Published online May 27, 2022. doi: 10.4254/wjh.v14.i5.956
Peer-review started: December 31, 2021
First decision: February 21, 2022
Revised: March 1, 2022
Accepted: May 7, 2022
Article in press: May 7, 2022
Published online: May 27, 2022
Processing time: 143 Days and 17.1 Hours
Liver cancer ranks the third cause of cancer-related death worldwide. The most common type of liver cancer is hepatocellular carcinoma (HCC). The survival time for HCC patients is very limited by years due to the lack of efficient treatment, failure of early diagnosis, and poor prognosis. Ubiquitination plays an essential role in the biochemical processes of a variety of cellular functions.
To investigate three ubiquitination-associated genes in HCC.
Herein, the expression levels of ubiquitin-conjugating enzymes 2 (UBE2) including UBE2C, UBE2T, and UBE2S in tumor samples of HCC patients and non-tumor controls at the Cancer Genome Atlas (TCGA) database, was comprehensively analyzed. The relationship of UBE2 gene expression level with cancer stage, prognostic outcome, and TP53 mutant status was studied.
Our results showed that UBE2C, UBE2T, and UBE2S genes were overexpressed in HCC samples compared to non-tumor tissues. Dependent on the cancer progression stage, three UBE2 genes showed higher expression in tumor tissues at all four stages compared to non-tumor control samples. Furthermore, a sign
In summary, this study shed light on the potential roles of UBE2C, UBE2T, UBE2S on diagnostic and prognostic biomarkers for HCC. Moreover, based on our findings, it is appealing to further explore the correlation of those genes with TP53 mutation in HCC and the related mechanisms.
Core Tip: Liver cancer ranks the third cause of cancer-related death worldwide. The most common type of liver cancer is hepatocellular carcinoma (HCC). Lack of effective treatment options and early diagnostic biomarkers results in a short survival time of HCC patients. Ubiquitination plays an essential role in the biochemical processes in cells. In this study, using bioinformatic analysis of the online TCGA database we found that three ubiquitin-conjugating enzyme 2 (UBE2) genes were overexpressed in HCC samples compared to normal samples in a stage-dependent manner, including UBE2C, UBE2T, and UBE2S. Additionally, overexpression of those genes was negatively associated with prognostic outcomes and overall survival times. Patients with TP53 mutation showed a higher level of expression of three UBE2 genes, indicating an association between UBE2 expression with p53 function. This study shed light on the potential roles of UBE2C, UBE2T, and UBE2S on diagnostic and prognostic biomarkers for HCC, as well as the therapeutic strategy.
- Citation: Zhang CY, Yang M. Functions of three ubiquitin-conjugating enzyme 2 genes in hepatocellular carcinoma diagnosis and prognosis. World J Hepatol 2022; 14(5): 956-971
- URL: https://www.wjgnet.com/1948-5182/full/v14/i5/956.htm
- DOI: https://dx.doi.org/10.4254/wjh.v14.i5.956
Liver hepatocellular carcinoma (HCC, or LIHC) is the most common type of primary liver cancer, which is the third most common cause of cancer-related death worldwide[1,2]. Hepatitis viral infections, abuse of alcohol, liver fibrosis, and cirrhosis are the major factors that cause liver cancer. HCC is closely associated with many metabolic diseases, such as non-alcoholic fatty liver disease (NAFLD), diabetes, obesity, cardiovascular diseases[3]. Surgical operation is a curative treatment for early-stage of liver cancer[4]. However, most HCC cases were found at the late stage due to a lack of effective diagnostic biomarkers, which are not suitable for surgical procedures[5]. Therefore, early diagnostic and exploration of novel treatment options are urgently needed. The mechanism-based investigation both on the genetic and molecular levels is necessary to further facilitate the exploration of diagnosis and treatment.
Ubiquitin is a highly conserved regulatory protein in all eukaryotic organisms, and it is covalently tagged to proteins, severing as a signal for further proteasome degradation[6]. Ubiquitination is an essential biochemical process, which contributes to a variety of cellular functions, such as cell signaling pathway regulation, cell death, protein degradation, innate and adaptive immune response. Due to the significant role of ubiquitination in cell survival and death, it is closely associated with host health and disease[7,8]. When exploring the top overexpressed genes of E2 ubiquitin-conjugating enzymes (UBE2) in HCC patient tumor samples from the Cancer Genome Atlas (TCGA) database, UBE2C, UBE2T, and UBE2S were ranked as top 4, top 8, and top 31, respectively. Those highly expressed genes draw our attention in exploring their roles in cancers, specifically for HCC. Therefore, in this study, we focused on the investigation of highly expressed UBE2C, UBE2T, UBE2S as potential biomarkers for HCC diagnosis and prognosis, as well as their expression levels at different cancer stages.
UBE2 is a large enzyme family that plays a fundamental role in the second step of ubiquitination that connects the first step of ubiquitin activation by UBE1 enzyme and together with the third step ubiquitin-protein ligation via UBE3 enzyme to conduct the complex ubiquitination process[7]. UBE2C protein, encoded by gene UBE2C, was reported to exacerbate cell apoptosis[9] and contribute to chromosome mis-segregation during the formation of tumors[10]. Overexpression of gene UBE2C has been found in tumor cells of HCC patients compared to noncancerous liver cells in 62 out of the studied 65 clinical cases[11]. Most recently, another study also showed that UBE2C can promote cancer cell growth and migration[12].
UBE2T can bind to Fanconi anemia complementation group L and meditate the monoubiquitinating of Fanconi anemia complementation group D2 (FANCD2), a critical process of regulation associated with damaged-DNA repair in the Fanconi anemia pathway[13,14]. It was also reported that UBE2T played an important role in the carcinogenesis in different cancer types[15-18], such as human breast cancer cells[17,18], lung cancer[19], gastric cancer[20], etc.
UBE2S serves as the key component on the degradation of anaphase-promoting complex/cyclosome substrate via mitosis, in which process UBE2C is also involved[21]. In breast cancer cells, UBE2S deficiency can suppress their migration, invasion, and growth via disruption of actin cytoskeleton and focal adhesion[22]. In addition, silencing UBE2S reduced cell proliferation and colony formation of lung cancer cells, resulting in cell apoptosis[23]. In HCC, UBE2S was upregulated and showed oncogenic activity by increasing p53 ubiquitination[24]. Overall, these three genes play pivotal roles in cancer cell progression and invasion, indicating the potential as biomarkers for HCC.
However, the bioinformatic-based systematic analysis of UBE2C, UBE2T, and UBE2S in liver cancers from clinical patients have not been reported. Herein, this study carried a comprehensive bioinformatic-based analysis of clinical data from the online database to illustrate the significant roles of three UBE2 genes in HCC, by analyzing their gene expression levels between non-tumor and tumor tissues, their association with cancer progression stage, and their prognostic values, and by investigating their expression in different cell types, association with TP53 mutation status, co-expression efficiency, involved signaling pathways, and associated proteins in interaction networks.
All the data for this study originated and was generated from the online open resource database and published literature.
The RNA-seq data and LIHC/HCC patient clinic information were originated and generated from TCGA. (TCGA research network: https://www.cancer.gov/tcga). The genomic alterations for UBE2C, UBE2T, UBE2S in different liver cancer types were analyzed using cBioPortal (https://www.cbioportal.org/)[25,26]. A survival heatmap was generated using GEPIA[27].
The expression level of the genes of interest on pan-cancer was analyzed using UALCAN web-based tool. The quired gene expression level in pan-cancer was analyzed using Student’s t-test to compare the normal and tumor group with P < 0.05 as significant differential expression. Heatmap was generated to display the RNA-seq data using the median value of expression Log2 (TPM+1). Analysis of gene expression level on HCC cancer progression stages, expression level on TP53 mutant status and non-TP53 mutant status was performed using online resources UALCAN[28].
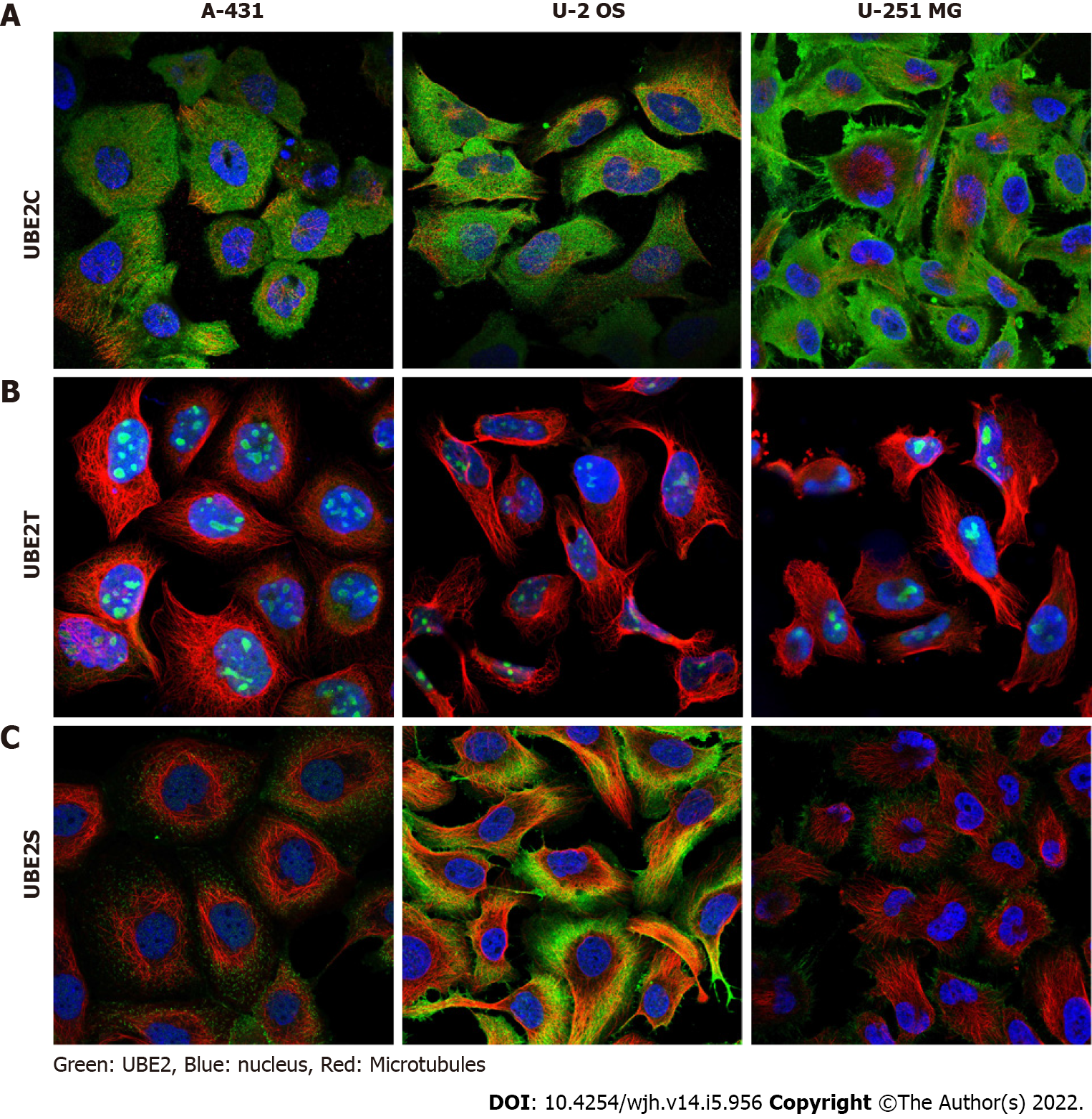
The analysis of RNA expression in different cell types and the expression location of UBE2 genes in three different cells including epidermoid carcinoma cell line A-431, human osteosarcoma cell line U-2 OS, and human glioblastoma cell line U-251 MG were explored using the Human Protein Atlas database (Human Protein Atlas, https://www.proteinatlas.org, Protein Atlas version 21.0), an online public resource for the investigation of protein-coding genes of variable cancers in cell and tissue samples[29].
In this study, the STRING online tool[30] was used to show the interaction of functional enrichment of the generated network with queried input: UBE2C, UBE2T, UBE2S, and TP53. (Default threshold for interaction specificity score > 0.4 was defined as significant). The functional property was generated and summarized based on the online information and literature.
The survival curve was generated using Kaplan Meier-plotter that was commonly used for assessment of the gene expression on survival from a large database such as TCGA samples. The significance of survival impact was measured by a log-rank test. A log-rank P < 0.05 was set for statistically significant cut-off.
Expression of genes in tumor samples and normal samples using Student’s t-test (considering unequal variance) for comparison between different groups. A P value less than 0.05 is considered statistically significant.
Overall survival heat map of the patients across multiple cancer types was generated with the input of 95% confidence interval and the calculation of the hazard ratio based on the Cox PH model. Pearson's correlation coefficient (r) was used to plot the co-expression between quired input paired genes.
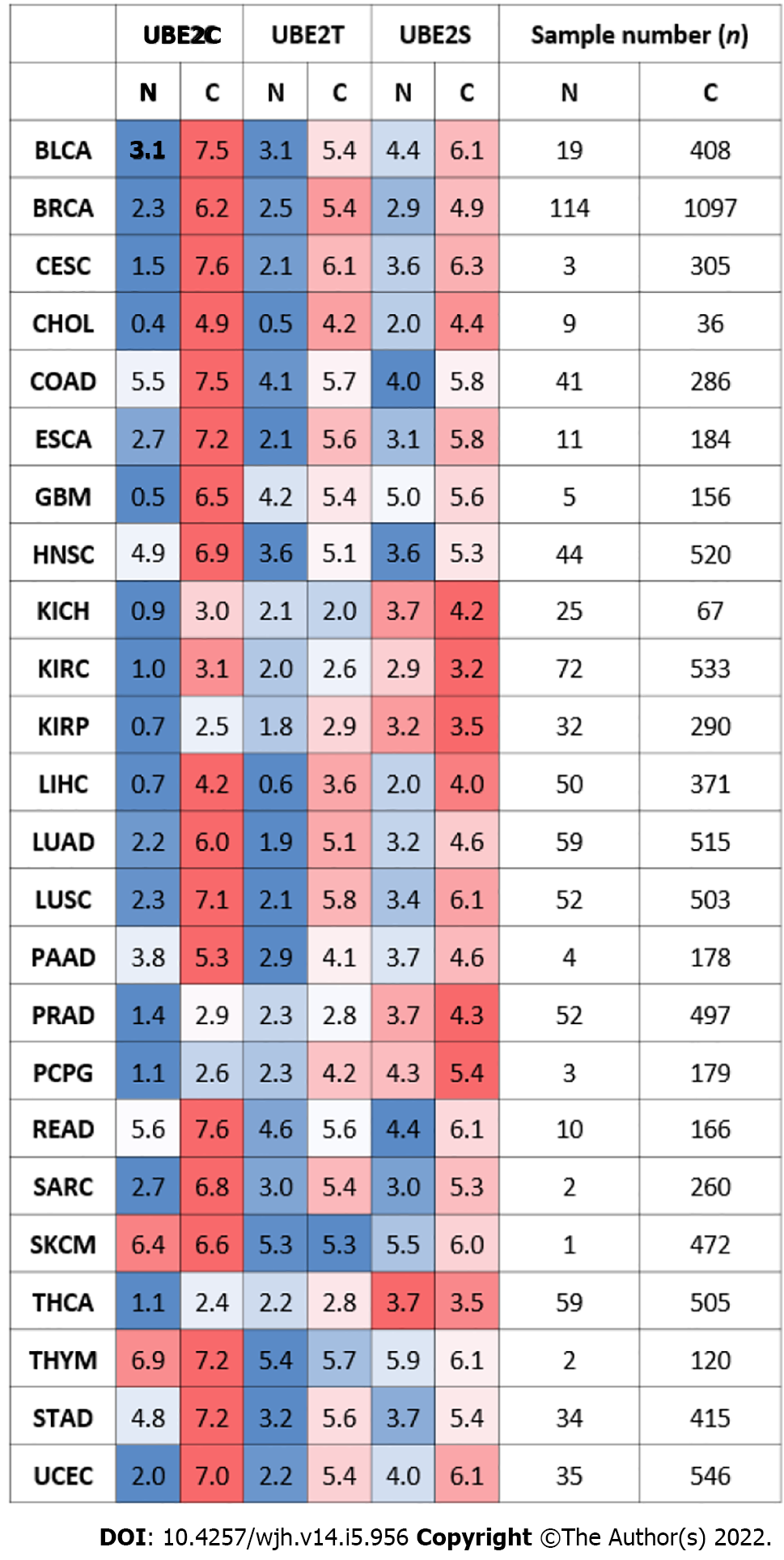
As shown in Figure 1, three UBE2 family members including UBE2C, UBE2T, and UBE2S were highly expressed in most tumor samples compared to the corresponding normal (non-tumor) samples. A formula of log2 (TPM+1) was used to calculate the UBE2 gene expression level, where TPM is transcripts per million. The case numbers for each cancer were included in the figure. The expression level among normal and tumor tissues are heterogeneous, with relatively high expression in both normal and tumors, such as colon and rectum adenocarcinoma (COAD and READ), skin cutaneous melanoma (SKCM), and thymoma (THYM). For HCC (or LIHC), UBE2C showed a high expression level in tumor samples (n = 371) compared with normal samples (n = 50), with the mean value of the log2 (TPM + 1) of 4.2 for tumor samples and 0.7 for normal samples. UBE2T also showed an increased expression level in tumor samples with the mean value of log2 (TPM + 1), compared to 0.6 for normal samples. Similarly, a higher expression level was shown for UBE2S, with the value of the log2 (TPM + 1) of 4 for tumor samples and the value of log2 (TPM + 1) of 2 for normal samples.
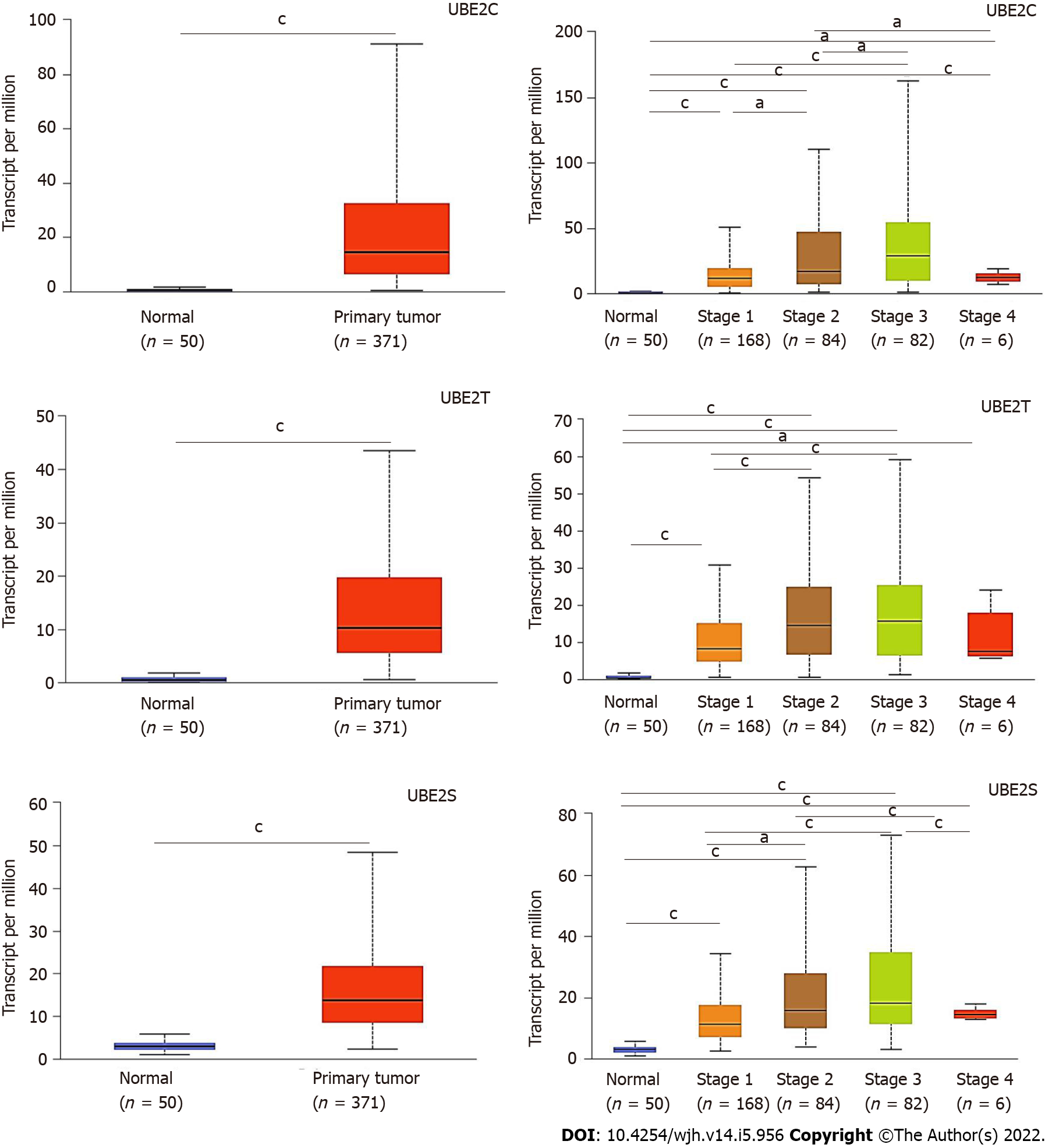
Similarly, the distribution of the expression UBE2C, UBE2T, and UBE2S indicated that they were expressed in HCC tumor samples compared to normal samples (Figure 2A-C), indicating the potential as a diagnostic biomarker for HCC. Furthermore, the expression of each gene at different tumor stages was analyzed to investigate the association of gene expression with the tumor progression stage. The results demonstrated that along with the progression of cancer, the expression patterns of genes UBE2C, UBE2T, and UBE2S kept increasing from stage 1 to stage 3 (Figure 2D-F). There was a pike level at stage 3, while at stage 4, the expression level significantly decreased for UBE2C and UBE2S compared to that from stage 2 and stage 3, even though it was increased compared to that in normal tissue (Figure 2D and F). In addition, the expression of UBE2T was also decreased at stage 4 of HCC, but without significant change compared to that in stages 2 and 3, which might be impacted by the sample size (Figure 2E). Even though, the significantly increased expression level of those genes in HCC stage 1, 2, and 3 when compared to normal samples or compared between stage 1 and stage 2&3, which suggests that it is valuable for further exploring them as potential biomarkers or key genes mediating HCC progression.
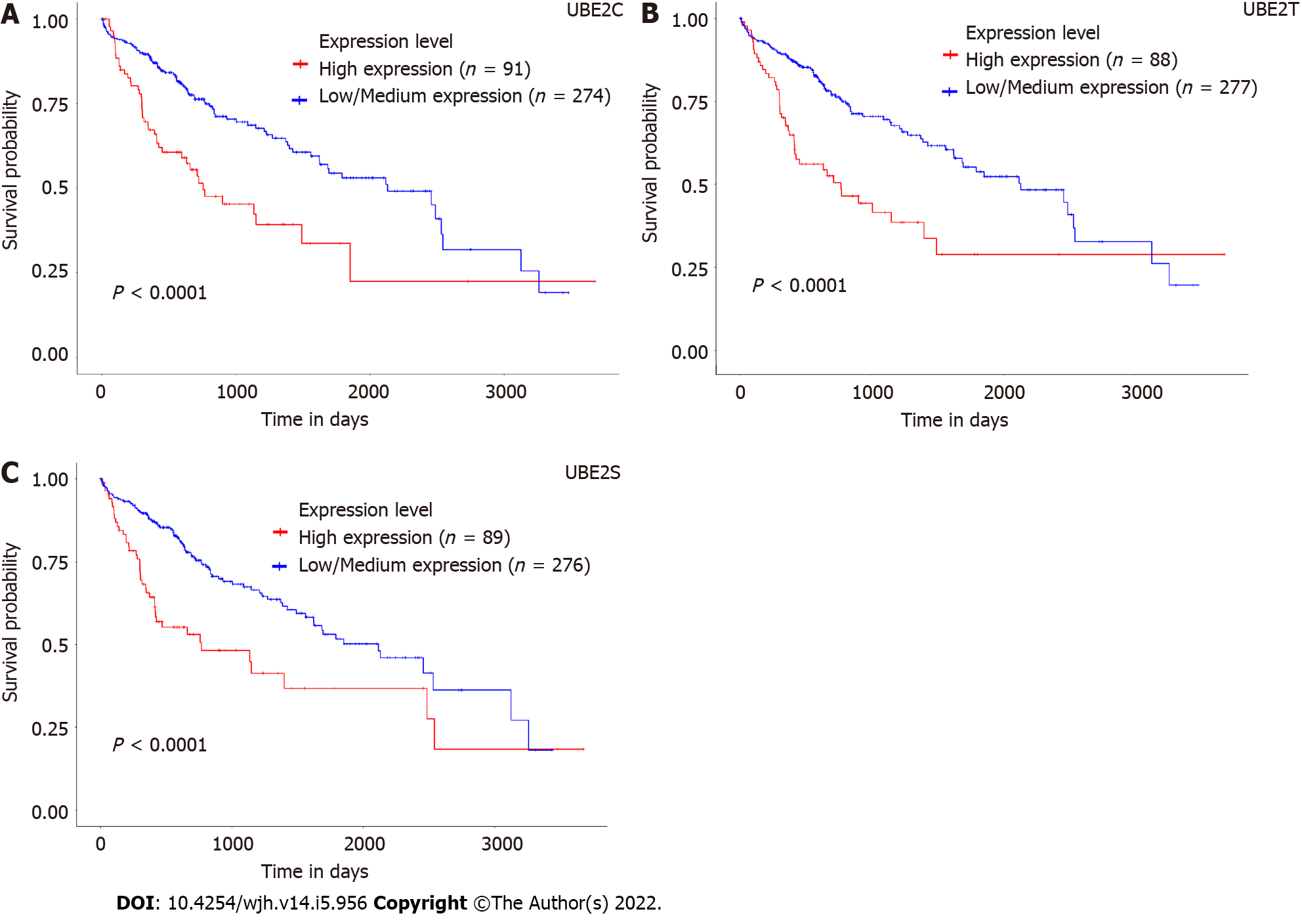
To explore the prognostic value of these gene expression levels in HCC patients, the survival outcome of patients, the expression of UBE2 genes, and the survival curve were analyzed. Remarkably, high expression levels of UBE2C, UBE2T, and UBE2S were associated with a negative prognostic outcome in HCC patients. The patients with overexpression of UBE2C (n = 91) showed significantly less survival time compared to the patients with low or medium expression levels (n = 274) (Figure 3A, P < 0.0001). For UBE2T, 277 patients with low or medium expression levels showed significantly higher survival days compared to the patients (n = 88) with high expression levels (Figure 3B, P < 0.0001). Similar results were also found for UBE2S, a significantly shorten survival time was associated with the higher UBE2S expression level (n = 89) compared with longer survival patients with low or medium expression levels of UBE2S (n = 276) (Figure 3C, P < 0.0001).
In addition, with the analysis of prognostic markers for pan-cancers, the results indicated that a higher expression level of UBE2C, UBE2T, and UBE2S was associated with the poor prognostic outcome for most of the cancers, such as adrenocortical carcinoma (ACC), kidney chromophobe (KICH), brain lower grade glioma (LGG) (Figure 3D).
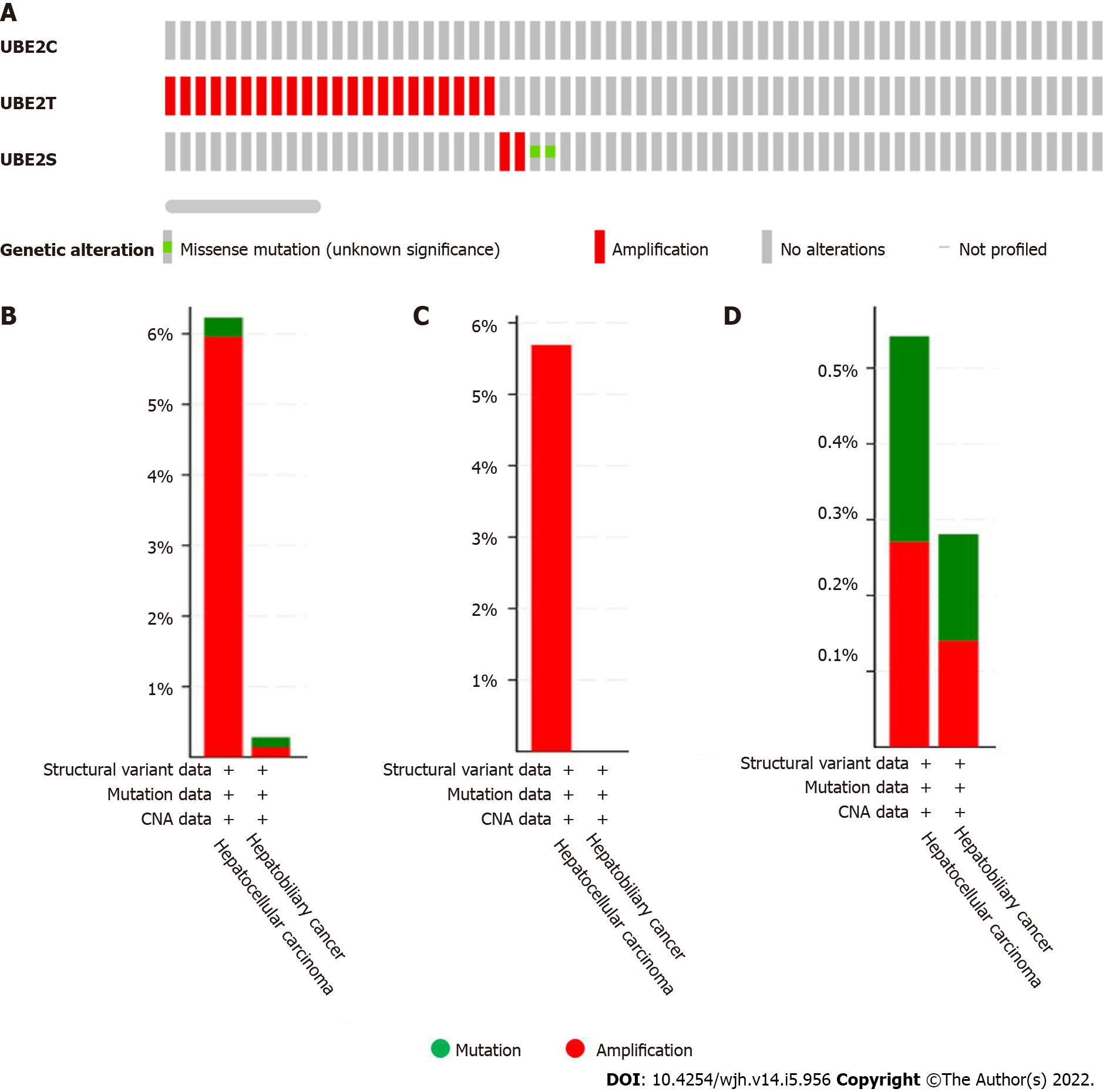
We further explored the underlying mechanisms of three UBE2 genes in HCC development and progression. A total of 1238 samples were analyzed using the cBioPortal from the TCGA pan-cancer database for genetic alterations including mutations, structural variants, and copy number alteration of three UBE2 genes. The OncoPrint results analyzed from cBioPortal showed the queried genes genomic alteration frequency from TCGA studied HCC samples is 2.1% (26 out of 1238). Among them, the genetic alteration occurred on genes UBE2T and UBE2S, and no alteration was found for UBE2C (Figure 4A). Then, the genetic alteration based on liver cancer subtypes was further analyzed. Among 1238 samples, 369 samples were HCC and 712 samples were hepatobiliary cancer. The genetic alteration was mostly found in HCC and hepatobiliary cancer. For HCC, the overall genetic alteration frequency of genes UBE2T and UBE2S was 6.23% (23 out of 369 samples) that largely resulted from amplification (5.96%, 22 cases, red color) and less from mutation (0.27%, 1 case, green color) out of 369 cases. For hepatobiliary cancer, the overall genetic alteration frequency of genes UBE2T and UBE2S was 0.28% in total 712 cases with 1 case amplification (0.14%) and 1 case mutation (0.14%) (Figure 4B). Specifically, for gene UBE2T, the only genetic alteration type is amplification which was found in 21 out of 369 cases of HCC samples, while no mutation had occurred for UBE2T (Figure 4C). For UBE2S, genetic alteration occurred in 2 cases out of 369 cases (0.54%) in HCC, including 1 case of mutation (0.27%) and 1 case of amplification (0.27%). Similarly, genetic alteration of UBE2S occurred in 2 cases out of 712 cases (0.28%) in hepatobiliary cancer, including 1 case mutation (0.14%) and 1 case amplification (0.14%) (Figure 4D). Overall, the most of genetic alteration occurred in HCC samples with 23 out of 369 samples (6.23%), which was mainly from gene UBE2T.
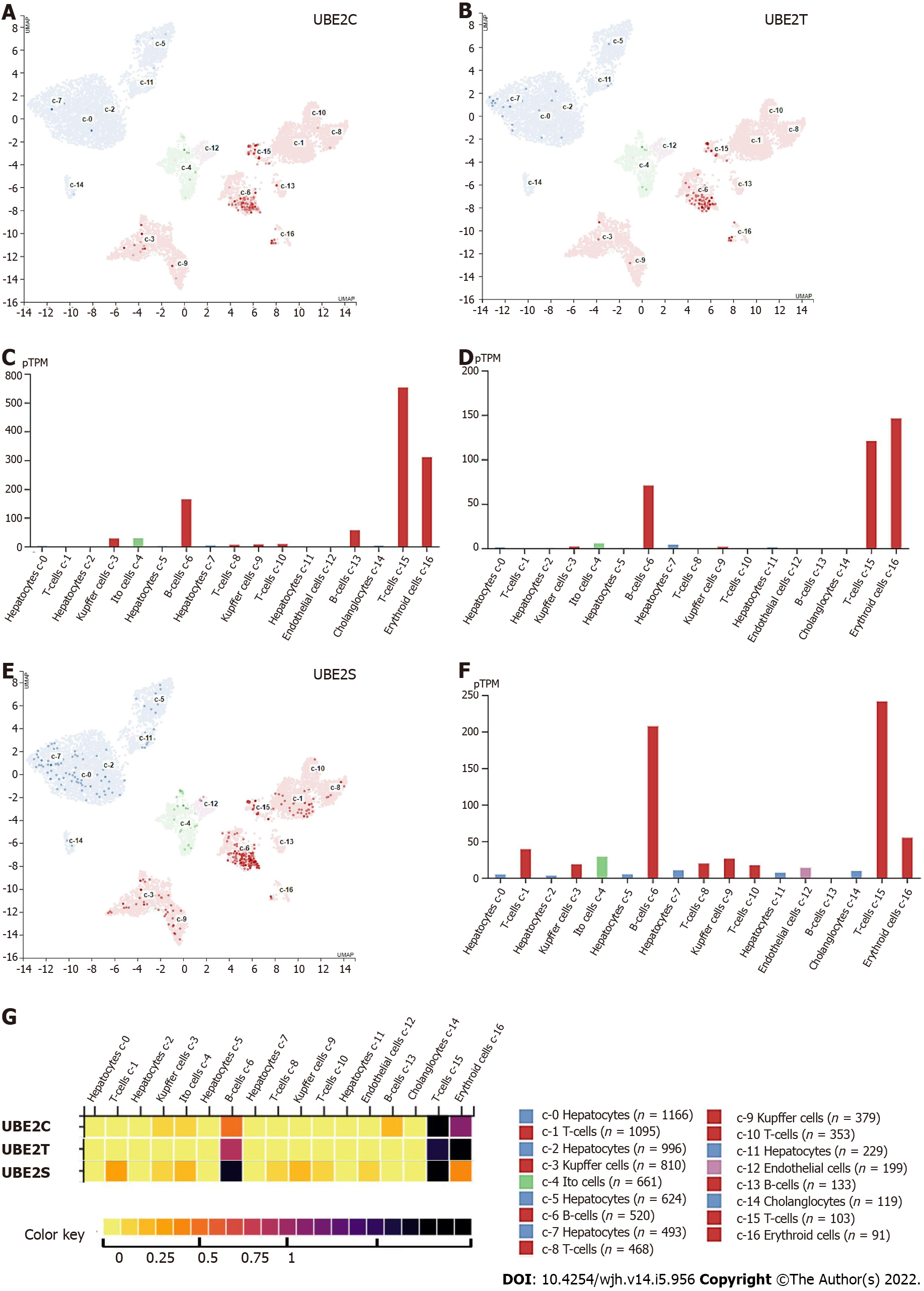
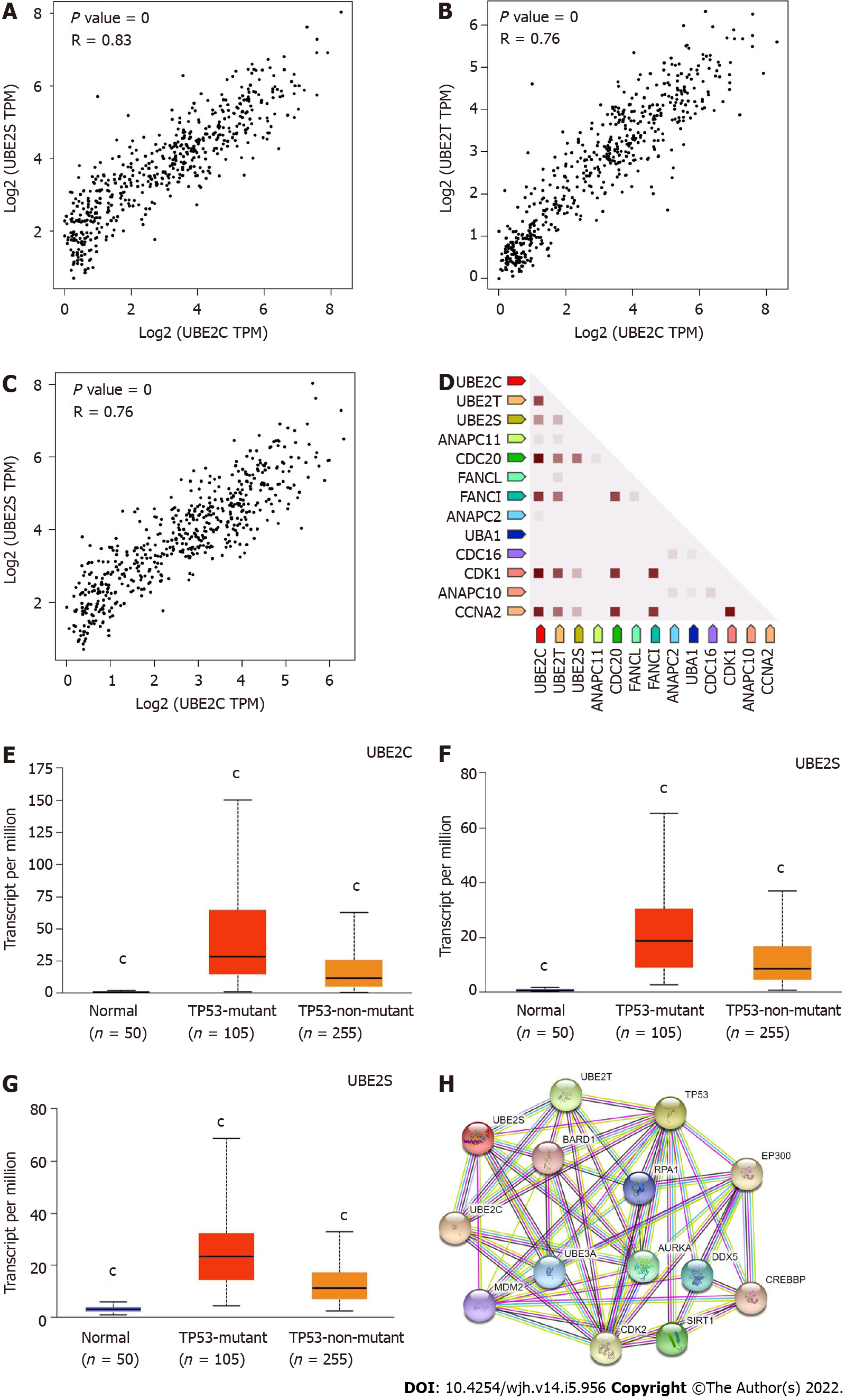
Single-cell RNA sequencing (scRNA-seq) revealed three clusters c-6 (B-cells), c-15 (T-cells), and c-16 (Erythroid cells) exhibited high mRNA expression of all three UBE2C, UBE2T, and UBE2S (Figure 5A-G). In addition, the protein expression location in three cell lines including A-431, U-2 OS, and U-251 MG (Human Protein Atlas, https://www.proteinatlas.org) indicated that UBE2C was mainly expressed in the cytosol and plasma membrane, UBE2T was mainly expressed in nucleoli or nucleoplasm, and UBE2S was highly expressed in the cytosol and plasma membrane and less expressed in nucleoli (Figure 6). Furthermore, the co-expression relationship of these genes was analyzed, since all three genes UBE2C, UBE2T, and UBE2S were expressed by the same clusters in HCC. The correlation expression of pair-wise genes was analyzed using Pearson methods (Figure 7A, 7B, 7C). Results showed there was a co-expression between two paired genes (r = 0.83 UBE2C-UBE2S; r = 0.76 UBE2C-UBE2T; r = 0.76 UBE2S-UBE2T). In addition, the analysis of co-expression of their proteins also showed that there was a correlation among three proteins (Figure 7D), indicating a co-expressing pattern.
Notably, the expression levels of all three genes UBE2C, UBE2T, and UBE2S were significantly higher in TP53 mutant samples compared with both normal samples and non-TP53 mutant samples (Figure 7E-7G). However, the correlation or causation between the higher expression level of UBE2C, UBE2T, UBE2S, and TP53 mutation is unknown. This finding of a comprehensive analysis of clinical patient samples was supported by several studies. For example, overexpression of UBE2C in patients with endometrial cancer was associated with cancer progression and recurrence, by increasing endometrial cancer cell proliferation, migration, invasion, as well as the process of epithelial-mesenchymal transition through inhibition of p53 expression[31]. UBE2T can enhance the ubiquitination of p53 in HCC cells[32,33]. Similarly, UBE2S also can promote the ubiquitination of p53 and mediate its protein degradation in HCC cells[24]. Therefore, recovering or enhancing p53 function can partially attenuate UBE2 genes-induced malignant phenotypes of tumor cells.
To further investigate the association among three genes UBE2S, UBE2T, UBE2S, and TP53 function, we generated the protein-protein interaction network using their proteins and TP53. The most closely associated proteins related to these protein interactions were analyzed using STRING, shown in Figure 7H. The functional annotation and associated signaling pathways of the queried genes were summarized in Table 1.
| UBE2 genes | Biological process | ||
| UBE2T | UBE2S | Protein K27-linked ubiquitination | |
| UBE2T | UBE2S | Protein K29-linked ubiquitination | |
| UBE2C | UBE2S | Free ubiquitin chain polymerization | |
| UBE2T | UBE2S | Protein K6-linked ubiquitination | |
| UBE2C | UBE2S | Exit from mitosis | |
| UBE2C | UBE2S | Positive regulation of ubiquitin protein ligase activity | |
| UBE2C | UBE2T | UBE2S | Protein K11-linked ubiquitination |
| UBE2C | UBE2T | Protein K48-linked ubiquitination | |
| UBE2T | UBE2S | Protein K63-linked ubiquitination | |
| UBE2C | UBE2S | Anaphase-promoting complex-dependent catabolic proceed | |
| UBE2 | Cellular component and uniport annotated keywords | ||
| UBE2C | UBE2S | Anaphase-promoting complex | |
| UBE2C | UBE2T | UBE2S | Nucleoplasm |
| UBE2C | UBE2T | UBE2S | ATP-binding |
| UBE2C | UBE2S | Cell cycle | |
| UBE2C | UBE2T | UBE2S | Transferase |
| UBE2C | UBE2T | UBE2S | Ubl conjugation |
| UBE2 | Reactome pathways | ||
| UBE2C | UBE2S | Aberrant regulation of mitotic exit in cancer due to RB1 defects | |
| UBE2C | UBE2S | APC-Cdc20 mediated degradation of Nek2A | |
| UBE2C | UBE2S | APC/C:Cdc20 mediated degradation of Cyclin B | |
| UBE2C | UBE2S | APC/C: Cdh1 mediated degradation of Cdc20 and other APC/C | |
| UBE2C | UBE2S | Cell cycle checkpoints | |
| UBE2C | UBE2S | Cell cycle, mitotic | |
| UBE2C | UBE2S | Cellular responses to stress | |
| UBE2C | UBE2S | Cellular Senescence | |
| UBE2C | UBE2S | Conversion from APC/C: Cdc20 to APC/C: Cdh1 in late anaphase | |
| UBE2C | UBE2S | Generic transcription pathway | |
| UBE2C | UBE2S | Inactivation of APC/C via direct inhibition of the APC/C compel | |
| UBE2C | UBE2S | Phosphorylation of the APC/C | |
| UBE2C | UBE2T | UBE2S | Post-translational protein modification |
| UBE2C | UBE2T | UBE2S | Synthesis of active ubiquitin: roles of E1 and E2 enzymes |
| UBE2C | UBE2S | Synthesis of DNA | |
| UBE2C | UBE2S | Transcriptional regulation by VENTX | |
The early diagnostic of HCC or LIHC is critically important for cancer treatment and selection of therapeutic methods[34-36]. In addition, a better understanding of the development and progression of the cancer stage is helpful to choose the therapy that can result in a good outcome. In this study, with clinical data analysis, we found that the expression of UBE2C, UBE2T, and UBE2S was increased in tumor tissue compared to normal tissue, and their expression was associated with the procession of cancer stage from stage 1 to stage 3, although there was a decrease at last stage (stage 4). These results suggest there is a potential to use those genes as biomarkers to assist the diagnosis of HCC at the early-stage point. What’s more, the results also showed that there was a significantly increased expression level of those genes at stage 2 and stage 3 compared with stage 1 during HCC progression. This may shed light on the potential usage of those genes as biomarkers to better predict the cancer progression stage.
UBE2 family members play a role in the development and prognosis of cancers[37,38], such as ovarian cancer. It has been shown that the mRNA expression of UBE2A in liver cancer cell lines (e.g., HepG2 and Huh-7) was significantly higher compared to that in normal liver cancer line HL-7702. Meanwhile, UBE2A mRNA and protein were highly expressed in HCC tumor tissues than those in the adjacent normal tissues[39]. In HCC, qPCR data showed that the expression of UBE2S was significantly increased in HCC samples compared to non-tumor liver tissues[40]. Another study showed that the expression of UBE2T mRNA and protein was significantly increased in HCC tissues compared to adjacent non-tumor tissues. A molecular mechanism study showed that UBE2T can suppress the G2/M transition of hepatoma cells by regulating cyclin B1 and cyclin-dependent kinase 1 expression[41]. A recent study showed that the expression of UBE2T can be regulated by microRNA miR-212-5p, and overexpression of UBE2T can promote HCC cell proliferation and migration[42]. In this study, we have demonstrated that overexpression of UBE2C, UBE2T, and UBE2S was associated with poor prognosis and shorter survival time. These results indicate the gene expression levels of three genes might be useful to assist to predict the outcome of HCC. Remarkably, our analysis revealed that HCC tumors with TP53 mutant status exhibited significantly higher expression levels of those genes compared with TP53 non-mutant status in tumor samples. This finding shows the potential correlation between the overexpression of investigated genes and TP53 mutation status, as well as their contribution to HCC progression. A further mechanistic study needs to be investigated in the field.
From the therapeutic and treatment standpoint, considering the significant roles of UBE2C[43,44], UBE2T[33,45], and UBE2S[46,47] in the ubiquitination process, which contributes to the cellular function and their close association with tumor cell’s function, UBE2C, UBE2T, and UBE2S could be used as a diagnostic biomarker to assist the diagnosis and prediction of the progression of HCC as mentioned above. Most importantly, the causation of the higher level of UBE2 expression, as well as the contributing effect of those highly expressed UBE2 genes on the disease outcome should be thoroughly investigated for further exploration of the effective therapeutic strategy discovery.
Further studies from following aspects, such as (1) Identification of causing factors of UBE2 overexpression; (2) investigation of the underlying mechanism on overexpression of UBE2 genes causing disease severity and poor survival outcome of patients; (3) exploration of the associated therapeutic targets of UBE2; (4) the roles of co-expressed genes from the analysis of protein-protein network in HCC; and (5) the relationship of p53 mutation with UBE2 expression; will be studied in the future research to better understand the role of three UBE2 genes in liver cancer.
This bioinformatics study sheds light on the important roles of UBE2C, UBE2T, UBE2S for HCC diagnostic and prognostic as potential biomarkers. In addition, it is appealing to further explore the correlation of those genes with TP53 mutation in HCC and the related mechanisms.
The expression of three ubiquitin-conjugating enzymes 2 (UBE2) including UBE2C, UBE2T, and UBE2S was significantly increased in HCC samples compared to non-tumor tissues.
To explore potential diagnostic and prognostic markers for HCC.
To identify the potential of UBE2C, UBE2T, and UBE2S as potential biomarkers as HCC.
Online database was analyzed with different bioinformatic tools.
Our data showed that UBE2C, UBE2T, and UBE2S genes were overexpressed in hepatocellular carcinoma (HCC) samples compared to non-tumor tissues. Dependent on the cancer progression stage, three UBE2 genes showed higher expression in tumor tissues at all four stages compared to non-tumor control samples. Furthermore, a significantly higher expression of these genes was found in stage 2 and stage 3 cancers compared to stage 1 cancer. Additionally, overexpression of those genes was negatively associated with prognostic outcome and overall survival time. Patients with TP53 mutation showed a higher expression level of three UBE2 genes, indicating an association between UBE2 expression with p53 function.
This bioinformatics study sheds light on the important roles of UBE2C, UBE2T, UBE2S for HCC diagnostic and prognostic as potential biomarkers. In addition, it is appealing to further explore the correlation of those genes with TP53 mutation in HCC and the related mechanisms.
Further studies from following aspects, such as (1) Identification of causing factors of UBE2 overexpression; (2) investigation of the underlying mechanism on overexpression of UBE2 genes causing disease severity and poor survival outcome of patients; (3) exploration of the associated therapeutic targets of UBE2; (4) the roles of co-expressed genes from the analysis of protein-protein network in HCC; and (5) the relationship of p53 mutation with UBE2 expression; will be studied in the future research to better understand the role of three UBE2 genes in liver cancer.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Ke X, China; Ling Q, China S-Editor: Liu JH L-Editor: A P-Editor: Liu JH
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64456] [Article Influence: 16114.0] [Reference Citation Analysis (176)] |
| 2. | Zhang C, Yang M. The Emerging Factors and Treatment Options for NAFLD-Related Hepatocellular Carcinoma. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 3. | Zhang C, Liu S, Yang M. Hepatocellular Carcinoma and Obesity, Type 2 Diabetes Mellitus, Cardiovascular Disease: Causing Factors, Molecular Links, and Treatment Options. Front Endocrinol (Lausanne). 2021;12:808526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 4. | Zhang C, Yang M, Ericsson AC. The Potential Gut Microbiota-Mediated Treatment Options for Liver Cancer. Front Oncol. 2020;10:524205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 5. | Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4432] [Cited by in RCA: 3852] [Article Influence: 963.0] [Reference Citation Analysis (3)] |
| 6. | Shi D, Grossman SR. Ubiquitin becomes ubiquitous in cancer: emerging roles of ubiquitin ligases and deubiquitinases in tumorigenesis and as therapeutic targets. Cancer Biol Ther. 2010;10:737-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 93] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 7. | Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20:1242-1253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 982] [Article Influence: 89.3] [Reference Citation Analysis (0)] |
| 8. | Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 924] [Cited by in RCA: 1008] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 9. | Jiang L, Bao Y, Luo C, Hu G, Huang C, Ding X, Sun K, Lu Y. Knockdown of ubiquitin-conjugating enzyme E2C/UbcH10 expression by RNA interference inhibits glioma cell proliferation and enhances cell apoptosis in vitro. J Cancer Res Clin Oncol. 2010;136:211-217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 10. | van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188:83-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 178] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 11. | Ieta K, Ojima E, Tanaka F, Nakamura Y, Haraguchi N, Mimori K, Inoue H, Kuwano H, Mori M. Identification of overexpressed genes in hepatocellular carcinoma, with special reference to ubiquitin-conjugating enzyme E2C gene expression. Int J Cancer. 2007;121:33-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 83] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 12. | Xiong Y, Lu J, Fang Q, Lu Y, Xie C, Wu H, Yin Z. UBE2C functions as a potential oncogene by enhancing cell proliferation, migration, invasion, and drug resistance in hepatocellular carcinoma cells. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 13. | Schultz-Rogers L, Lach FP, Rickman KA, Ferrer A, Mangaonkar AA, Schwab TL, Schmitz CT, Clark KJ, Dsouza NR, Zimmermann MT, Litzow M, Jacobi N, Klee EW, Smogorzewska A, Patnaik MM. A homozygous missense variant in UBE2T is associated with a mild Fanconi anemia phenotype. Haematologica. 2021;106:1188-1192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 14. | Hira A, Yoshida K, Sato K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Shimamoto A, Tahara H, Ito E, Kojima S, Kurumizaka H, Ogawa S, Takata M, Yabe H, Yabe M. Mutations in the gene encoding the E2 conjugating enzyme UBE2T cause Fanconi anemia. Am J Hum Genet. 2015;96:1001-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 90] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 15. | Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 216] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 16. | Alpi A, Langevin F, Mosedale G, Machida YJ, Dutta A, Patel KJ. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol. 2007;27:8421-8430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 73] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 17. | Ueki T, Park JH, Nishidate T, Kijima K, Hirata K, Nakamura Y, Katagiri T. Ubiquitination and downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T overexpression in human breast cancer cells. Cancer Res. 2009;69:8752-8760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 18. | Liu J, Liu X. UBE2T silencing inhibited non-small cell lung cancer cell proliferation and invasion by suppressing the wnt/β-catenin signaling pathway. Int J Clin Exp Pathol. 2017;10:9482-9488. [PubMed] |
| 19. | Perez-Peña J, Corrales-Sánchez V, Amir E, Pandiella A, Ocana A. Ubiquitin-conjugating enzyme E2T (UBE2T) and denticleless protein homolog (DTL) are linked to poor outcome in breast and lung cancers. Sci Rep. 2017;7:17530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 20. | Luo C, Yao Y, Yu Z, Zhou H, Guo L, Zhang J, Cao H, Zhang G, Li Y, Jiao Z. UBE2T knockdown inhibits gastric cancer progression. Oncotarget. 2017;8:32639-32654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 21. | Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363-1369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 217] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 22. | Ayesha AK, Hyodo T, Asano E, Sato N, Mansour MA, Ito S, Hamaguchi M, Senga T. UBE2S is associated with malignant characteristics of breast cancer cells. Tumour Biol. 2016;37:763-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 23. | Liu Z, Xu L. UBE2S promotes the proliferation and survival of human lung adenocarcinoma cells. BMB Rep. 2018;51:642-647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 24. | Pan YH, Yang M, Liu LP, Wu DC, Li MY, Su SG. UBE2S enhances the ubiquitination of p53 and exerts oncogenic activities in hepatocellular carcinoma. Biochem Biophys Res Commun. 2018;503:895-902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 25. | Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9144] [Cited by in RCA: 12752] [Article Influence: 980.9] [Reference Citation Analysis (0)] |
| 26. | Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8187] [Cited by in RCA: 11222] [Article Influence: 935.2] [Reference Citation Analysis (0)] |
| 27. | Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1991] [Cited by in RCA: 3057] [Article Influence: 509.5] [Reference Citation Analysis (0)] |
| 28. | Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2365] [Cited by in RCA: 4209] [Article Influence: 526.1] [Reference Citation Analysis (0)] |
| 29. | Karlsson M, Zhang C, Méar L, Zhong W, Digre A, Katona B, Sjöstedt E, Butler L, Odeberg J, Dusart P, Edfors F, Oksvold P, von Feilitzen K, Zwahlen M, Arif M, Altay O, Li X, Ozcan M, Mardinoglu A, Fagerberg L, Mulder J, Luo Y, Ponten F, Uhlén M, Lindskog C. A single-cell type transcriptomics map of human tissues. Sci Adv. 2021;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 307] [Cited by in RCA: 975] [Article Influence: 243.8] [Reference Citation Analysis (0)] |
| 30. | Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10161] [Cited by in RCA: 11712] [Article Influence: 1952.0] [Reference Citation Analysis (1)] |
| 31. | Liu Y, Zhao R, Chi S, Zhang W, Xiao C, Zhou X, Zhao Y, Wang H. UBE2C Is Upregulated by Estrogen and Promotes Epithelial-Mesenchymal Transition via p53 in Endometrial Cancer. Mol Cancer Res. 2020;18:204-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 32. | Liu LP, Yang M, Peng QZ, Li MY, Zhang YS, Guo YH, Chen Y, Bao SY. UBE2T promotes hepatocellular carcinoma cell growth via ubiquitination of p53. Biochem Biophys Res Commun. 2017;493:20-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 33. | Wu M, Li X, Huang W, Chen Y, Wang B, Liu X. Ubiquitin-conjugating enzyme E2T(UBE2T) promotes colorectal cancer progression by facilitating ubiquitination and degradation of p53. Clin Res Hepatol Gastroenterol. 2021;45:101493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 34. | Wang W, Wei C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 2020;7:308-319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 292] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 35. | Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301-1314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2800] [Cited by in RCA: 4092] [Article Influence: 584.6] [Reference Citation Analysis (6)] |
| 36. | Zhang C, Yang M. Targeting T Cell Subtypes for NAFLD and NAFLD-Related HCC Treatment: An Opinion. Front Med (Lausanne). 2021;8:789859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 37. | Zou R, Xu H, Li F, Wang S, Zhu L. Increased Expression of UBE2T Predicting Poor Survival of Epithelial Ovarian Cancer: Based on Comprehensive Analysis of UBE2s, Clinical Samples, and the GEO Database. DNA Cell Biol. 2021;40:36-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Hu W, Xiao L, Cao C, Hua S, Wu D. UBE2T promotes nasopharyngeal carcinoma cell proliferation, invasion, and metastasis by activating the AKT/GSK3β/β-catenin pathway. Oncotarget. 2016;7:15161-15172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 39. | Shen JD, Fu SZ, Ju LL, Wang YF, Dai F, Liu ZX, Ji HZ, Shao JG, Bian ZL. High expression of ubiquitin-conjugating enzyme E2A predicts poor prognosis in hepatocellular carcinoma. Oncol Lett. 2018;15:7362-7368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 40. | Ma Y, Li K, Li S, Liang B, Liu Q, Mo Z. Prognostic value of ubiquitin-conjugating enzyme E2 S overexpression in hepatocellular carcinoma. Int J Biol Macromol. 2018;119:225-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 41. | Liu LL, Zhu JM, Yu XN, Zhu HR, Shi X, Bilegsaikhan E, Guo HY, Wu J, Shen XZ. UBE2T promotes proliferation via G2/M checkpoint in hepatocellular carcinoma. Cancer Manag Res. 2019;11:8359-8370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 42. | Ren X, Li A, Ying E, Fang J, Li M, Yu J. Upregulation of ubiquitin-conjugating enzyme E2T (UBE2T) predicts poor prognosis and promotes hepatocellular carcinoma progression. Bioengineered. 2021;12:1530-1542. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 43. | Pan B, Yin S, Peng F, Liu C, Liang H, Su J, Hsiao WLW, Cai Y, Luo D, Xia C. Vorinostat targets UBE2C to reverse epithelial-mesenchymal transition and control cervical cancer growth through the ubiquitination pathway. Eur J Pharmacol. 2021;908:174399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 44. | Li R, Pang XF, Huang ZG, Yang LH, Peng ZG, Ma J, He RQ. Overexpression of UBE2C in esophageal squamous cell carcinoma tissues and molecular analysis. BMC Cancer. 2021;21:996. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 45. | Yu Z, Jiang X, Qin L, Deng H, Wang J, Ren W, Li H, Zhao L, Liu H, Yan H, Shi W, Wang Q, Luo C, Long B, Zhou H, Sun H, Jiao Z. A novel UBE2T inhibitor suppresses Wnt/β-catenin signaling hyperactivation and gastric cancer progression by blocking RACK1 ubiquitination. Oncogene. 2021;40:1027-1042. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 76] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 46. | Zhang RY, Liu ZK, Wei D, Yong YL, Lin P, Li H, Liu M, Zheng NS, Liu K, Hu CX, Yang XZ, Chen ZN, Bian H. UBE2S interacting with TRIM28 in the nucleus accelerates cell cycle by ubiquitination of p27 to promote hepatocellular carcinoma development. Signal Transduct Target Ther. 2021;6:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 47. | Tang H, Fang T, Ji M, Wang JP, Song LL, Zhang QY, Wu JS. UBE2S exerts oncogenic activities in urinary bladder cancer by ubiquitinating TSC1. Biochem Biophys Res Commun. 2021;578:7-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |