Published online May 27, 2022. doi: 10.4254/wjh.v14.i5.866
Peer-review started: November 30, 2021
First decision: December 26, 2021
Revised: January 31, 2022
Accepted: April 25, 2022
Article in press: April 25, 2022
Published online: May 27, 2022
Processing time: 174 Days and 23.4 Hours
Chronic infection with hepatitis B virus (HBV) remains a major global health problem, especially in developing countries. It may lead to prolonged liver damage, fibrosis, cirrhosis, and hepatocellular carcinoma. Persistent chronic HBV infection is related to host immune response and the stability of the covalently closed circular DNA (cccDNA) in human hepatocytes. In addition to being essential for viral transcription and replication, cccDNA is also suspected to play a role in persistent HBV infections or hepatitis relapses since cccDNA is very stable in non-dividing human hepatocytes. Understanding the pathogenicity and oncogenicity of HBV components would be essential in the development of new diagnostic tools and treatment strategies. This review summarizes the role and molecular mechanisms of HBV cccDNA in hepatocyte transformation and hepatocarcinogenesis and current efforts to its detection and targeting.
Core Tip: The covalently closed circular DNA (cccDNA) of hepatitis B virus (HBV) in human hepatocytes serves as the template for viral replication machinery. HBV cccDNA is also related to host immune response and persistent HBV infection leading to the development of hepatocellular carcinoma. This review summarizes current knowledge on cccDNA in hepatocarcinogenesis and comprehensive efforts to its detection and targeting.
- Citation: Bianca C, Sidhartha E, Tiribelli C, El-Khobar KE, Sukowati CHC. Role of hepatitis B virus in development of hepatocellular carcinoma: Focus on covalently closed circular DNA . World J Hepatol 2022; 14(5): 866-884
- URL: https://www.wjgnet.com/1948-5182/full/v14/i5/866.htm
- DOI: https://dx.doi.org/10.4254/wjh.v14.i5.866
Hepatitis B virus (HBV) is a DNA virus belonging to the Hepadnaviridae family. In humans, HBV may cause both acute and chronic infections in the liver that can lead to an increased risk of hepatocellular carcinoma (HCC) following persistent chronic infection[1].
The genome of HBV is a relaxed-circular DNA (rcDNA) that is 3.2 kbp in length. The small genome size of HBV causes the genome to be extremely compact, encoding four open reading frames (ORFs) that are overlapping: C, P, S, and X. These ORFs produced functional viral proteins: HBc and HBe antigens (HBcAg and HBeAg) and precore protein from C, polymerase (Pol) from P, surface antigens L-HBs, M-HBs, and S-HBs from S, and HBV X protein (HBx) from X[2].
HBV genome also contains four unidirectional promoters, core, SPI, SPII, and X, that are responsible for the initiation of transcription at different positions. Upon entry into cells, the rcDNA is converted into covalently closed circular DNA (cccDNA), which will be converted into different lengths of RNA (3.5 kb, 2.4 kb, 2.1 kb, and 0.7 kb) depending on transcription initiation from the different promoters.
The 3.5 kb RNA is the preC RNA that encodes for HBe and Pol. Another 3.5 kb RNA is the pregenomic RNA (pgRNA) that encodes for HBc protein and Pol. pgRNA serves as a template for both translation and reverse transcription[1]. Pol contains four different domains: A terminal protein (TP) domain that acts as primer to initiate minus strand DNA synthesis and for binding to pgRNA; a spacer domain; a reverse transcriptase (RT) domain that is necessary for reverse transcription and DNA-dependent DNA polymerization; and a ribonuclease H (RNase H) domain that is responsible for the digestion of pgRNA following reverse transcription[1,2]. The 2.4 kb preS1 RNA encodes for L-HBs protein, while the 2.1 kb RNA preS2 or S RNA encodes for the overlapping M-HBs and S-HBs. These HBs proteins form the HBV surface antigens that surround the viral nucleocapsid, and promote receptor binding during viral entry into target cells. Meanwhile, the 0.7 kb X RNA encodes for HBx protein. HBx protein promotes the production of new viral particles via the promotion of viral transcription and replication, and plays a role in the development of HBV-related HCC[1,2]. In addition, HBx protein has also been reported to have a role in cccDNA formation[3,4].
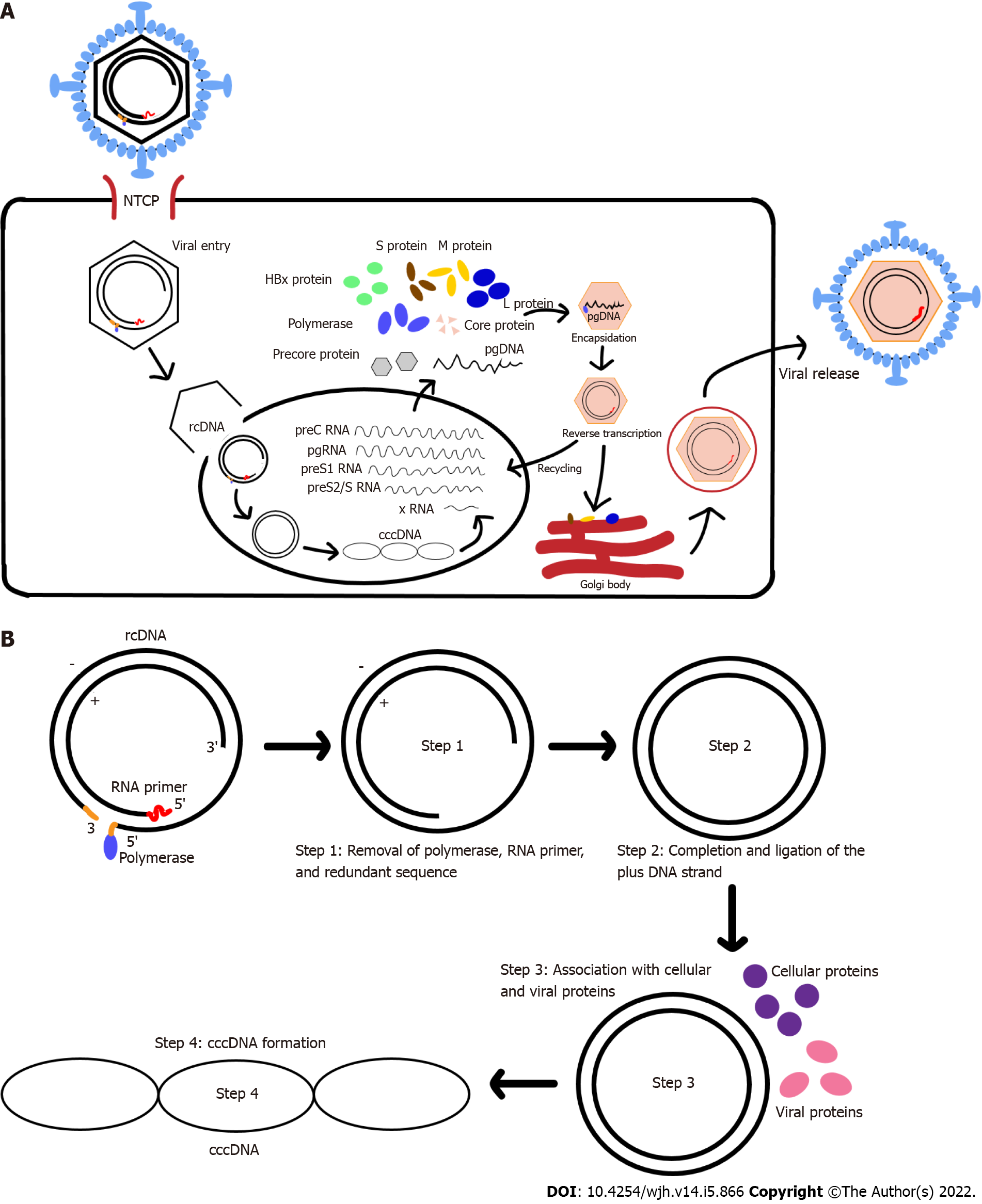
The HBV life cycle starts with its entry to hepatocytes (Figure 1A). HBV attaches to the host cell surface via binding to heparan sulfate proteoglycans (HSPGs) in a non-specific and low-affinity manner. This process is then followed by more specific and high-affinity interaction between the virus surface proteins and their respective receptors on the cell surface[5]. Multiple studies[2,6-10] have shown that sodium taurocholate co-transporting peptide (NTCP/SLC10A1) specifically expressed in the liver is the main receptor for HBV entry.
Recent studies[11,12] have reported that HBV internalization into hepatocytes, via endocytosis, is triggered by the direct interaction between epidermal growth factor receptor, a tyrosine kinase receptor, and NTCP. Upon endocytosis, the viral envelope and the host cell-derived vesicular membrane fuse to release the nucleocapsid to the cytoplasm. This process might be facilitated by the N-terminus of HBV preS1 domain which could contain the fusogenic sequence[13]. Meanwhile, another study had suggested an alternative process that HBV nucleocapsid delivery into the cytoplasm is based on membrane translocation instead of membrane fusion[14].
The nucleocapsid in the cytoplasm is translocated into the nucleus by intracellular trafficking, mainly via microtubules and importin[15-17]. Successful HBV infection is achieved when the HBV genome is delivered into the host cell nucleus. Once in the nucleus, the HBV rcDNA is converted into cccDNA with the help of Pol and other protein factors[18-21].
The cccDNA is generated (Figure 1B) by removing the Pol-linked terminal sequence at the 5’-end of the minus-DNA strand and the RNA oligonucleotide attached to the 5’-end of the plus-DNA strand. The gaps in both the minus- and plus-DNA strands are filled and ligated to produce the cccDNA[4]. The resulting cccDNA acts as the template for the transcription of the four viral mRNAs (Figure 1A), which are regulated by four different promoters.
HBV replication is performed through reverse transcription of pgRNA, an RNA intermediate generated from cccDNA. The pgRNA contains the ε signal and poly-A tail, which serves as template for the synthesis of minus-DNA strand by reverse transcription and translation of viral polymerase, core protein, and precursor of early antigen[1]. The encapsidation process, in which the pgRNA, Pol, and core protein are assembled to form a nucleocapsid, marks the start of HBV replication[22]. This process is mediated by RNA-binding motif protein 24 (RBM24) via the interaction of RBM24 with Pol and ε signal[23]. Furthermore, several host factors, such as eukaryotic translation initiation factor 4E (eIF4E), DEAD-box RNA helicase DDX3, and APOBEC3G, have been reported to also be incorporated into the viral nucleocapsid[24-26].
The polymerase enzyme interacts with the ε signal of pgRNA and forms the nucleocapsid in association with the core protein. HBV nucleocapsid formation begins when pgRNA, Pol, and HBcAg dimers are formed. The reverse transcription begins with packing of pgRNA-polymerase complexes into the lumen of assembling capsids. Initiation of the minus-DNA strand synthesis is done through the binding of polymerase that is covalently linked to a short DNA oligonucleotide to ε signal. Another initiation factor is a protein-priming mechanism. RNase H domain will also simultaneously degrade the pgRNA template during the minus-DNA strand synthesis, eventually resulting in a short RNA fragment that contains a capped 5’ terminal region. This capped 5’ terminal region RNA fragment will be translocated to the 3’ terminus and extended to the 5’ end of the minus-DNA strand[2,3]. Plus-DNA strand synthesis leads to the formation of rcDNA. After the DNA genome is synthesized, the nucleocapsid will interact with envelope protein in the endoplasmic reticulum (ER) to form new mature virions that will be released[1,22]. Alternatively, the nucleocapsid can also re-deliver their rcDNA to repeat the viral replication, which may cause the build-up of cccDNA within the nucleus[27].
Chronic HBV infection is a growing public health issue with around 300 million people infected worldwide[28]. Depending on the age and route of infection, around 25% of individuals with chronic HBV infection could develop HBV-related HCC[29-31]. Indeed, chronic HBV infection is one of the leading risk factors for HCC development in most parts of the globe[28].
An important intermediate factor for HCC development from chronic HBV infection is the development of HBV-associated cirrhosis[32]. Studies have observed a strong relationship between cirrhosis from chronic HBV infection and the development of HCC, in which around 70%-90% of all HCCs developed from decompensated cirrhosis[29,33]. The repeated hepatitis flares or continuous recruitment of inflammatory cells and cytotoxic T cells (CTLs) to the liver may eventually lead to fibrosis and cirrhosis, and increase the risk of developing HCC[34-36]. It is a general understanding that HCC development from chronic HBV infection is the result of multifactorial mechanisms[28]. However, most studies had identified three major contributing factors for HBV-related hepatocarcinogenesis: (1) Chronic inflammation with continuous cycles of destruction and regeneration of hepatocytes; (2) cccDNA persistence and HBV DNA integration into the host genome; and (3) expression of oncogenic viral proteins and/or consequence of HBV-mediated alterations of various cellular pathways[30,31,37-40].
cccDNA is generated from rcDNA as a plasmid-like episome that is retained in the nucleus of host cells. cccDNA forms a minichromosome with around 3 to 50 copies per infected cell, which decrease as the host cell divides[41]. However, cccDNA distribution among daughter cells is presumed to be unequal during cell division, which allows the cccDNA to form distinct pools that differ in their degradation susceptibility[42,43]. Therefore, the number of cccDNA copies during cell division can be maintained from the newly synthesized rcDNA-containing nucleocapsids that are imported into the nucleus. The intracellular amplification of cccDNA occurs during intracellular recycling and plays a major role in the early phases of HBV infection[41,44].
Fundamentally, cccDNA acts as a template for the transcription of all viral RNAs[44], including pgRNAs and other viral RNAs that are essential for viral proteins production. As such, cccDNA is very important for viral replication and progeny generation[45,46]. The pgRNA generated from cccDNA may also be reverse transcribed to form rcDNA for viral replication. The cccDNA function is heavily regulated by HBx protein, and inhibition of HBx protein will decrease HBV replication[47].
In addition, cccDNA is also speculated to play a role in persistent HBV infection or hepatitis relapse since cccDNA is very stable in non-dividing human hepatocytes. Furthermore, cccDNA can survive for the entire life span of the hepatocytes, thus acting as a persistent viral reservoir[46]. As cccDNA is established in the infected hepatocytes, viral replication can occur without stimulating the intrinsic antiviral defense mechanisms[46], hence making it possible for chronically infected individuals to develop hepatitis relapse after stopping the antiviral treatment. Moreover, cccDNA also mediated both HBV persistence and occult HBV infection (OBI), with OBIs resulting from epigenetic inactivation of cccDNA[48,49].
During HBV life cycle, the rcDNA conversion into cccDNA occurred through a repair process via an intermediate form called protein-free rcDNA[44,50]. The plus-strand DNA is synthesized via a gap-filling mechanism, and viral polymerase and RNA primers that are attached to the 5’-termini of the minus-strand and plus-strand DNAs were removed to generate cccDNA[48]. Several host factors have been demonstrated to be involved in cccDNA formation. An in vitro study has shown that the host tyrosyl-DNA-phosphodiesterase 2 (TDP2) may be involved in the removal of viral polymerase that is covalently linked to the 5’-end of the minus-strand DNA[21]. However, TDP2 gene knockout[18] resulted in uninhibited cccDNA formation in HBV infection of permissive hepatoma cells and intr
The transcription of cccDNA is controlled in a similar way to the regulation of host chromatin. cccDNA transcription is regulated by two enhancer elements and four distinct promoters, which relies on the dynamic interplay of various transcription factors, co-activators, co-repressors, and chromatin-modifying enzymes[48]. cccDNA also contains many binding sites for ubiquitous and liver-specific transcription factors, which have been demonstrated to be involved in the transcription of viral RNAs[52].
cccDNA forms minichromosomes in the nucleus by associating with histone proteins H2a, H2b, H3, and H4 as well as linker H1, and non-histone proteins such as viral core and HBx protein[44,45,53]. Studies in hepatoma cell lines have indicated that cccDNA transcription is regulated by the acetylation status of cccDNA-bound H3 and H4 histones, as well as the acetylation status of non-histone proteins[54]. Furthermore, studies in HBV-infected patients also showed that histone hypoacetylation and histone deacetylase 1 (HDAC1) recruitment into the cccDNA were correlated with low HBV viremia[54]. Similar in vitro and in vivo studies have also demonstrated that HBV transcriptional activity and viral load were affected by the acetylation degree of cccDNA-bound histones H3/H4 and the association between cccDNA and histone-modifying enzymes[49].
cccDNA formation occurs early in the viral cycle, and due to its stability, it may persist inside the nucleus even without active viral replication as long as the infected cells survive[55,56]. cccDNA acts as the transcription template for the other viral RNAs. However, in the latter stage of infection, cccDNA activity is greatly regulated by HBx, which is required for efficient cccDNA transcription[57]. HBx protein as an oncoprotein plays crucial roles in the pathogenesis of HBV infection[58] and in the development of HCC[59]. HBx protein mainly affects the cell cycle regulation and DNA repair mechanisms to stimulate oncogenic transformation of the liver cells[60,61].
HBx protein, as the viral component of the cccDNA minichromosome, is required to initiate cccDNA-driven transcription of HBV RNA[58,62]. HBx protein regulates cccDNA function and activity by binding to cccDNA and modifying its epigenetic regulation[44]. The key roles of HBx include the degradation of structural maintenance of chromosome 5/6 (Smc5/6) restriction factors[58], the prevention of cccDNA transcriptional repressor recruitment[62,63], and the regulation of coding and non-coding RNA promoters[64]. HBx protein-mediated degradation of Smc5/6 may act as a host restriction factor that suppresses the transcription of cccDNA. HBx protein also binds to damaged DNA-binding protein 1 (DDB1), promoting the interaction of Smc5/6 with Cul4, a component of E3 ubiquitin ligase. This interaction triggers the ubiquitination and degradation of Smc5/6 complex, thus promoting cccDNA transcription[65].
In addition, HBx protein has also been demonstrated to recruit chromatin regulators such as p300 and other acetyltransferases to cccDNA, which further enhances viral transcription[62]. Consequently, mutations in HBx protein caused impaired viral replication and severely impaired acetyltransferases recruitment. Failure in recruiting acetyltransferases causes cccDNA to be deacetylated by histone deacetylases, thus reducing both viral transcription and replication[62].
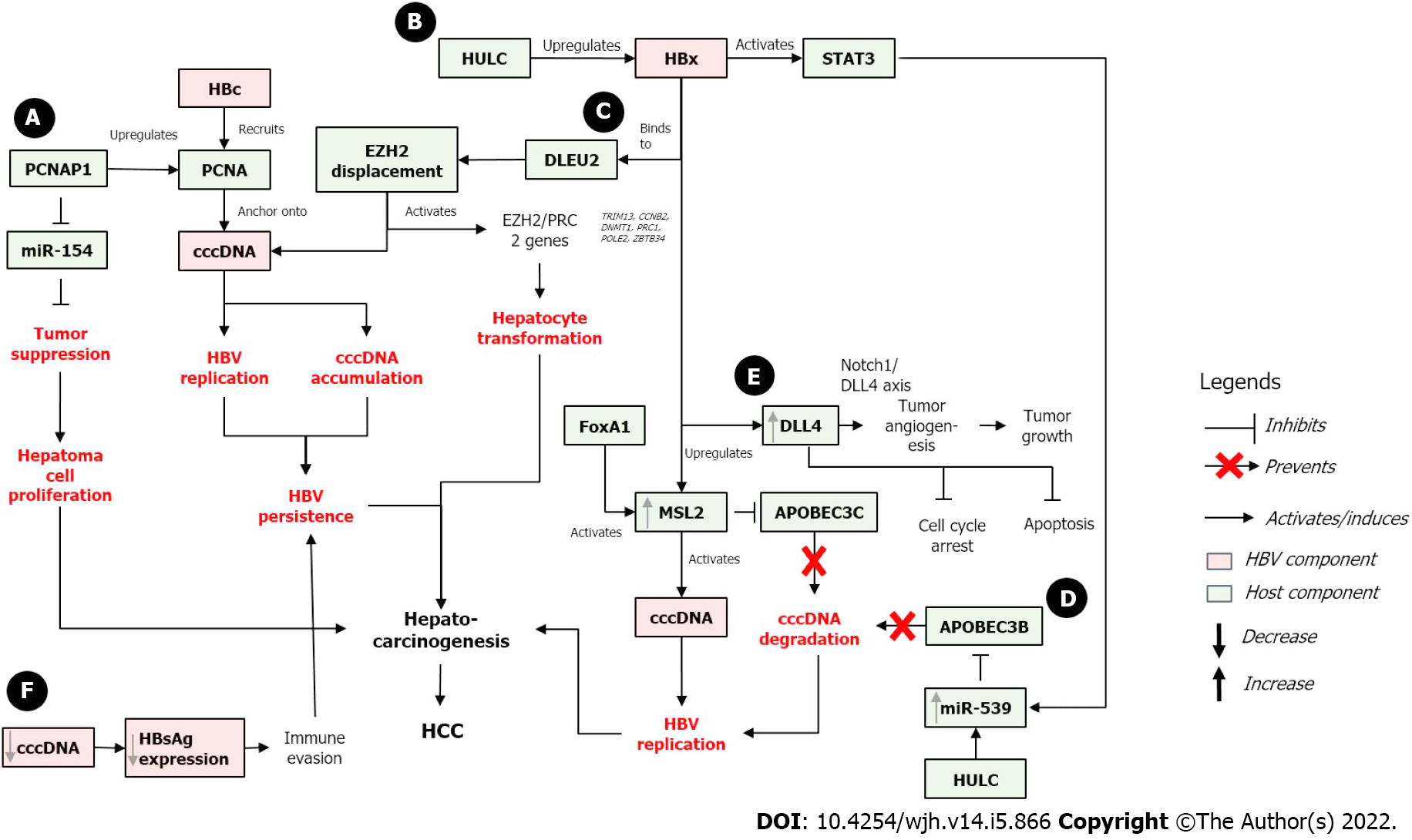
On the other hand, the absence of HBx protein results in rapid silencing of cccDNA which is then maintained in a close state. HBx protein promotes the de-silencing of cccDNA, which converts the cccDNA into the open state and activates the cccDNA. This de-silencing process is usually done by stimulating the activating modifications, blocking the repressive modifications, or both[45]. As cccDNA activity is tightly regulated by HBx, the effect of cccDNA on HBV-related carcinogenesis may occur directly through its stability and persistence in infected cells and indirectly through HBx-related effect and interaction with numerous host factors that regulate cell cycle and cell death. The proposed effects of cccDNA in HBV-related hepatocarcinogenesis including the involvement of HBx and several host proteins are described below and are presented in Figure 2.
Modulation of HBx/STAT3/miR-539/APOBEC3B: Long non-coding RNAs (lncRNAs) are transcripts of more than 200 bp that are not translated into proteins. Recent transcriptome sequencing has revealed that some lncRNAs may contain pseudogenes, which are ancestral copies of protein-coding genes[66,67]. HULC (Highly Upregulated in Liver Cancer) is one of the first and most studied lncRNAs in HCC. It is found highly expressed in HCC tissues[68].
In HBV-infected HCC cells, HULC enhanced the stability of cccDNA by preventing its degradation by APOBEC3B, thus activating HBV and promoting the growth of cancer cells. Further, HULC also significantly increased the levels of HBeAg, HBsAg, HBx, and cccDNA to activate more HBV replication. At the same time, HULC upregulated miR-539, which targeted APOBEC3B mRNA for deactivation. APOBEC3B is also responsible for cccDNA elimination by inducing its deamination. Therefore, APOBEC3B inhibition allows for active cccDNA and promotes HBV replication. Simultaneously, HULC also upregulated HBx which co-activated STAT3. The activation of STAT3 stimulated activation of miR-539 promoter, which further downregulated APOBEC3B and enhanced hepatocarcinogenesis by promoting hepatoma cell growth[69].
Positive feedback loop of HBx/MSL2/cccDNA: HBV cccDNA is deaminated by APOBEC3A and APOBEC3B, and overexpression of these APOBECs resulted in decreased cccDNA levels[58]. HBx protein has also been documented to modulate degradation of Smc5/6 complex by hijacking DDB1-containing E3 ubiquitin ligase[65].
An E3 ubiquitin ligase, male-specific lethal 2 (MSL2), is upregulated in HCC compared to adjacent non-tumorous liver tissues, suggesting that MSL2 might be involved in hepatocarcinogenesis. MSL2-induced cells have elevated levels of cccDNA, while MSL2 knockdown resulted in the opposite. These suggest that MSL2 may activate cccDNA in hepatoma cells to accelerate HBV replication leading to hepatocarcinogenesis. Moreover, MSL2 induced the degradation of APOBEC3C, thus further suggesting that MSL2 can activate and maintain the levels of HBV cccDNA in hepatoma cells[70].
HBx protein also contributes to the upregulation of MSL2 in hepatoma cells. In clinical HCC samples, high level of HBx mRNA is accompanied with high level of MSL2. MSL2 upregulation was associated via YAP/FoxA1 signaling, where upregulation of FoxA1 further activated the MSL2 promoter. Altogether, the positive feedback between HBx, MSL2, and cccDNA may contribute to HCC development by further enhancing the growth of hepatoma cells[70].
HBx/DLEU2 interaction and cccDNA activation: HBx protein can sustain the transcription of cccDNA and HCC-related genes by binding to DLEU2 (Deleted in lymphocytic leukemia 2), an lncRNA expressed in the liver. DLEU2 was upregulated in HBV-related HCC and in HBV/HBx-expressing cells[71]. HBx protein binding to DLEU2 activated DLEU2, resulted in increased cccDNA transcription and HBV replication. Furthermore, HBx-mediated DLEU2 upregulation and HBx recruitment to the target gene regulatory sequence increased chromatin accessibility and activated a subset of EZH2/PRC2 targets in both HBV-replicating cells and HBV-related HCCs. EZH2 (enhancer of zeste homolog 2) is the major cellular H3K27 trimethyl-transferase that catalyzes the addition of methyl groups at lysine 27 of H3 histone[72]. EZH2 is found to be overexpressed in many cancers including HCC.
Furthermore, in silico modeling and biochemical evidence suggested that HBx and EZH2 compete for the same binding sites in DLEU2 intron 1, and co-recruitment of HBx and DLEU2 on cccDNA displaces EZH2 from the viral chromatin to boost both viral transcription and replication. DLEU2-HBx association with the target host promoters relieved EZH2 repression, which eventually led to the activation of a subset of EZH2/PRC2 targets in HBV-replicating cells and in HBV-related HCCs. Several regulatory genes (TRIM13, CCNB2, DNMT1, PRC1, POLE2, and ZBTB34) that play roles in DNA replication, cell cycle, and mitosis were also co-regulated by HBx, DLEU2, and EZH2[73]. These data suggested that co-recruitment of HBx and DLEU2 may modulate the infected hepatocytes cell cycle, which may induce hepatocyte transformation and hepatocarcinogenesis.
HBx/DLL4/Notch 1 signaling pathway: The role of HBx-mediated DLL4 (Delta like canonical notch ligand 4) upregulation and Notch signaling in hepatocarcinogenesis has been reported[74]. DLL4 is a Notch ligand that plays a role in angiogenesis including tumor angiogenesis. It can act as both an oncogene and a tumor suppressor gene[75]. Overexpression of HBx protein in HCC cell line upregulates the expression of all Notch ligands, suggesting the role of the Notch pathway in oncogenesis[76]. Silencing of DLL4 led to cell cycle arrest and increased apoptosis of HCC cells. Meanwhile, HBx overexpression resulted in DLL4 upregulation in HCC cells. The HBx-mediated DLL4 upregulation activates Notch signaling via Notch1/DLL4 axis to induce angiogenesis, thus promoting tumor growth[74,77].
PCNA (Proliferating Cell Nuclear Antigen) has been identified to play a role in hepatocarcinogenesis[67]. PCNA is a coordinator of DNA polymerase that plays a role in genomic integrity maintenance at both genetic and epigenetic levels, thus having multiple roles in DNA replication and repair by interacting with various proteins[78]. PCNA has been associated with the expression of miR-154[79], a tumor suppressor that inhibits tumor cells proliferation and metastasis[80]. Notably, miR-154 appears to be downregulated in multiple types of cancers, including HCC.
Similarly, the lncRNA PCNAP1 has been shown to promote HBV replication and cccDNA accumulation. PCNAP1 expression in HBV+ cells was 10- to 20-fold higher compared to HBV- hepatoma cells, and its level were significantly higher in HCC relative to the adjacent non-tumorous liver tissues. HBV DNA and cccDNA were upregulated in vitro in PCNAP1-transfected cells, while PCNAP1 knockdown resulted in the opposite effect. This in vitro result was supported by the clinical observation that PCNAP1 expression was significantly higher in cccDNA+ HCC tissues compared to cccDNA- HCC tissues[81]. Interestingly, PCNA (the ancestor of PCNAP1) was found anchored onto cccDNA via the interaction with HBc protein. HBc recruited and anchored PCNA onto cccDNA to induce HBV replication and cccDNA accumulation, thus further contributing to HBV persistence that may lead to hepatocarcinogenesis[81].
In relation with microRNA, PCNAP1 competed with miR-154 to enhance PCNA expression, resulting in the inhibition of miR-154. The inhibition of miR-154 led to unregulated cell proliferation and could induce hepatocarcinogenesis. PCNAP1 and PCNA significantly promoted the growth of hepatoma cells both in vitro and in vivo, suggesting the effect of PCNAP1/PCNA on the growth of HBV-related HCC[81].
A recent study has investigated the relationship between serum HBsAg and intrahepatic cccDNA in HBV-associated HCC. This study showed that the levels of serum HBsAg and intrahepatic cccDNA were significantly reduced in HBV-associated HCC tissues. The cccDNA reduction is speculated as the result of host tumor suppressor activity which controls the proliferation of cancerous cells by inducing the eradication of intrahepatic cccDNA. This cccDNA reduction also led to reduced expression of HBsAg, which could consequently contribute to immune evasion of the cancerous cells. Thus, this immune evasion strategy may further contribute to HBV persistence and eventually induce hepatocarcinogenesis[82]. However, the actual significance of these cccDNA and HBsAg reductions, and the exact mechanisms and the host factors involved in the cccDNA reduction in HBV-associated HCC tissues remain unclear and require further investigation.
The detection of cccDNA in a patient's serum and/or liver biopsies is important for the treatment of CHB. There have been numerous detection methods for cccDNA developed throughout the years. Southern blot is the gold standard for quantitative cccDNA detection; however, it is quite complicated and not suitable for high-throughput screening. Several more sensitive and simpler methods have been utilized, such as PCR-based methods, invader assays, in situ hybridization, and surrogates or substituted markers as described in Table 1[83].
| Method | Advantages | Disadvantages | Ref. |
| Blotting | |||
| Southern blot | Specific detection of DNA sequences; Able to distinguish cccDNA from other viral DNA species; Reliable and reproducible | Complicated, time-consuming, and costly | [101-102] |
| PCR-based methods | |||
| Conventional qPCR | Simple, accurate, and sensitive; Suitable for high-throughput screening; Rapid and economical | Specificity is reduced if high concentration of rcDNA is present | [100,102] |
| Semi-nested and nested qPCR | Sensitive and specific; Allows for cccDNA quantification | [100,102,104,106] | |
| Competitive qPCR | Sensitive; Can readily distinguish cccDNA from rcDNA; Allows for cccDNA quantification | [100,102,107] | |
| Droplet-digital PCR | Very sensitive and accurate; Can detect a single copy of cccDNA precisely; Allows for cccDNA quantification | [102,108-114] | |
| Rolling circle amplification qPCR | Very sensitive; cccDNA is visible at single-cell resolution | Effective amplification may be hindered by cross-linked proteins or diffusion of DNA | [100,102,115-117] |
| Magnetic capture hybridization PCR | Sensitive and specific; Allows for selective isolation of cccDNA; Reproducible | Unable to capture all cccDNA completely; Complicated and costly | [100,102,118-119] |
| Invader Assay | Specific, simple, and reproducible | [100,102,121-124] | |
| In situ hybridization | Specific; Can distinguish different types of nucleic acids; Visible at single-cell resolution | Complicated probe design | [100,102,125-126] |
| Indirect method | |||
| Surrogate markers | Non-invasive; Convenient and cost-effective; Suitable for high-throughput screening | Indirect | [100,114,126-128,131-135] |
Southern blot is a molecular biology method for the detection of a specific DNA sequence in DNA samples. It is a straightforward and reliable method for cccDNA assay using cell culture samples[84]. Due to its specificity, cccDNA detection using Southern blot may also distinguish the cccDNA from other viral DNA species by the differential migration rate during electrophoresis. Southern blot is performed in sequential steps including probe preparation, electrophoresis, transmembrane hybridization, and detection[84]. It is a reliable and reproducible method, with a limit of cccDNA detection of around 2 × 106 copies. However, Southern blot procedures can be complicated, time-consuming, and costly[85].
PCR-based methods for cccDNA detection include conventional qPCR, competitive qPCR, real-time PCR, droplet-digital PCR, rolling circle amplification qPCR, and magnetic capture hybridization qPCR. Conventional qPCR is a simple method that has been used for cccDNA detection with a limit of detection of 2 × 103 copies/mL[85]. It is a rapid, accurate, economical, and sensitive method, which makes it suitable for high-throughput screening. However, the specificity of conventional qPCR is compromised if a high concentration of rcDNA is present in the sample. This is due to the shared partial homology of rcDNA and cccDNA, which reduces the specificity of conventional qPCR towards cccDNA[83]. Thus, chimeric sequences may be used to improve the specificity of conventional qPCR. These chimeric sequences consist of two different segments: Segment A which is complementary to HBV DNA plus-strand from nucleotide number 1615 to 1604, and segment B which is consensual to the HIV LTR region and dissimilar from HBV DNA[86].
A similar detection technique is the semi-nested and nested qPCR, which are sensitive and specific with a limit of cccDNA detection of 3.0 × 102 copies/mL[83,85]. Semi-nested qPCR includes two PCR reactions where the second PCR reaction uses the generated products from the first PCR as the templates. The first PCR reaction can only generate products when the template cccDNA is above a certain concentration as the primer pairs were only partially complementary[87]. Similarly, nested qPCR also includes two PCR reactions, in which the first PCR reaction uses outer primers, while the second PCR reaction uses inner primers with the first PCR products as the template. Nested PCR has been used to quantify cccDNA in peripheral blood mononuclear cells and bone marrow mononuclear cells[88]. The PCR sensitivity and accuracy can be increased by using two hybridization fluorescence resonance energy transfer (FRET) probes in real-time PCR. This modification maintains a cccDNA:rcDNA specificity ratio greater than 1:10000[89].
Competitive qPCR is a more sensitive method for cccDNA detection compared to Southern blot, as it can readily distinguish between rcDNA and cccDNA with a limit of detection of 2 × 104 copies[85]. However, its specificity is compromised in the presence of a high concentration of rcDNA[83]. Competitive qPCR involves two templates: A competitor template with known quantity and a target template with an unknown quantity. These two templates will combine and compete for the same cccDNA-specific primers with comparable amplification efficiency during PCR. Thus, the length of the PCR product templates will be different and can be quantifiable[90].
Droplet-digital PCR (ddPCR) is a super sensitive and accurate method of detecting cccDNA with a limit of detection of only one copy[85]. The number of cccDNAs that persist in infected cells after antiviral therapy is usually very scarce, thus a more sensitive and reliable system is needed to detect and quantify cccDNA in these cells. This issue can be overcome by utilizing ddPCR[91-96], as it uses specific primers that can precisely detect one single copy of HBV cccDNA. In ddPCR, samples are partitioned in water-in-oil droplets into tens of thousands of droplets, and each droplet acts as an independent reactant for a conventional PCR. Thus, a droplet that contains a detectable fluorescent signal is scored as a positive event, while droplets with no detectable signal are scored as a negative event[83]. This approach allows ddPCR[91-96] to accurately detect low copy number of HBV cccDNA in the samples. The high specificity and accuracy of ddPCR for cccDNA detection had been confirmed using a cohort of OBI patients, where cccDNA was able to be detected and quantified in half of the examined OBI cases[97].
Rolling circle amplification qPCR is developed to increase the sensitivity and specificity of cccDNA detection in formalin-fixed paraffin-embedded (FFPE) liver biopsy tissues, since regular qPCR is unusable in this type of sample. It is also designed to minimize the interference of integrated HBV DNA in FFPE liver biopsy tissue, as cccDNA quantity in FFPE liver tissue is usually 100-fold less than that in cryo-preserved liver tissue[98,99]. This method is very sensitive and cccDNA is visible at single-cell resolution with a limit of detection of two copies per cell[85]. However, effective amplification may be hindered by diffusion of the amplified DNA into neighboring cells or by cross-linked proteins[83]. A modified rolling circle amplification-in situ qPCR technique has since been developed to accurately visualize the distribution and localization and also quantify the number of cccDNA copies in the liver tissue[100].
The magnetic capture hybridization qPCR allows for selective cccDNA isolation as well as enrichment for specific cccDNA quantification[101], with a limit of detection of 90 IU/mL[85]. This method is sensitive, specific, and reproducible. However, it is not able to completely capture all cccDNA, and can be complicated and costly[83]. The magnetic beads used to capture the cccDNA are synthesized using the reverse microemulsion method and further modified with streptavidin[102]. The captured cccDNA is released through denaturation and further processed using conventional qPCR[101].
Invader assay is a non-PCR signal amplification assay that is used for genotyping and gene expression monitoring, and able to detect only one strand of double-stranded DNA[103]. This assay was first used to quantify cccDNA in CHB patient serum[104]. It is specific, simple, and reproducible[83], with a limit of detection of 104 copies/mL[85]. Invader assay requires two oligonucleotides (a primary probe and an invader probe) and a FRET cassette. The two oligonucleotides will hybridize to the target DNA to form a partially overlapping structure. This overlapping structure is cleaved by a cleavase enzyme to generate a 5’-flap from the primary probe. Meanwhile, another primary probe will cycle to the target DNA and hybridize with the invader probe to form an overlapping structure. The released 5’-flaps will increase proportionally to the concentration of the target DNA. The FRET cassette will react with the released 5’-flaps to generate fluorescent signals that can be measured using real-time PCR. The differences between cccDNA and other forms of HBV DNA are used for the design of primary and invader probe sequences, resulting in positive signals for cccDNA and negative signals for non-cccDNA[105-107].
In situ hybridization was first performed[108] using digoxigenin-labeled single-stranded probe in HBV producing HepG2 cells, but was not done in liver tissues. In situ hybridization is specific, as it can distinguish and locate different DNAs, RNAs, and proteins without diffusion of the amplified products, and is visible at a single-cell resolution[83]. However, this method has a complicated probe design and its limit of detection is only one copy even under optimal conditions[85]. Recently, a highly sensitive and specific modification of in situ hybridization from ViewRNA assay was designed by using a probe set that spans the gap in rcDNA[109].
Indirect methods can also be used for cccDNA detection by using different surrogate markers that correlate with the quantity of cccDNA in an infected cell. This approach allows for non-invasive method that is more convenient and cost-effective, and is also suitable for high-throughput screening[83]. Numerous markers have been correlated with cccDNA concentration, including hepatitis B core-related antigen (HBcrAg), HBsAg, HBeAg, and anti-HBc-IgG[83,97,109,110].
HBcrAg is detectable in HBsAg-negative CHB patients with undetectable HBV DNA. The decrease of HBcrAg levels is significantly associated with a hopeful HCC prognosis, as HBcrAg was shown as a reliable marker to predict HCC occurrence. Furthermore, HBcrAg is also correlated with both serum HBV DNA and intrahepatic cccDNA levels[110]. A study using chemiluminescent enzyme immunoassay to measure the HBcrAg levels in 130 CHB patients has found that HBcrAg level was correlated with serum HBV DNA, intrahepatic HBV DNA, pgRNA, and cccDNA levels[111]. Furthermore, patients who were negative for HBcrAg had less liver cccDNA and lower cccDNA activities than patients who were HBcrAg-positive. These finding suggest that HBcrAg may be used as a reliable surrogate marker for intrahepatic cccDNA and its transcriptional activities[111].
HBsAg has also been correlated with cccDNAs. The decline of cccDNA in liver biopsies was correlated with the decline in serum HBsAg during therapy, suggesting that quantification of serum HBsAg may represent a non-invasive surrogate marker for intrahepatic cccDNA pools[112,113]. A study has shown that serum HBsAg levels are significantly correlated with both intrahepatic HBsAg and cccDNA levels in matched non-cancerous tissues[109]. This finding is also supported by another study with similar results[114]. However, serum HBsAg levels were found not correlated with intrahepatic HBsAg and cccDNA levels in HCC tissues, while intrahepatic HBsAg levels were significantly correlated with intrahepatic cccDNA both in matched non-cancerous tissues and in HCC tissues. Further, the intrahepatic cccDNA levels in HCC tissues were significantly lower than those in matched non-cancerous tissue[109]. These findings were also supported by other similar studies[114,115]. In contrast, two different studies had showed opposite findings, and found no significant differences in intrahepatic cccDNA levels between tumor and non-tumor liver tissues in HBV-related HCC patients[116,117].
In addition to HBcrAg and HBsAg, HBeAg levels have also been shown to be correlated with cccDNA concentrations. Thus, HBeAg reporter assay may be a convenient and cost-effective tool for high-throughput screening for cccDNA targeting drugs[83,118]. Serum anti-HBc-IgG level has also been associated with intrahepatic cccDNA, as such titer of anti-HBc-IgG may be useful as a surrogate marker to predict the risk of OBI reactivation especially in immunosuppressed patients[97].
The virological key to persistent HBV infection is cccDNA that persists in the nucleus of infected cells. However, current therapies for CHB infection, interferons and nucleoside analog inhibitors, are unable to effectively remove and/or eliminate cccDNA. cccDNA persistence also resulted in HBV reactivation when antiviral treatment is stopped and in immunosuppressed condition. It has been proposed that any mutations occurring in the cccDNA may be highly conserved during HBV life cycle and can quickly give rise to circulating mutant viruses that may result in antiviral resistance[119]. This may lead to both virological and clinical breakthrough in patients and faster progression to cancer development. Even so, cccDNA reduction or loss has been reported in small numbers of patients, through yet unclear mechanisms, but most likely achieved through a combined processes of reduced cccDNA formation due to rcDNA depletion, degradation of pre-exisiting cccDNAs, and loss/turnover of infected cells[56]. Thus, effective elimination of cccDNA is required to reduce HBV-related liver disease progression and to achieve complete cure.
Recently, more studies have been conducted to find effective strategies to eliminate HBV cccDNA, which include: Silencing of cccDNA expression by gene editing techniques or silencing of cccDNA transcription via epigenetic modifications[41,120]. These approaches are still in early stages of development and has to tackle many issues before any viable clinical application. Nevertheless, some have shown good potential as an effective approach for cccDNA elimination. The summary of gene editing and epigenetic modification techniques to target and eliminate cccDNA is listed in Table 2.
| Technique | Study model | Study results | Ref. |
| Gene editing | |||
| Synthetic RNAi | Clinical trial | ARC-520 was well tolerated, with only two serious adverse effects. ARC-520 was active in both HBeAg-neg and HBeAg-pos patients, but only moderate reduction in HBsAg was observed | [122] |
| Zinc finger nucleases (ZFNs) | In vitro (AAV-mediated delivery of ZFNs in HepAD38 cells) | Completely inhibited HBV DNA replication and decreased HBV pgRNA level | [124] |
| In vitro | Decreased pgRNA level, thus having the potential to target cccDNA | [123] | |
| Transcription activator-like effector nucleases (TALENs) | In vitro and in vivo [murine hydrodynamic injection (HDI)] | Efficient disruption of target sites and suppression of viral replication markers; targeted mutation in 35% of cccDNAs was observed in vitro under mildly hyphothermic conditions and further confirmed in vivo | [125] |
| CRISPR/Cas9 System | In vitro (A64 cells) | Inhibited both HBV antigen expression and replication, excised the entire full-length of integrated HBV genome, and disrupted cccDNA | [126-131] |
| Epigenetic modification | |||
| Dicoumarol | In vitro (HBV-infected cells HepG2-NTCP cells) and in vivo (humanised liver mouse) | Reduced HBx protein expression, therefore having a potent antiviral activity against HBV RNAs, DNA, HBsAg, and HBc protein; cccDNA-ChIP decreased active histone marks and increased repressive histone marks | [132] |
| In vitro (NTCP-expressing HepG2 and primary hepatocytes) | Inhibited HBV replication in HBV-infected primary human hepatocytes by inhibiting the activity of cccDNA | [133] | |
| Interferon-alpha (IFN) | In vitro and in vivo (chimeric uPA/SCID mice) | Hypoacetylation of cccDNA-bound histone and active recruitment of transcriptional corepressors to the cccDNA; Inhibited HBV replication and cccDNA transcription | [134] |
| In vitro | Induced a prolonged suppression of human and duck HBV cccDNA transcription | [135] | |
| Zinc finger proteins (ZFPs) | In vitro (male longhorn hepatoma cells) | ZFPs binding to HBV enhancer region inhibited viral replication by inhibiting cccDNA transcriptional activity | [136] |
| Curcumin | In vitro (HepG2.2.15) | Reduced HBsAg and cccDNA levels up to 58% and 76%, respectively | [137] |
Synthetic RNAi: RNAi is an endogenous gene regulatory pathway that can be reprogrammed by exogenous RNA molecules to create synthetic RNAi that targets specific sequences. As viral RNA transcripts contain overlapping sequences, a single RNAi trigger can, in theory, be utilized to degrade all viral transcripts, resulting in prevention of viral proteins production by eliminating cccDNA and other viral RNA transcripts. RNAi triggers may also be used to target pgRNAs, thus contributing to the reduction of cccDNA reservoir in infected cells via inhibition of viral replication[121]. A clinical trial of an RNAi-based drug, ARC-520, showed that ARC-520 was active in both HBeAg-negative and -positive patients. However, the reduction in HBsAg level seemed to be hindered by the concomitant expression of HBsAg from the integrated HBV DNA. This result indicated that specifically designed RNAi that can also target the viral transcripts from the integrated HBV DNA will be crucial for the total elimination of CHB infection[122].
Zinc finger nucleases: Zinc finger nucleases (ZFNs) are custom DNA endonucleases that are utilized to create DNA double-strand breaks in a specified target site and repair that double-strand break by creating sequence alterations at the cleavage sites[41]. ZFN treatment in HBV-infected cells has been shown to decrease the HBV pgRNA levels[123]. Further, HBV-targeted ZFNs were able to produce a sustained HBV level suppression for around 3 wk after the ZFN treatment[124]. This was achieved by using three specifically designed ZFNs to target HBV P, X, and C genes, which were delivered to HepAD38 cells via self-complementary adeno-associated viral (AAV) vectors[124].
Transcription activator-like effector nucleases: Transcription activator-like effector nucleases (TALENs) are also nucleases, similar to ZFNs. However, TALENs comprise a nonspecific nuclease that is fused to a sequence-specific DNA-binding domain, in which the DNA-binding domain is highly repeated and derived from transcription activator-like III effectors[41,120]. The efficiency of HBV cccDNA-targeting TALENs in reducing HBV replication in cell culture was first reported in 2013[125], using four TALENs that target specific sites within the S/pol, C/pol, and pol ORFs of the HBV genome. The S and C TALENs disrupted the intended target sites efficiently and suppressed other markers of viral replication. Subsequent experiment in HepG2.2.15 cells, which were triple transfected with the S TALEN under mildly hyphothermic conditions, resulted in targeted mutation in around 35% of cccDNAs. These results were further confirmed in vivo, where mice subjected to hydrodynamic injection of the S and C TALENs showed overall reduced markers of viral replication and accumulation of viral mutation in the targeted sites. Together, these results demonstrated the potential use of TALENs for targeted disruption of both HBV DNA and cccDNA[125].
CRISPR/Cas9 system: CRISPR/Cas9 system is the adaptive immune system of bacteria and archaea that acts against foreign DNA via the RNA-guided DNA cleavage[41,120]. The CRISPR-Cas9 system has been shown to successfully inhibit both HBV antigen expression and HBV replication in transfected A64 cells[126]. Furthermore, CRISPR/Cas9 system was able to excise the entire full-length of integrated HBV genome as well as disrupt cccDNAs. These findings were also supported by several other studies[127-130]. Due to its effectiveness, CRISPR/Cas9 system is currently regarded as the best method for successfully inactivating HBV cccDNA and eliminating the entire length of integrated HBV genome in the liver cells[131].
Dicoumarol: Dicoumarol is a competitive NADPH quinone oxidoreductase (NQO1) inhibitor that has been identified to inhibit the expression of HBx protein. Under normal circumstances, NQO1 binds to and stabilizes HBx protein by inhibiting the activity of 20S proteasome, thus preventing the proteasome-mediated degradation of HBx protein. Dicoumarol has been demonstrated to significantly reduce HBx protein expression, and has potent antiviral activity against HBV RNAs, DNA, HBsAg, and HBc protein in HBV-associated cells as well as in humanized liver mouse model[132]. Using cccDNA-ChIP (chromatin-immunoprecipitation) assay, dicoumarol treatment resulted in decreased active histone marks, but increased repressive histone marks in HBV-infected HepG2-NTCP cells. Therefore, dicoumarol exhibits a repressive effect on cccDNA transcription. This finding was also supported in an in vivo model that showed decreased levels of cccDNA-associated HBx protein in the dicoumarol-treated group[132]. A similar study[133] also showed that dicoumarol inhibited HBV replication in HBV-infected primary human hepatocytes by inhibiting the cccDNA activity.
Interferon-alpha: Interferon-alpha (IFN) has been shown to affect the epigenetic control of HBV cccDNA minichromosome by inducing persistent recruitment of co-repressors and components of the polycomb repressive complex 2 (PRC2) that target the acetylation and methylation of the histone tail. Therefore, IFN may provide an additional molecular mechanism for the repression of HBV transcription[134] to its immune modulating effect when used as antiviral treatment. IFN administration resulted in hypoacetylation of cccDNA-bound histone and active recruitment of transcriptional co-repressors to the cccDNA. These were achieved through the IFN effect on the reduced binding of STAT1 and STAT2 transcription factors to active cccDNA, thus mediating the epigenetic repression of cccDNA activity. IFN treatment also inhibited HBV replication and cccDNA transcription in both HCC cells and chimeric uPA/SCID mice[134].
A similar study[135] has also found that IFN treatment induced a prolonged suppression of both human and duck HBV cccDNA transcription, which was associated with a reduction of cccDNA-associated histone modifications that play a role in the activation of cccDNA transcription activity. On the other hand, downregulation of STAT1, structural maintenance of chromosome flexible hinge domain containing 1 (SMCHD1), or promyelocytic leukemia (PML) proteins resulted in increased basal level of cccDNA transcription activity and partially hindered the suppression activity of IFN towards cccDNA transcription. Meanwhile, ectopic expression of STAT1, SMCHD1, or PML can significantly reduce the activity of cccDNA. These findings indicate that IFN may modulate the epigenetic control of cccDNA function by affecting the recruitment of chromatin-modifying enzymes[134,135].
Zinc finger proteins: Zinc finger proteins (ZFPs) binding to the HBV enhancer region may inhibit viral replication by inhibition of cccDNA transcriptional activity. This was demonstrated by using six different ZFPs designed to bind to DNA sequences in the duck HBV enhancer regions[136]. The enhancer regions are the accessible parts of the cccDNA minichromosome which control the HBV core and surface promoters. Thus, ZFPs binding to these regions will interfere with viral transcription. The ZFPs were cloned into a eukaryotic expression vector and co-transfected into longhorn male hepatoma cells. The results demonstrated that ZFP treatment caused a significant reduction in viral RNA and HBV protein levels, indicating the effect of ZFPs on the transcription of viral proteins[136].
Curcumin: Curcumin is another compound that has been demonstrated to inhibit HBV infection by downregulation of cccDNA-bound histone acetylation in HepG2.2.15 cell line[137]. Additionally, treatment with 20 μmol/L curcumin for 2 d resulted in reduced HBsAg and cccDNA levels in HepG2.2.15 cells by up to 58% and 76%, respectively. Moreover, treatment with curcumin resulted in both time- and dose-dependent reductions of H3 acetylation levels, thus contributing to the reduction of H3- and H4-bound cccDNA. These findings indicate the potential use of curcumin as a cccDNA-targeting antiviral agent[41,137].
Chronic HBV infection remains a global health problem since it may lead to prolonged inflammation and subsequently more advanced liver diseases, including liver cancer. Furthermore, direct oncogenic properties of HBV viral components have been associated with their abilities to interact and alter the functions of various host genes, further contributing to HBV pathogenesis.
cccDNA is one of the most important HBV components. cccDNAs may persist in infected hepatocytes and serve as template for viral replication machinery. This highlights the need for an effective method for cccDNA detection and removal. Various methods have been developed for cccDNA detection and targeted removal; however, their overall sensitivity and specificity are still far from satisfactory. The use of antiviral therapy and/or interferon has been shown to effectively reduce the viral load, improve the general health status, and prevent the development of HCC in chronic HBV patients. However, current antiviral therapy does not eliminate the cccDNA in the liver. Based on our review, we presume that the versatility of PCR-based technologies may be a potential approach for advancing effective methods for cccDNA detection and quantification. As for cccDNA targeting, the vast application of CRISPR/Cas9 system might be the most optimum resort to modify cccDNA function, and more importantly to inactivate the cccDNA activity.
Nevertheless, concentrated effort should be focused more on prevention of HBV infection and not on HBV treatment and elimination. As shown in our previous review[138], this preventative approach, which may be achieved through immunization, is crucial to prevent viral transmission to the new generations, particularly in endemic areas. At the same time, it is also necessary to improve the awareness of the general public for the consequences of the disease and to expand the national and regional surveillance program.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: European Association for the Study of the Liver, No. 61174.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jiang W, China; Tu H, China S-Editor: Liu JH L-Editor: Wang TQ P-Editor: Liu JH
| 1. | Schädler S, Hildt E. HBV life cycle: entry and morphogenesis. Viruses. 2009;1:185-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 84] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 2. | Tsukuda S, Watashi K. Hepatitis B virus biology and life cycle. Antiviral Res. 2020;182:104925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 231] [Article Influence: 46.2] [Reference Citation Analysis (0)] |
| 3. | Seeger C, Mason WS. HBV replication, pathobiology and therapy: Unanswered questions. J Hepatol. 2016;64:S1-S3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | Wei L, Ploss A. Hepatitis B virus cccDNA is formed through distinct repair processes of each strand. Nat Commun. 2021;12:1591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 73] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 5. | Verrier ER, Colpitts CC, Bach C, Heydmann L, Weiss A, Renaud M, Durand SC, Habersetzer F, Durantel D, Abou-Jaoudé G, López Ledesma MM, Felmlee DJ, Soumillon M, Croonenborghs T, Pochet N, Nassal M, Schuster C, Brino L, Sureau C, Zeisel MB, Baumert TF. A targeted functional RNA interference screen uncovers glypican 5 as an entry factor for hepatitis B and D viruses. Hepatology. 2016;63:35-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 139] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 6. | Barrera A, Guerra B, Notvall L, Lanford RE. Mapping of the hepatitis B virus pre-S1 domain involved in receptor recognition. J Virol. 2005;79:9786-9798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 109] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 7. | Glebe D, Urban S, Knoop EV, Cag N, Krass P, Grün S, Bulavaite A, Sasnauskas K, Gerlich WH. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 Lipopeptides and tupaia hepatocytes. Gastroenterology. 2005;129:234-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 211] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 8. | Gripon P, Cannie I, Urban S. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J Virol. 2005;79:1613-1622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 279] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 9. | Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Fälth M, Stindt J, Königer C, Nassal M, Kubitz R, Sültmann H, Urban S. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 540] [Cited by in RCA: 621] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 10. | Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1280] [Cited by in RCA: 1599] [Article Influence: 123.0] [Reference Citation Analysis (1)] |
| 11. | Iwamoto M, Saso W, Nishioka K, Ohashi H, Sugiyama R, Ryo A, Ohki M, Yun JH, Park SY, Ohshima T, Suzuki R, Aizaki H, Muramatsu M, Matano T, Iwami S, Sureau C, Wakita T, Watashi K. The machinery for endocytosis of epidermal growth factor receptor coordinates the transport of incoming hepatitis B virus to the endosomal network. J Biol Chem. 2020;295:800-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Iwamoto M, Saso W, Sugiyama R, Ishii K, Ohki M, Nagamori S, Suzuki R, Aizaki H, Ryo A, Yun JH, Park SY, Ohtani N, Muramatsu M, Iwami S, Tanaka Y, Sureau C, Wakita T, Watashi K. Epidermal growth factor receptor is a host-entry cofactor triggering hepatitis B virus internalization. Proc Natl Acad Sci U S A. 2019;116:8487-8492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 182] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 13. | Núñez E, Yélamos B, Delgado C, Gómez-Gutiérrez J, Peterson DL, Gavilanes F. Interaction of preS domains of hepatitis B virus with phospholipid vesicles. Biochim Biophys Acta. 2009;1788:417-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 14. | Stoeckl L, Funk A, Kopitzki A, Brandenburg B, Oess S, Will H, Sirma H, Hildt E. Identification of a structural motif crucial for infectivity of hepatitis B viruses. Proc Natl Acad Sci U S A. 2006;103:6730-6734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 15. | Rabe B, Vlachou A, Panté N, Helenius A, Kann M. Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc Natl Acad Sci U S A. 2003;100:9849-9854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 216] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 16. | Rabe B, Glebe D, Kann M. Lipid-mediated introduction of hepatitis B virus capsids into nonsusceptible cells allows highly efficient replication and facilitates the study of early infection events. J Virol. 2006;80:5465-5473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 77] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 17. | Gallucci L, Kann M. Nuclear Import of Hepatitis B Virus Capsids and Genome. Viruses. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 18. | Cui X, McAllister R, Boregowda R, Sohn JA, Cortes Ledesma F, Caldecott KW, Seeger C, Hu J. Does Tyrosyl DNA Phosphodiesterase-2 Play a Role in Hepatitis B Virus Genome Repair? PLoS One. 2015;10:e0128401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 19. | Kitamura K, Que L, Shimadu M, Koura M, Ishihara Y, Wakae K, Nakamura T, Watashi K, Wakita T, Muramatsu M. Flap endonuclease 1 is involved in cccDNA formation in the hepatitis B virus. PLoS Pathog. 2018;14:e1007124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 86] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 20. | Tong S, Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol. 2016;64:S4-S16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 312] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 21. | Königer C, Wingert I, Marsmann M, Rösler C, Beck J, Nassal M. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc Natl Acad Sci U S A. 2014;111:E4244-E4253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 186] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 22. | Liang TJ. Hepatitis B: the virus and disease. Hepatology. 2009;49:S13-S21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 747] [Cited by in RCA: 635] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 23. | Yao Y, Yang B, Chen Y, Wang H, Hu X, Zhou Y, Gao X, Lu M, Niu J, Wen Z, Wu C, Chen X. RNA-Binding Motif Protein 24 (RBM24) Is Involved in Pregenomic RNA Packaging by Mediating Interaction between Hepatitis B Virus Polymerase and the Epsilon Element. J Virol. 2019;93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 24. | Kim S, Wang H, Ryu WS. Incorporation of eukaryotic translation initiation factor eIF4E into viral nucleocapsids via interaction with hepatitis B virus polymerase. J Virol. 2010;84:52-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 25. | Nguyen DH, Hu J. Reverse transcriptase- and RNA packaging signal-dependent incorporation of APOBEC3G into hepatitis B virus nucleocapsids. J Virol. 2008;82:6852-6861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 62] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 26. | Wang H, Kim S, Ryu WS. DDX3 DEAD-Box RNA helicase inhibits hepatitis B virus reverse transcription by incorporation into nucleocapsids. J Virol. 2009;83:5815-5824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 113] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 27. | Zhang YY, Zhang BH, Theele D, Litwin S, Toll E, Summers J. Single-cell analysis of covalently closed circular DNA copy numbers in a hepadnavirus-infected liver. Proc Natl Acad Sci U S A. 2003;100:12372-12377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 114] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 28. | Lamontagne RJ, Bagga S, Bouchard MJ. Hepatitis B virus molecular biology and pathogenesis. Hepatoma Res. 2016;2:163-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 121] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 29. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4264] [Article Influence: 236.9] [Reference Citation Analysis (2)] |
| 30. | Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093-5107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 374] [Article Influence: 17.0] [Reference Citation Analysis (1)] |
| 31. | McClune AC, Tong MJ. Chronic hepatitis B and hepatocellular carcinoma. Clin Liver Dis. 2010;14:461-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 32. | Gish RG, Given BD, Lai CL, Locarnini SA, Lau JY, Lewis DL, Schluep T. Chronic hepatitis B: Virology, natural history, current management and a glimpse at future opportunities. Antiviral Res. 2015;121:47-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 195] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 33. | Chen CJ, Yang HI. Natural history of chronic hepatitis B REVEALed. J Gastroenterol Hepatol. 2011;26:628-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 226] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 34. | Feitelson MA. Pathogenesis of Hepatitis B Virus Associated Chronic Liver Disease. IntechOpen. . [DOI] [Full Text] |
| 35. | Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem. 2000;275:2247-2250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1567] [Cited by in RCA: 1597] [Article Influence: 63.9] [Reference Citation Analysis (0)] |
| 36. | Canbay A, Taimr P, Torok N, Higuchi H, Friedman S, Gores GJ. Apoptotic body engulfment by a human stellate cell line is profibrogenic. Lab Invest. 2003;83:655-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 311] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 37. | Chisari FV, Isogawa M, Wieland SF. Pathogenesis of hepatitis B virus infection. Pathol Biol (Paris). 2010;58:258-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 299] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 38. | Keng VW, Tschida BR, Bell JB, Largaespada DA. Modeling hepatitis B virus X-induced hepatocellular carcinoma in mice with the Sleeping Beauty transposon system. Hepatology. 2011;53:781-790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 39. | Bouchard MJ, Navas-Martin S. Hepatitis B and C virus hepatocarcinogenesis: lessons learned and future challenges. Cancer Lett. 2011;305:123-143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 40. | Arbuthnot P, Capovilla A, Kew M. Putative role of hepatitis B virus X protein in hepatocarcinogenesis: effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J Gastroenterol Hepatol. 2000;15:357-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 151] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 41. | Zhu A, Liao X, Li S, Zhao H, Chen L, Xu M, Duan X. HBV cccDNA and Its Potential as a Therapeutic Target. J Clin Transl Hepatol. 2019;7:258-262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 42. | Lechardeur D, Sohn KJ, Haardt M, Joshi PB, Monck M, Graham RW, Beatty B, Squire J, O'Brodovich H, Lukacs GL. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6:482-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 464] [Cited by in RCA: 419] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 43. | Wang X, Le N, Denoth-Lippuner A, Barral Y, Kroschewski R. Asymmetric partitioning of transfected DNA during mammalian cell division. Proc Natl Acad Sci U S A. 2016;113:7177-7182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 44. | Kim DH, Kang HS, Kim KH. Roles of hepatocyte nuclear factors in hepatitis B virus infection. World J Gastroenterol. 2016;22:7017-7029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 40] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (3)] |
| 45. | Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 692] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 46. | Allweiss L, Dandri M. The Role of cccDNA in HBV Maintenance. Viruses. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 134] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 47. | Kim JW, Lee SH, Park YS, Jeong SH, Kim N, Lee DH. [Inhibition of in vitro hepatitis B virus replication by lentivirus-mediated short-hairpin RNA against HBx]. Korean J Hepatol. 2009;15:15-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 48. | Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol. 2009;51:581-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 431] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 49. | Mak LY, Wong DK, Pollicino T, Raimondo G, Hollinger FB, Yuen MF. Occult hepatitis B infection and hepatocellular carcinoma: Epidemiology, virology, hepatocarcinogenesis and clinical significance. J Hepatol. 2020;73:952-964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 134] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 50. | Gao W, Hu J. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J Virol. 2007;81:6164-6174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 166] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 51. | Wang HP, Rogler CE. Topoisomerase I-mediated integration of hepadnavirus DNA in vitro. J Virol. 1991;65:2381-2392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 75] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 52. | Quasdorff M, Protzer U. Control of hepatitis B virus at the level of transcription. J Viral Hepat. 2010;17:527-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 178] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 53. | Newbold JE, Xin H, Tencza M, Sherman G, Dean J, Bowden S, Locarnini S. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J Virol. 1995;69:3350-3357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 239] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 54. | Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 374] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 55. | Zhao K, Liu A, Xia Y. Insights into Hepatitis B Virus DNA Integration-55 Years after Virus Discovery. Innovation (N Y). 2020;1:100034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 56. | Hu J, Liu K. Complete and Incomplete Hepatitis B Virus Particles: Formation, Function, and Application. Viruses. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 212] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 57. | Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;479-480:672-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 569] [Cited by in RCA: 625] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 58. | Lucifora J, Arzberger S, Durantel D, Belloni L, Strubin M, Levrero M, Zoulim F, Hantz O, Protzer U. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol. 2011;55:996-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 357] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 59. | Ng SA, Lee C. Hepatitis B virus X gene and hepatocarcinogenesis. J Gastroenterol. 2011;46:974-990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 136] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 60. | Zhang X, Zhang H, Ye L. Effects of hepatitis B virus X protein on the development of liver cancer. J Lab Clin Med. 2006;147:58-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 156] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 61. | Zhang WY, Cai N, Ye LH, Zhang XD. Transformation of human liver L-O2 cells mediated by stable HBx transfection. Acta Pharmacol Sin. 2009;30:1153-1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 62. | Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci U S A. 2009;106:19975-19979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 399] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 63. | Rivière L, Gerossier L, Ducroux A, Dion S, Deng Q, Michel ML, Buendia MA, Hantz O, Neuveut C. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J Hepatol. 2015;63:1093-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 166] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 64. | Guerrieri F, Belloni L, D'Andrea D, Pediconi N, Le Pera L, Testoni B, Scisciani C, Floriot O, Zoulim F, Tramontano A, Levrero M. Genome-wide identification of direct HBx genomic targets. BMC Genomics. 2017;18:184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 65. | Decorsière A, Mueller H, van Breugel PC, Abdul F, Gerossier L, Beran RK, Livingston CM, Niu C, Fletcher SP, Hantz O, Strubin M. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 414] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 66. | Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grandér D, Morris KV. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20:440-446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 328] [Cited by in RCA: 358] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 67. | St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 688] [Cited by in RCA: 880] [Article Influence: 88.0] [Reference Citation Analysis (0)] |
| 68. | Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 587] [Cited by in RCA: 634] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 69. | Liu Y, Feng J, Sun M, Yang G, Yuan H, Wang Y, Bu Y, Zhao M, Zhang S, Zhang X. Long non-coding RNA HULC activates HBV by modulating HBx/STAT3/miR-539/APOBEC3B signaling in HBV-related hepatocellular carcinoma. Cancer Lett. 2019;454:158-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 70. | Gao Y, Feng J, Yang G, Zhang S, Liu Y, Bu Y, Sun M, Zhao M, Chen F, Zhang W, Ye L, Zhang X. Hepatitis B virus X protein-elevated MSL2 modulates hepatitis B virus covalently closed circular DNA by inducing degradation of APOBEC3B to enhance hepatocarcinogenesis. Hepatology. 2017;66:1413-1429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 71. | Moyo B, Nicholson SA, Arbuthnot PB. The role of long non-coding RNAs in hepatitis B virus-related hepatocellular carcinoma. Virus Res. 2016;212:103-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 72. | Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343-349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2760] [Cited by in RCA: 2521] [Article Influence: 180.1] [Reference Citation Analysis (0)] |
| 73. | Salerno D, Chiodo L, Alfano V, Floriot O, Cottone G, Paturel A, Pallocca M, Plissonnier ML, Jeddari S, Belloni L, Zeisel M, Levrero M, Guerrieri F. Hepatitis B protein HBx binds the DLEU2 LncRNA to sustain cccDNA and host cancer-related gene transcription. Gut. 2020;69:2016-2024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 113] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 74. | Kongkavitoon P, Tangkijvanich P, Hirankarn N, Palaga T. Hepatitis B Virus HBx Activates Notch Signaling via Delta-Like 4/Notch1 in Hepatocellular Carcinoma. PLoS One. 2016;11:e0146696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 75. | Mailhos C, Modlich U, Lewis J, Harris A, Bicknell R, Ish-Horowicz D. Delta4, an endothelial specific notch ligand expressed at sites of physiological and tumor angiogenesis. Differentiation. 2001;69:135-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 216] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 76. | Wang F, Zhou H, Yang Y, Xia X, Sun Q, Luo J, Cheng B. Hepatitis B virus X protein promotes the growth of hepatocellular carcinoma by modulation of the Notch signaling pathway. Oncol Rep. 2012;27:1170-1176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 77. | Wang Z, Kawaguchi K, Honda M, Hashimoto S, Shirasaki T, Okada H, Orita N, Shimakami T, Yamashita T, Sakai Y, Mizukoshi E, Murakami S, Kaneko S. Notch signaling facilitates hepatitis B virus covalently closed circular DNA transcription via cAMP response element-binding protein with E3 ubiquitin ligase-modulation. Sci Rep. 2019;9:1621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 78. | Park SY, Jeong MS, Han CW, Yu HS, Jang SB. Structural and Functional Insight into Proliferating Cell Nuclear Antigen. J Microbiol Biotechnol. 2016;26:637-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 79. | Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X, Asangani IA, Kothari V, Prensner JR, Lonigro RJ, Iyer MK, Barrette T, Shanmugam A, Dhanasekaran SM, Palanisamy N, Chinnaiyan AM. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell. 2012;149:1622-1634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 217] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 80. | Niu JT, Zhang LJ, Huang YW, Li C, Jiang N, Niu YJ. MiR-154 inhibits the growth of laryngeal squamous cell carcinoma by targeting GALNT7. Biochem Cell Biol. 2018;96:752-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 81. | Feng J, Yang G, Liu Y, Gao Y, Zhao M, Bu Y, Yuan H, Yuan Y, Yun H, Sun M, Gao H, Zhang S, Liu Z, Yin M, Song X, Miao Z, Lin Z, Zhang X. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics. 2019;9:5227-5245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 82. | Tantiwetrueangdet A, Panvichian R, Sornmayura P, Sueangoen N, Leelaudomlipi S. Reduced HBV cccDNA and HBsAg in HBV-associated hepatocellular carcinoma tissues. Med Oncol. 2018;35:127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 83. | Li X, Zhao J, Yuan Q, Xia N. Detection of HBV Covalently Closed Circular DNA. Viruses. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 84. | Cai D, Nie H, Yan R, Guo JT, Block TM, Guo H. A southern blot assay for detection of hepatitis B virus covalently closed circular DNA from cell cultures. Methods Mol Biol. 2013;1030:151-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 118] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 85. | WHO Guidelines Approved by the Guidelines Review Committee. Guidelines for the Prevention, Care and Treatment of Persons with Chronic Hepatitis B Infection. Geneva: World Health Organization; 2015 Mar . [PubMed] |
| 86. | Jun-Bin S, Zhi C, Wei-Qin N, Jun F. A quantitative method to detect HBV cccDNA by chimeric primer and real-time polymerase chain reaction. J Virol Methods. 2003;112:45-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 87. | Mason AL, Xu L, Guo L, Kuhns M, Perrillo RP. Molecular basis for persistent hepatitis B virus infection in the liver after clearance of serum hepatitis B surface antigen. Hepatology. 1998;27:1736-1742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 167] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 88. | Xu CH, Li ZS, Dai JY, Zhu HY, Yu JW, Lü SL. Nested real-time quantitative polymerase chain reaction assay for detection of hepatitis B virus covalently closed circular DNA. Chin Med J (Engl). 2011;124:1513-1516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 89. | Singh M, Dicaire A, Wakil AE, Luscombe C, Sacks SL. Quantitation of hepatitis B virus (HBV) covalently closed circular DNA (cccDNA) in the liver of HBV-infected patients by LightCycler real-time PCR. J Virol Methods. 2004;118:159-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 90. | Addison WR, Wong WW, Fischer KP, Tyrrell DL. A quantitative competitive PCR assay for the covalently closed circular form of the duck hepatitis B virus. Antiviral Res. 2000;48:27-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 91. | Mu D, Yan L, Tang H, Liao Y. A sensitive and accurate quantification method for the detection of hepatitis B virus covalently closed circular DNA by the application of a droplet digital polymerase chain reaction amplification system. Biotechnol Lett. 2015;37:2063-2073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 92. | Miotke L, Lau BT, Rumma RT, Ji HP. High sensitivity detection and quantitation of DNA copy number and single nucleotide variants with single color droplet digital PCR. Anal Chem. 2014;86:2618-2624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 169] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 93. | Morisset D, Štebih D, Milavec M, Gruden K, Žel J. Quantitative analysis of food and feed samples with droplet digital PCR. PLoS One. 2013;8:e62583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 190] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 94. | Whale AS, Huggett JF, Cowen S, Speirs V, Shaw J, Ellison S, Foy CA, Scott DJ. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res. 2012;40:e82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 337] [Cited by in RCA: 327] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 95. | Weaver S, Dube S, Mir A, Qin J, Sun G, Ramakrishnan R, Jones RC, Livak KJ. Taking qPCR to a higher level: Analysis of CNV reveals the power of high throughput qPCR to enhance quantitative resolution. Methods. 2010;50:271-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 171] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 96. | Qin J, Jones RC, Ramakrishnan R. Studying copy number variations using a nanofluidic platform. Nucleic Acids Res. 2008;36:e116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 90] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 97. | Caviglia GP, Abate ML, Tandoi F, Ciancio A, Amoroso A, Salizzoni M, Saracco GM, Rizzetto M, Romagnoli R, Smedile A. Quantitation of HBV cccDNA in anti-HBc-positive liver donors by droplet digital PCR: A new tool to detect occult infection. J Hepatol. 2018;69:301-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 122] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 98. | Zhong Y, Han J, Zou Z, Liu S, Tang B, Ren X, Li X, Zhao Y, Liu Y, Zhou D, Li W, Zhao J, Xu D. Quantitation of HBV covalently closed circular DNA in micro formalin fixed paraffin-embedded liver tissue using rolling circle amplification in combination with real-time PCR. Clin Chim Acta. 2011;412:1905-1911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 99. | Margeridon S, Carrouée-Durantel S, Chemin I, Barraud L, Zoulim F, Trépo C, Kay A. Rolling circle amplification, a powerful tool for genetic and functional studies of complete hepatitis B virus genomes from low-level infections and for directly probing covalently closed circular DNA. Antimicrob Agents Chemother. 2008;52:3068-3073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 47] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 100. | Zhong Y, Hu S, Xu C, Zhao Y, Xu D, Zhao J, Li Z, Zhang X, Zhang H, Li J. A novel method for detection of HBVcccDNA in hepatocytes using rolling circle amplification combined with in situ PCR. BMC Infect Dis. 2014;14:608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 101. | Guo Y, Sheng S, Nie B, Tu Z. Development of magnetic capture hybridization and quantitative polymerase chain reaction for hepatitis B virus covalently closed circular DNA. Hepat Mon. 2015;15:e23729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 102. | Rossi LM, Shi L, Quina FH, Rosenzweig Z. Stöber synthesis of monodispersed luminescent silica nanoparticles for bioanalytical assays. Langmuir. 2005;21:4277-4280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 175] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 103. | de Arruda M, Lyamichev VI, Eis PS, Iszczyszyn W, Kwiatkowski RW, Law SM, Olson MC, Rasmussen EB. Invader technology for DNA and RNA analysis: principles and applications. Expert Rev Mol Diagn. 2002;2:487-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 104. | Wong DK, Yuen MF, Yuan H, Sum SS, Hui CK, Hall J, Lai CL. Quantitation of covalently closed circular hepatitis B virus DNA in chronic hepatitis B patients. Hepatology. 2004;40:727-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 107] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 105. | Bourne EJ, Dienstag JL, Lopez VA, Sander TJ, Longlet JM, Hall JG, Kwiatkowski RW, Wright T, Lai CL, Condreay LD. Quantitative analysis of HBV cccDNA from clinical specimens: correlation with clinical and virological response during antiviral therapy. J Viral Hepat. 2007;14:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 43] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 106. | Wong DK, Yuen MF, Poon RT, Yuen JC, Fung J, Lai CL. Quantification of hepatitis B virus covalently closed circular DNA in patients with hepatocellular carcinoma. J Hepatol. 2006;45:553-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 107. | Yuen MF, Wong DK, Sum SS, Yuan HJ, Yuen JC, Chan AO, Wong BC, Lai CL. Effect of lamivudine therapy on the serum covalently closed-circular (ccc) DNA of chronic hepatitis B infection. Am J Gastroenterol. 2005;100:1099-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 108. | Yeh CT, Chiu HT, Chu CM, Liaw YF. G1 phase dependent nuclear localization of relaxed-circular hepatitis B virus DNA and aphidicolin-induced accumulation of covalently closed circular DNA. J Med Virol. 1998;55:42-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 109. | Zhang X, Lu W, Zheng Y, Wang W, Bai L, Chen L, Feng Y, Zhang Z, Yuan Z. In situ analysis of intrahepatic virological events in chronic hepatitis B virus infection. J Clin Invest. 2016;126:1079-1092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 110. | Inoue T, Tanaka Y. Novel biomarkers for the management of chronic hepatitis B. Clin Mol Hepatol. 2020;26:261-279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 92] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 111. | Testoni B, Lebossé F, Scholtes C, Berby F, Miaglia C, Subic M, Loglio A, Facchetti F, Lampertico P, Levrero M, Zoulim F. Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J Hepatol. 2019;70:615-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 217] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 112. | Zoulim F. New insight on hepatitis B virus persistence from the study of intrahepatic viral cccDNA. J Hepatol. 2005;42:302-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 191] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 113. | Werle-Lapostolle B, Bowden S, Locarnini S, Wursthorn K, Petersen J, Lau G, Trepo C, Marcellin P, Goodman Z, Delaney WE 4th, Xiong S, Brosgart CL, Chen SS, Gibbs CS, Zoulim F. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750-1758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 691] [Cited by in RCA: 733] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 114. | Wang Q, Luan W, Warren L, Fiel MI, Blank S, Kadri H, Tuvin D, Hiotis SP. Serum hepatitis B surface antigen correlates with tissue covalently closed circular DNA in patients with hepatitis B-associated hepatocellular carcinoma. J Med Virol. 2016;88:244-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 115. | Luo X, Huang Y, Chen Y, Tu Z, Hu J, Tavis JE, Huang A, Hu Y. Association of Hepatitis B Virus Covalently Closed Circular DNA and Human APOBEC3B in Hepatitis B Virus-Related Hepatocellular Carcinoma. PLoS One. 2016;11:e0157708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 116. | Fu S, Li N, Zhou PC, Huang Y, Zhou RR, Fan XG. Detection of HBV DNA and antigens in HBsAg-positive patients with primary hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2017;41:415-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 117. | Bai F, Yano Y, Fukumoto T, Takebe A, Tanaka M, Kuramitsu K, Anggorowati N, Rinonce HT, Widasari DI, Saito M, Hirano H, Hayakumo T, Seo Y, Azuma T, Ku Y, Hayashi Y. Quantification of pregenomic RNA and covalently closed circular DNA in hepatitis B virus-related hepatocellular carcinoma. Int J Hepatol. 2013;2013:849290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 118. | Ogura N, Watashi K, Noguchi T, Wakita T. Formation of covalently closed circular DNA in Hep38.7-Tet cells, a tetracycline inducible hepatitis B virus expression cell line. Biochem Biophys Res Commun. 2014;452:315-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 119. | Rybicka M, Woziwodzka A, Romanowski T, Stalke P, Dręczewski M, Bielawski KP. Differences in sequences between HBV-relaxed circular DNA and covalently closed circular DNA. Emerg Microbes Infect. 2017;6:e55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 120. | Bloom K, Maepa MB, Ely A, Arbuthnot P. Gene Therapy for Chronic HBV-Can We Eliminate cccDNA? Genes (Basel). 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 121. | Gish RG, Yuen MF, Chan HL, Given BD, Lai CL, Locarnini SA, Lau JY, Wooddell CI, Schluep T, Lewis DL. Synthetic RNAi triggers and their use in chronic hepatitis B therapies with curative intent. Antiviral Res. 2015;121:97-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 122. | Yuen MF, Schiefke I, Yoon JH, Ahn SH, Heo J, Kim JH, Lik Yuen Chan H, Yoon KT, Klinker H, Manns M, Petersen J, Schluep T, Hamilton J, Given BD, Ferrari C, Lai CL, Locarnini SA, Gish RG. RNA Interference Therapy With ARC-520 Results in Prolonged Hepatitis B Surface Antigen Response in Patients With Chronic Hepatitis B Infection. Hepatology. 2020;72:19-31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 123. | Cradick TJ, Keck K, Bradshaw S, Jamieson AC, McCaffrey AP. Zinc-finger nucleases as a novel therapeutic strategy for targeting hepatitis B virus DNAs. Mol Ther. 2010;18:947-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 146] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 124. | Weber ND, Stone D, Sedlak RH, De Silva Feelixge HS, Roychoudhury P, Schiffer JT, Aubert M, Jerome KR. AAV-mediated delivery of zinc finger nucleases targeting hepatitis B virus inhibits active replication. PLoS One. 2014;9:e97579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 88] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 125. | Bloom K, Ely A, Mussolino C, Cathomen T, Arbuthnot P. Inactivation of hepatitis B virus replication in cultured cells and in vivo with engineered transcription activator-like effector nucleases. Mol Ther. 2013;21:1889-1897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 169] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 126. | Li H, Sheng C, Wang S, Yang L, Liang Y, Huang Y, Liu H, Li P, Yang C, Yang X, Jia L, Xie J, Wang L, Hao R, Du X, Xu D, Zhou J, Li M, Sun Y, Tong Y, Li Q, Qiu S, Song H. Removal of Integrated Hepatitis B Virus DNA Using CRISPR-Cas9. Front Cell Infect Microbiol. 2017;7:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 127. | Seeger C, Sohn JA. Targeting Hepatitis B Virus With CRISPR/Cas9. Mol Ther Nucleic Acids. 2014;3:e216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 227] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 128. | Kennedy EM, Bassit LC, Mueller H, Kornepati AVR, Bogerd HP, Nie T, Chatterjee P, Javanbakht H, Schinazi RF, Cullen BR. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 185] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 129. | Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan DY, Dong LH, Song HF, Gao X. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015;22:404-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 187] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 130. | Li H, Sheng C, Liu H, Liu G, Du X, Du J, Zhan L, Li P, Yang C, Qi L, Wang J, Yang X, Jia L, Xie J, Wang L, Hao R, Xu D, Tong Y, Zhou Y, Zhou J, Sun Y, Li Q, Qiu S, Song H. An Effective Molecular Target Site in Hepatitis B Virus S Gene for Cas9 Cleavage and Mutational Inactivation. Int J Biol Sci. 2016;12:1104-1113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 131. | Seeger C, Sohn JA. Complete Spectrum of CRISPR/Cas9-induced Mutations on HBV cccDNA. Mol Ther. 2016;24:1258-1266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 109] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 132. | Cheng ST, Hu JL, Ren JH, Yu HB, Zhong S, Wai Wong VK, Kwan Law BY, Chen WX, Xu HM, Zhang ZZ, Cai XF, Hu Y, Zhang WL, Long QX, Ren F, Zhou HZ, Huang AL, Chen J. Dicoumarol, an NQO1 inhibitor, blocks cccDNA transcription by promoting degradation of HBx. J Hepatol. 2021;74:522-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 87] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 133. | Takeuchi F, Ikeda S, Tsukamoto Y, Iwasawa Y, Qihao C, Otakaki Y, Ryota O, Yao WL, Narita R, Makoto H, Watashi K, Wakita T, Takeuchi K, Chayama K, Kogure A, Kato H, Fujita T. Screening for inhibitor of episomal DNA identified dicumarol as a hepatitis B virus inhibitor. PLoS One. 2019;14:e0212233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 134. | Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M, Levrero M. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest. 2012;122:529-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 484] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 135. | Cheng J, Zhao Q, Zhou Y, Tang L, Sheraz M, Chang J, Guo JT. Interferon Alpha Induces Multiple Cellular Proteins That Coordinately Suppress Hepadnaviral Covalently Closed Circular DNA Transcription. J Virol. 2020;94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 136. | Zimmerman KA, Fischer KP, Joyce MA, Tyrrell DL. Zinc finger proteins designed to specifically target duck hepatitis B virus covalently closed circular DNA inhibit viral transcription in tissue culture. J Virol. 2008;82:8013-8021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 137. | Wei ZQ, Zhang YH, Ke CZ, Chen HX, Ren P, He YL, Hu P, Ma DQ, Luo J, Meng ZJ. Curcumin inhibits hepatitis B virus infection by down-regulating cccDNA-bound histone acetylation. World J Gastroenterol. 2017;23:6252-6260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 68] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 138. | Sukowati CH, El-Khobar KE, Ie SI, Anfuso B, Muljono DH, Tiribelli C. Significance of hepatitis virus infection in the oncogenic initiation of hepatocellular carcinoma. World J Gastroenterol. 2016;22:1497-1512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 49] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |