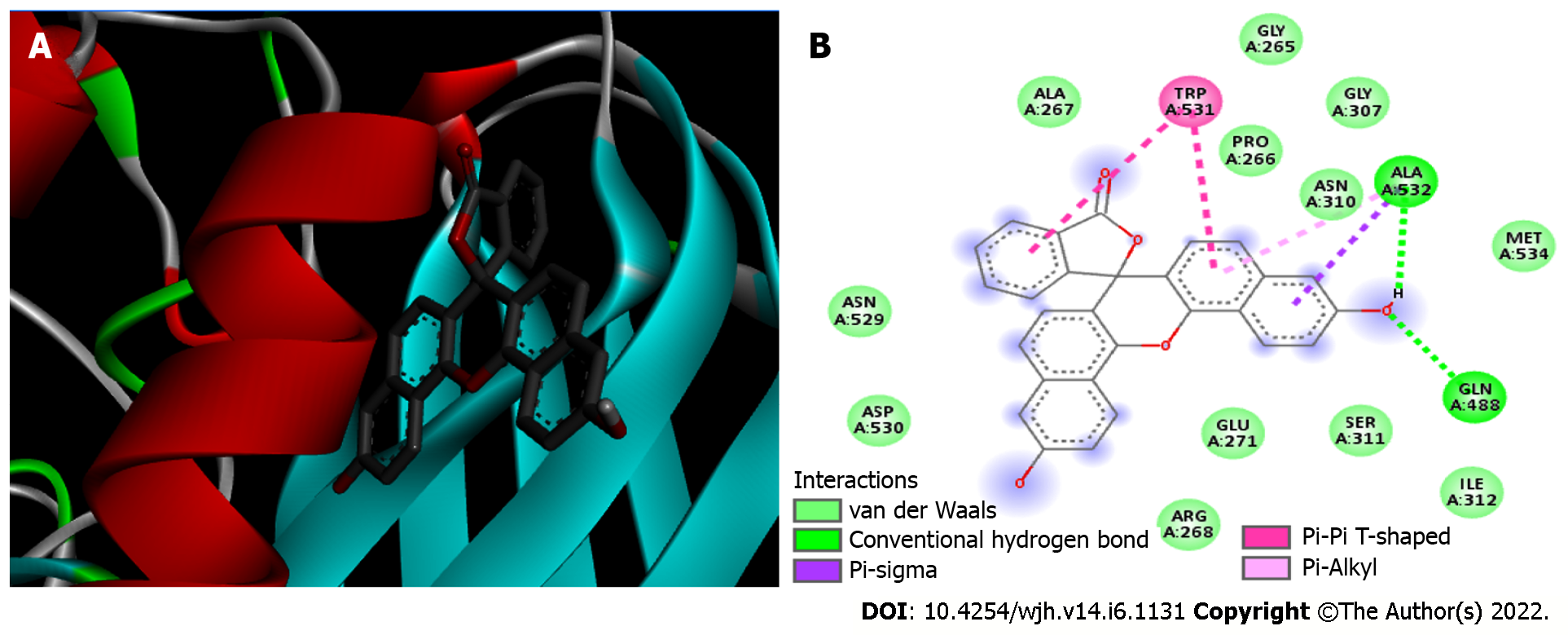
| Furin | TRP 531 | Pi-Pi T-shaped | 4.97 |
| TRP 531 | Pi-Pi T-shaped | 4.95 | |
| ALA 532 | Hydrogen bond | 2.57 | |
| ALA 532 | Pi-Alkyl | 4.98 | |
| ALA 532 | Pi-Sigma | 3.94 | |
| GLN 488 | Hydrogen bond | 3.12 | |
| 12 residues | Van der Waals | - |
Akt/MK-2206 complex: Analysis of the interaction between the Akt enzyme and its protonated inhibitor-MK-2206, showed that the complex was maintained by 12 chemical bonds, in addition to three Van der Waals interactions (Figure 3 and Table 1). All five rings of the MK-2206 structure were involved in bonds with amino acid residues of Akt enzyme. Seven residues of Akt were involved in chemical bonds; ASP 388 and GLN 381 residues were bound to an oxygen atom and a nitrogen atom of MK-2206 by a hydrogen bond, respectively. Hydrogen bonds were the shortest of the Akt/MK-2206 complex (2.61 and 3.08 Å). ASP 388 was bound to MK-2206 by a Pi-Alkyl bond (hydrophobic character). Both ASP 388 and GLN 391 were bound to the same MK-2206 ring, the smaller one. LYS 386 had four chemical bonds with the inhibitor (3.08-5.25 Å), one with each ring structure; three of them were Pi-Cation and the other was Pi-Alkyl. LYS 387 was bound to two distinct rings of the MK-2206 inhibitor by Pi-Alkyl bonds (5.19 and 5.22). One of the MK-2206 inhibitor's rings that bound LYS 387 also bound simultaneously to two other residues-ILE 367 (Pi-Alkyl) and LEU 385 (Amide-Pi). These bonds involved four of five rings of MK-2206. The fifth ring of the structure was involved in a single Pi-Alkyl bond with the ALA 382 residue. All bonds and their distribution conferred an affinity energy of -8.8 kcal/moL (Figure 1).
Furin/naphthofluorescein complex: Molecular docking was used to analyze whether there was an interaction between Naphthofluorescein and human Furin. In this way, the simulation helped to better understand the interaction dynamics between the protein and the inhibitor. After analyzing the protonation state of Naphthofluorescein, the interaction affinity was calculated by AutoDock Vina, and was -9.8 kcal/moL for the complex formed. It was possible to observe that Naphthofluorescein interacted with human Furin through six interactions with a length ranging from 2.57 to 4.98 Å. Two distinct hydrogen bonds were formed between Furin and its inhibitor involving GLN 388 and ALA 532 residues. Among them, two were polar and the others were non-polar, in addition to twelve more Van der Waals interactions. ALA 532 was also bound to two distinct rings of Naphthofluorescein by a Pi-Sigma bond and Pi-Alkyl bond. TRP 531 residue showed two simultaneous bonds with the inhibitor (Figure 4 and Table 1).
Liver is the main organ involved in drug metabolism and xenobiotic detoxification; therefore, its proper functioning is essential for the effectiveness of pharmacological treatments. As altered liver function is reported in up to half of patients with COVID-19, it is important to clearly understand the possible mechanisms involved in liver injury in order to optimize the treatment outcome of this disease[22].
Moderate microvesicular steatosis and mild inflammation in the lobular and portal area are pathological findings in liver tissue in patients with COVID-19[23]; thus, this may contribute to the incidence of elevated levels of hepatic transaminases reported in this disease[24]. In the early stage of COVID-19, infected individuals have positive SARS-CoV-2 RNA in fecal and blood samples, and present gastrointestinal symptoms such as diarrhea, abdominal pain, nausea, and vomiting[25] suggesting that SARS-CoV-2 could infect liver cells[26].
In SARS-CoV-2 infection, it is known that the viral Spike glycoprotein interacts with the ACE2 present in humans, leading to entry of the virus. However, for its entry to occur, there is a need for activation of the glycoprotein by host cell proteases which occurs between the S1/S2 subunits of Spike generating a conformational change in the S2 subunit and allowing the interaction of SARS-CoV-2 with ACE2, completing virus entry[12].
As SARS-CoV-2 interacts with the ACE2 receptor of host cells to invade them[27]; thus, cells that have this receptor are susceptible to infection. The level of ACE2 expression is low in hepatocytes (2.6%), but in bile duct cells (cholangiocytes) this expression is high (59.7%)[28]. Therefore, SARS-CoV-2 does not necessarily directly infect liver cells, but causes bile duct dysfunction that plays an important role in liver regeneration and immune response[26].
In fact, according to Xu et al[23] (2020), no direct cytopathic effect of SARS-CoV-2 on the liver was found in pathological autopsy findings. On the other hand, the findings of Pirola and Sookoian[29] support the possibility that SARS-CoV-2 may cause direct liver injury by the viral cytopathic effect. These authors showed that the three host cell proteins-ACE2, Furin and TMPRSS2, responsible for viral infection are expressed in liver tissue. Although ACE2 has low expression in hepatocytes compared with cholangiocytes, TMPRSS2[1] and Furin[30] are expressed more in hepatocytes.
The Spike glycoprotein of SARS-CoV-2 facilitates viral entry into host cells; the surface unit S1 binds to the cellular receptor-ACE2, while the transmembrane unit S2 facilitates fusion of both viral and cellular membrane. Membrane fusion depends on S protein cleavage by host cell proteases at the S1/S2 and the S2′ sites[31], and among these proteases, TMPRSS2 and Furin play major roles in proteolytic activation of a broad range of viruses[32] including SARS-CoV-2. After infection, the signaling path
An analysis of the summary of the SARS-CoV-2 infection process indicates that host cell proteases and signaling cascade proteins could be a potential target for therapeutic interventions for COVID-19 symptoms, including gastrointestinal symptoms due to liver damage.
This study analyzed the interactions of SARS-CoV-2 with molecules which are necessary for the success of infection and are expressed in the liver using bioinformatics tools and in silico enzymatic inhibition tests to try to associate such interactions with the gastrointestinal findings and liver injuries in COVID-19. It was shown that the interaction affinity of some proteins present in the human body with their respective inhibitors that can act in their pathways and prevent the development of infection. The target proteins were the Furin enzyme, involved in cell invasion, and mTORC1 and Akt enzymes belonging to the signaling pathway.
To verify the interaction between the mTORC1/CC-223, Akt/MK-2206 and Furin/naphthofluorescein complexes at the molecular level, molecular docking was used. Thus, if there is an interaction between proteins and inhibitors, simulation helps us to understand the dynamics that occur in silico[34].
The interaction affinity calculated by AutoDock Vina for the mTORC1/CC-223 complex was -7.7 kcal/moL, for the Akt/MK-2206 complex it was -8.8, and for the complex formed by Furin/naphthofluorescein it was -9.8 kcal/moL. These values are considered significant since values lower than -6.0 kcal/moL already constitute stable interactions in silico analysis[35].
The complex with the highest interaction affinity was that formed by Furin and its inhibitor. This significantly low affinity energy value indicates a more stable complex, in other words, indicates that the inhibitor will have higher biological activity[36]. Unlike other coronaviruses, SARS-CoV-2 has a potentially critical insertion of a Furin cleavage site upstream of the S1 cleavage site in the Spike glycoprotein reducing its dependence on host cell proteases for infection. The high affinity between ACE2 and the Spike glycoprotein cleaved by Furin allows SARS-CoV-2 to maintain its efficient entry into cells while preventing the action of the immune system which can contribute to the widespread infection capacity of the virus[14,37]. As ACE2 is present in type 2 alveolar cells, the gastrointestinal tract and the liver, these tissues would be more affected by COVID-19[38]; therefore, inhibiting the action of Furin would prevent infection of these tissues and consequently the associated symptoms.
Vankadari[39] in his study on Furin analyzed not only its structure but also how it would bind at S1/S2 subunits of the Spike glycoprotein. Thus, it was suggested that Furin binds to these subunits through the equatorial region present in the Spike glycoprotein which creates a 970 Å interface between the participants.
The Furin enzyme is required in various normal functions of the body[40]. Prolonged Furin blocking can therefore generate side effects or damage[41]. In this context, Furin's involvement in the viral invasion process could reduce the effectiveness of the action of this enzyme in normal physiological processes triggering pathological processes. In fact, studies suggest that Furin plays an important role in homeostasis and disease[42]; thus, it is possible that liver cell lesions would be reduced and AST and ALT levels would be normal. On the other hand, brief Furin inhibition can be well tolerated and has therapeutic benefit[41]; therefore, Naphthofluorescein has promising potential for treatment through this route given its high affinity for Furin in silico.
Another way of studying the symptoms resulting from liver injury triggered by SARS-CoV-2 infection is to analyze the signaling pathway affected by the virus. In fact, some studies point to the dysregulation in the Akt/mTOR signaling cascade, which could be a potential target in COVID-19 treatment[33].
It was observed in this study that the Akt/MK-2206 complex remains with 12 bonds, added to three Van der Waals interactions and all five rings of the MK-2206 inhibitor are bound to protein, suggesting stability of the complex, and the protein Akt would be unable to proceed with his cascade. Shi et al[43] in his findings on the inhibition of esophageal cancer growth through the PI3K/AKT/mTOR pathway, showed that MK-2206 would be a potential allosteric inhibitor of Akt, by decreasing cell proliferation, inducing cell cycle arrest and increasing apoptosis of cancer cells. Furthermore, Appelberg et al[33] observed that the Akt/mTOR/HIF-1 pathways participate in COVID-19 infection, in particular, Akt/mTOR are activated at the beginning of infection. It has also been shown that MK-2206 caused a decay in viral transcription in SARS-CoV-2 infected cells and supernatants by interacting with Akt. Based on these data and on the results obtained with molecular docking, in which significant affinity and stability of the bonds were observed, it is possible that the MK-2206 inhibitor may help to contain the development of COVID-19 by interacting with Akt, in a way that prevents continuity of the cascade triggered by this protein.
It is possible that in the mTORC1/CC-223 complex significant chemical bonds are established along the CC-223 structure involving three of four molecule rings. The same was observed for Van der Waals' interactions. The sum of all binding and interactions between mTORC1 and CC-223 contribute to the formation of a complex with significant affinity energy which contributes to its stability. Mortensen et al[44] showed that CC-223 as an inhibitor has high affinity for mTOR, and the pathway to which it belongs was unfeasible. It has been described that mTOR is relevant in cell growth, proliferation, survival and metabolism; in particular, mTORC-1 plays a role in protein synthesis and cell develop
Current understanding suggests that the possible interaction between SARS-CoV-2 and liver cells occurs with enhancer factors that facilitate ACE2-mediated SARS-CoV-2 infection, such as Furin and TMPRSS2. In this study, we demonstrate, by means of molecular docking, significant affinity and stability of the bonds concerning some human hepatic proteins and their inhibitors. These work in signaling pathways due to interactions between mTORC1/CC-223, Akt/MK-2206 and Furin/naphthofluorescein complexes, and are capable of preventing infection development. Our in silico analysis shows the possible inhibitory mechanisms of cell viral receptors in the liver, which is consistent with other studies. Therefore, multi-target therapeutic drugs based on these pathways may be an option in COVID-19 patients, especially in severe/critical cases, preventing multiple organ dysfunction and perhaps leading to a more positive prognosis.
Coronavirus disease 19 (COVID-19) has variable clinical manifestations, including gastrointestinal and hepatic disorders. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infects liver cells using angiotensin-converting enzyme 2, and the viral Spike glycoprotein must be cleaved by Furin or Type 2 Serine Protease. Following activation of the Akt/mTOR pathway several changes are triggered.
Liver damage in COVID-19 is not well understood; therefore, molecular analysis of the infection process and cell signaling, in silico, can help in the discovery of targets for treatment of the disease.
To analyze liver and gastrointestinal symptoms and cell signaling pathways triggered by SARS-CoV-2 infection due to virus-liver interactions in silico.
SARS-CoV-2/liver cell interactions, and signaling pathways activated by these interactions, were analyzed by inhibition studies using the molecular docking method.
The mTORC1/CC-223 complex, Akt/MK-2206 complex and Furin/naphthofluorescein complex showed significant affinity energy, indicating stability and consequent effectiveness in inhibiting target molecules for COVID-19 therapy.
Liver disease and signaling pathways altered by SARS-CoV-2 can be modulated by inhibitors that demonstrate significant affinity for interactions with human proteins, which could prevent progression of the infection and its symptoms.
Evaluate the inhibition complexes studied using molecular dynamics and verify the possibility of structural changes of the drug to increase its efficiency and avoid possible adverse effects.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Khan MKA, India; Zhu ZW, China A-Editor: Liu X S-Editor: Fan JR L-Editor: Webster JR P-Editor: Fan JR
| 1. | Dong M, Zhang J, Ma X, Tan J, Chen L, Liu S, Xin Y, Zhuang L. ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19. Biomed Pharmacother. 2020;131:110678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 182] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 2. | Nardo AD, Schneeweiss-Gleixner M, Bakail M, Dixon ED, Lax SF, Trauner M. Pathophysiological mechanisms of liver injury in COVID-19. Liver Int. 2021;41:20-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 281] [Cited by in RCA: 271] [Article Influence: 67.8] [Reference Citation Analysis (2)] |
| 3. | Zhang C, Shi L, Wang FS. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol Hepatol. 2020;5:428-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1348] [Cited by in RCA: 1295] [Article Influence: 259.0] [Reference Citation Analysis (4)] |
| 4. | Anirvan P, Bharali P, Gogoi M, Thuluvath PJ, Singh SP, Satapathy SK. Liver injury in COVID-19: The hepatic aspect of the respiratory syndrome - what we know so far. World J Hepatol. 2020;12:1182-1197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (1)] |
| 5. | Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35178] [Cited by in RCA: 30098] [Article Influence: 6019.6] [Reference Citation Analysis (3)] |
| 6. | Eastin C, Eastin T. Clinical Characteristics of Coronavirus Disease 2019 in China: Guan W, Ni Z, Hu Y, et al. J Emerg Med. 2020;58:711-712. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 7. | Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, Fan Y, Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425-434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2493] [Cited by in RCA: 2310] [Article Influence: 462.0] [Reference Citation Analysis (0)] |
| 8. | LiverTox: Clinical and Research Information on Drug-Induced Liver Injury [Internet]. Bethesda (MD): National Institute of Diabetes and Digestive and Kidney Diseases; 2012– . [PubMed] |
| 9. | Zampino R, Mele F, Florio LL, Bertolino L, Andini R, Galdo M, De Rosa R, Corcione A, Durante-Mangoni E. Liver injury in remdesivir-treated COVID-19 patients. Hepatol Int. 2020;14:881-883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 91] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 10. | Wander P, Epstein M, Bernstein D. COVID-19 Presenting as Acute Hepatitis. Am J Gastroenterol. 2020;115:941-942. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 11. | Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB, Diamond MS, Ciorba MA, Whelan SPJ, Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 809] [Cited by in RCA: 765] [Article Influence: 153.0] [Reference Citation Analysis (0)] |
| 12. | de Almeida JO, de Oliveira VRT, Avelar JL dos S, Moita BS, Lima LM. COVID-19: Physiopathology and Targets for Therapeutic Intervention. Revis Virt De Quimica. 2020;12:1464-1497. [DOI] [Full Text] |
| 13. | Johnson BA, Xie X, Kalveram B, Lokugamage KG, Muruato A, Zou J, Zhang X, Juelich T, Smith JK, Zhang L, Bopp N, Schindewolf C, Vu M, Vanderheiden A, Swetnam D, Plante JA, Aguilar P, Plante KS, Lee B, Weaver SC, Suthar MS, Routh AL, Ren P, Ku Z, An Z, Debbink K, Shi PY, Freiberg AN, Menachery VD. Furin Cleavage Site Is Key to SARS-CoV-2 Pathogenesis. bioRxiv. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 14. | Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1142] [Cited by in RCA: 1237] [Article Influence: 247.4] [Reference Citation Analysis (0)] |
| 15. | Ranadheera C, Coombs KM, Kobasa D. Comprehending a Killer: The Akt/mTOR Signaling Pathways Are Temporally High-Jacked by the Highly Pathogenic 1918 Influenza Virus. EBioMedicine. 2018;32:142-163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 16. | Ehrhardt C, Ludwig S. A new player in a deadly game: influenza viruses and the PI3K/Akt signalling pathway. Cell Microbiol. 2009;11:863-871. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 133] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 17. | Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. JNK and PI3k/Akt signaling pathways are required for establishing persistent SARS-CoV infection in Vero E6 cells. Biochim Biophys Acta. 2005;1741:4-10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 74] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 18. | Gassen NC, Papies J, Bajaj T, Dethloff F, Emanuel J, Weckmann K. Analysis of SARS-CoV-2-controlled autophagy reveals spermidine, MK-2206, and niclosamide as putative antiviral therapeutics. 2020 Preprints. Available from: bioRxiv:997254. [DOI] [Full Text] |
| 19. | Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455-461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26714] [Cited by in RCA: 14497] [Article Influence: 966.5] [Reference Citation Analysis (0)] |
| 20. | Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 826] [Cited by in RCA: 958] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 21. | Coutsias EA, Seok C, Dill KA. Using quaternions to calculate RMSD. J Comput Chem. 2004;25:1849-1857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 231] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 22. | Alqahtani SA, Schattenberg JM. Liver injury in COVID-19: The current evidence. United European Gastroenterol J. 2020;8:509-519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 166] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 23. | Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5228] [Cited by in RCA: 5781] [Article Influence: 1156.2] [Reference Citation Analysis (2)] |
| 24. | Bertolini A, van de Peppel IP, Bodewes FAJA, Moshage H, Fantin A, Farinati F, Fiorotto R, Jonker JW, Strazzabosco M, Verkade HJ, Peserico G. Abnormal Liver Function Tests in Patients With COVID-19: Relevance and Potential Pathogenesis. Hepatology. 2020;72:1864-1872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 193] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 25. | Yeo C, Kaushal S, Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol. 2020;5:335-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 546] [Cited by in RCA: 554] [Article Influence: 110.8] [Reference Citation Analysis (0)] |
| 26. | Wu J, Song S, Cao HC, Li LJ. Liver diseases in COVID-19: Etiology, treatment and prognosis. World J Gastroenterol. 2020;26:2286-2293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 105] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (1)] |
| 27. | Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271-280.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11946] [Cited by in RCA: 14254] [Article Influence: 2850.8] [Reference Citation Analysis (0)] |
| 28. | Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single-Cell RNA Expression Profiling of ACE2, the Receptor of SARS-CoV-2. Am J Respir Crit Care Med. 2020;202:756-759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 361] [Cited by in RCA: 599] [Article Influence: 119.8] [Reference Citation Analysis (0)] |
| 29. | Pirola CJ, Sookoian S. SARS-CoV-2 virus and liver expression of host receptors: Putative mechanisms of liver involvement in COVID-19. Liver Int. 2020;40:2038-2040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 30. | Essalmani R, Susan-Resiga D, Chamberland A, Asselin MC, Canuel M, Constam D, Creemers JW, Day R, Gauthier D, Prat A, Seidah NG. Furin is the primary in vivo convertase of angiopoietin-like 3 and endothelial lipase in hepatocytes. J Biol Chem. 2013;288:26410-26418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 31. | Hulswit RJ, de Haan CA, Bosch BJ. Coronavirus Spike Protein and Tropism Changes. Adv Virus Res. 2016;96:29-57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 275] [Cited by in RCA: 320] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 32. | Klenk HD, Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 1994;2:39-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 351] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 33. | Appelberg S, Gupta S, Svensson Akusjärvi S, Ambikan AT, Mikaeloff F, Saccon E, Végvári Á, Benfeitas R, Sperk M, Ståhlberg M, Krishnan S, Singh K, Penninger JM, Mirazimi A, Neogi U. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg Microbes Infect. 2020;9:1748-1760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 205] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 34. | Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit. 1996;9:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 35. | Shityakov S, Förster C. In silico predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter. Adv Appl Bioinform Chem. 2014;7:23-36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 106] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 36. | Ferreira LG, Dos Santos RN, Oliva G, Andricopulo AD. Molecular docking and structure-based drug design strategies. Molecules. 2015;20:13384-13421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1277] [Cited by in RCA: 1172] [Article Influence: 117.2] [Reference Citation Analysis (0)] |
| 37. | Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:11727-11734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1930] [Cited by in RCA: 2374] [Article Influence: 474.8] [Reference Citation Analysis (0)] |
| 38. | Jothimani D, Venugopal R, Abedin MF, Kaliamoorthy I, Rela M. COVID-19 and the liver. J Hepatol. 2020;73:1231-1240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 358] [Article Influence: 71.6] [Reference Citation Analysis (1)] |
| 39. | Vankadari N. Structure of Furin Protease Binding to SARS-CoV-2 Spike Glycoprotein and Implications for Potential Targets and Virulence. J Phys Chem Lett. 2020;11:6655-6663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 40. | Roebroek AJ, Umans L, Pauli IG, Robertson EJ, van Leuven F, Van de Ven WJ, Constam DB. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development. 1998;125:4863-4876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 214] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 41. | Sarac MS, Cameron A, Lindberg I. The furin inhibitor hexa-D-arginine blocks the activation of Pseudomonas aeruginosa exotoxin A in vivo. Infect Immun. 2002;70:7136-7139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 42. | Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 896] [Cited by in RCA: 945] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 43. | Shi N, Yu H, Chen T. Inhibition of esophageal cancer growth through the suppression of PI3K/AKT/mTOR signaling pathway. Onco Targets Ther. 2019;12:7637-7647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 44. | Mortensen DS, Fultz KE, Xu S, Xu W, Packard G, Khambatta G, Gamez JC, Leisten J, Zhao J, Apuy J, Ghoreishi K, Hickman M, Narla RK, Bissonette R, Richardson S, Peng SX, Perrin-Ninkovic S, Tran T, Shi T, Yang WQ, Tong Z, Cathers BE, Moghaddam MF, Canan SS, Worland P, Sankar S, Raymon HK. CC-223, a Potent and Selective Inhibitor of mTOR Kinase: In Vitro and In Vivo Characterization. Mol Cancer Ther. 2015;14:1295-1305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 45. | Mortensen DS, Perrin-Ninkovic SM, Shevlin G, Zhao J, Packard G, Bahmanyar S, Correa M, Elsner J, Harris R, Lee BG, Papa P, Parnes JS, Riggs JR, Sapienza J, Tehrani L, Whitefield B, Apuy J, Bisonette RR, Gamez JC, Hickman M, Khambatta G, Leisten J, Peng SX, Richardson SJ, Cathers BE, Canan SS, Moghaddam MF, Raymon HK, Worland P, Narla RK, Fultz KE, Sankar S. Discovery of mammalian target of rapamycin (mTOR) kinase inhibitor CC-223. J Med Chem. 2015;58:5323-5333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |