Published online Dec 27, 2021. doi: 10.4254/wjh.v13.i12.2039
Peer-review started: May 28, 2021
First decision: July 6, 2021
Revised: July 19, 2021
Accepted: November 15, 2021
Article in press: November 15, 2021
Published online: December 27, 2021
Processing time: 212 Days and 8.3 Hours
Hepatocellular carcinoma (HCC) is among the leading causes of cancer incidence and death. Despite decades of research and development of new treatment options, the overall outcomes of patients with HCC continue to remain poor. There are areas of unmet need in risk prediction, early diagnosis, accurate pro
Core Tip: There are emerging roles for deep learning technology in the field of hepatocellular carcinoma (HCC) including HCC risk prediction, as well as diagnosis, prognostication, and treatment planning leveraging readily available data from radiologic and histopathologic medical images. This article will provide a comprehensive review of the recently published studies that have applied deep learning for risk prediction, diagnosis, prognostication, and treatment planning for patients with HCC.
- Citation: Ahn JC, Qureshi TA, Singal AG, Li D, Yang JD. Deep learning in hepatocellular carcinoma: Current status and future perspectives. World J Hepatol 2021; 13(12): 2039-2051
- URL: https://www.wjgnet.com/1948-5182/full/v13/i12/2039.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i12.2039
Hepatocellular carcinoma (HCC) is an aggressive primary liver cancer that develops in the setting of chronic parenchymal liver diseases, and is among the top causes of cancer incidence and mortality worldwide[1,2]. While the burden of HCC has been declining with effective antiviral therapy against hepatitis B virus (HBV) and hepatitis C virus (HCV), HCC incidence related to metabolic syndrome will likely continue to rise due to the dramatic increase in the prevalence of non-alcoholic fatty liver disease (NAFLD) in the general population[3]. Decades of HCC research led to the deve
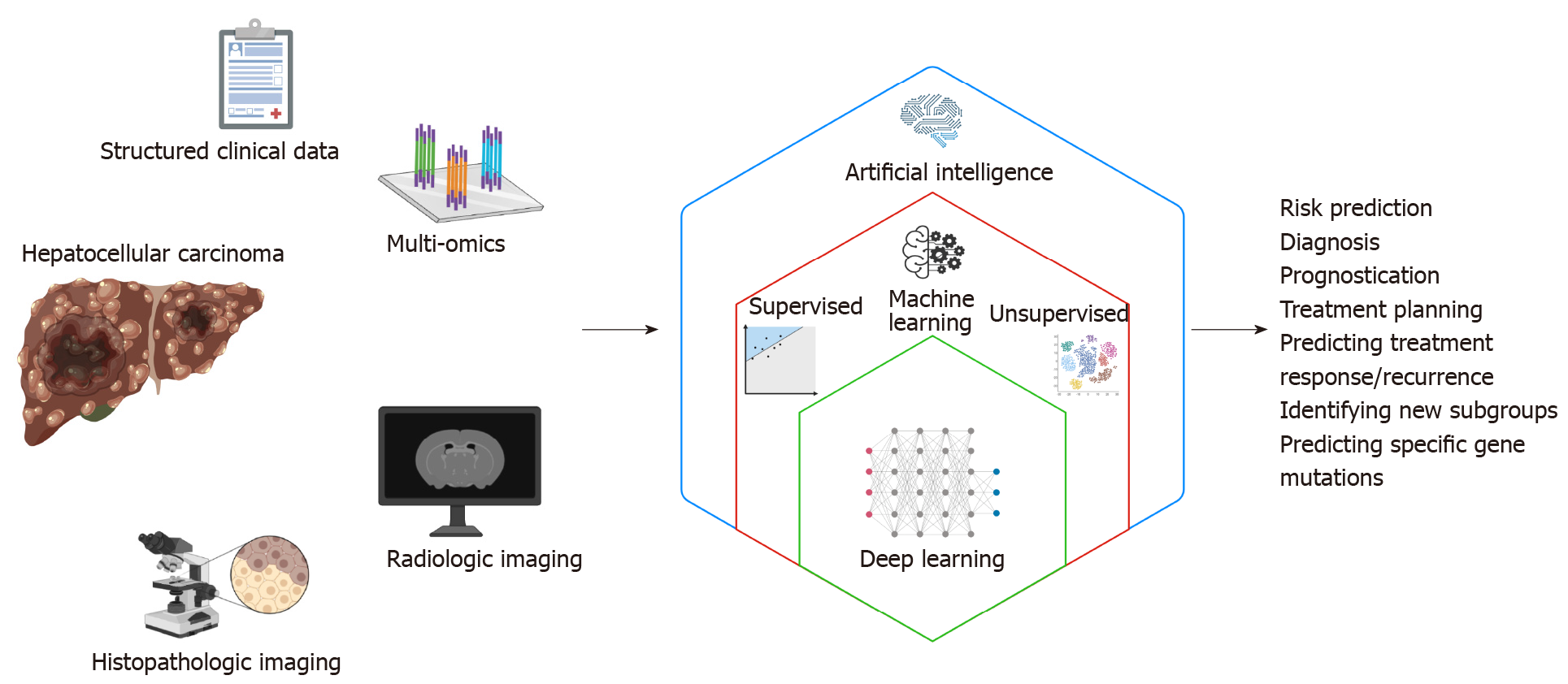
Patients with HCC generate enormous amounts of health data. While promising for researchers, ensuring that such high volumes of data are turned into actionable knowledge can be a significant challenge. Artificial intelligence (AI) is thought to be capable of synthesizing and analyzing multimodal data with superhuman degrees of accuracy or reliability, and recent years have seen a rapid growth in the application of AI to many fields of medicine including hepatology[6]. This “AI revolution” over the past decade has been possible due to the advent of deep learning technology. Deep learning algorithms can process a broad spectrum of medical data from structured numeric data such as vital signs and laboratory values, high dimensional data from multi-omics studies, as well as digitized high-resolution images from various radiologic and histopathologic studies. This review aims to provide an overview as well as highlight examples of the many potential applications of deep learning to improve the care of patients with HCC.
AI-based approaches provide a variety of methods for a range of tasks and clinical application including image classification, organ and lesion segmentation, accurate extraction of key imaging features and measurements, tumor detection, stratification of high-risk subjects, prediction of disease and treatment outcome (Figure 1). Advan
The term “artificial intelligence” encompasses a broad range of technology that enables machines to perform tasks typically thought to require human reasoning and problem-solving skills[7]. “Machine learning” is a branch of AI in which computer algorithms train on sample data to build a mathematical model that makes predictions or decisions without being explicitly programmed to do so[8]. Machine learning algorithms can be broadly divided into supervised and unsupervised learning. Supervised learning algorithms train on sample data with labeled outcome data, and their goal is to learn the relationship between the input data and the outcomes to make accurate predictions about the outcome when provided with a new set of input data[9]. Examples of supervised learning algorithms include traditional techniques such as linear regression and logistic regression, as well as more sophisticated techniques including support vector machines, random forest and gradient boosting. On the other hand, unsupervised learning algorithms train on unlabeled sample data and analyze the underlying structure or distribution within the data to discover new clusters or patterns[10]. Examples of unsupervised learning algorithms include K-means and principle component analysis among many others.
Among the various AI-based machine learning algorithms, artificial neural networks (ANNs) consist of layers of interconnected mathematical formulas that enable them to analyze complex non-linear relationships[11]. “Deep learning (DL)” refers to highly complex AI models utilizing multiple layers of ANNs and has recently emerged as a state-of-the-art AI technique for analyzing complex, high-dimensional healthcare data. Some of the commonly used DL techniques include convolutional neural networks (CNNs) and recurrent neural networks (RNNs)[12]. CNNs have connective patterns resembling those of an animal visual cortex and can detect inherent spatial features of high dimensional images. RNNs have connections forming a directed graph along a temporal sequence, and therefore can be highly useful in time series prediction.
It is crucial to recognize that any AI-based machine learning algorithms require external validation in an independent dataset as models could be overfitted and end up overestimating the performance. In this review article, the performance characteristics of the various DL models are from the validation cohorts, and not the original derivation cohorts used to train the algorithms.
Despite multiple available risk prediction tools for HCC, none have been rigorously validated or endorsed by major liver societies. Currently, HCC surveillance is recom
Serum AFP has been widely used as a predictive and prognostic biomarker for HCC[18], but AFP has limited sensitivity for detecting early-stage HCC and its levels do not reliably correlate with disease progression[19]. Recent advances in multi-omics related to HCC are expected to address this unmet need for novel biomarkers. Multi-omics refers to an approach to biological analysis which utilizes data sets from multiple "omics", such as the genome, epigenome, transcriptome, proteome, metabolome and microbiome. Multi-omics experiments generate an enormous amount of information, and various machine learning techniques including DL that can help with the computational challenges of processing and analyzing such high dimensional data. Xie et al[20] used gene expression profiling of peripheral blood to build an ANN model that classifies HCC patients from a control group. Using a nine-gene expression system, the ANN was able to distinguish HCC patients from controls with an AUC of 0.943, 98% sensitivity, and 85% specificity, although it should be noted that the control group was healthy individuals rather than patients with cirrhosis, which could have overestimated the performance of the model. Choi et al[21] proposed a novel network-based DL method to identify prognostic gene signatures via G2Vec, a modified Word2Vec model originally used for natural language processing (NLP). When applied to gene expression data for HCC from the Cancer Genome Atlas (TCGA), G2Vec showed superior prediction accuracy for patient outcomes compared to existing gene selection methods and was able to identify two distinct gene modules significantly associated with HCC prognosis. Chaudhary et al[22] used RNA se×quencing, miRNA, and methylation data of 360 HCC patients from TCGA to build an autoencoder, which is an unsupervised feed-forward neural network. Using this DL model, they were able to distinguish patients with survival differences and identify specific mutations and pathways as predictors of aggressive tumor behavior.
In recent years, there have been remarkable advances in the application of AI for the interpretation of medical imaging, primarily due to the use of DL algorithms using CNN[23]. CNN algorithms trained on ultrasound, computed tomography (CT), or magnetic resonance imaging (MRI) images have shown excellent performances in detection of lesions, classification of lesions, segmentation of organs or anatomic structures, and imaging reconstruction[24].
In 2012, Streba et al[25] prospectively studied contrast-enhanced ultrasound images of 112 patients to train an ANN that classified five different types of liver tumors. The ANN showed promising performances with accuracies of 94.5% in the training set and 87.1% in the testing set. In 2017, Hassan et al[26] reported using the stacked sparse auto-encoder, an unsupervised DL technique, to segment and classify liver lesions on ultrasound images with a classification accuracy of 97.2%. Additionally, Bharti et al[27] built a CNN using echotexture and roughness of liver surface on 754 segmented ultrasound images, which differentiated between normal liver, chronic liver disease, cirrhosis, and HCC with a classification accuracy of 96.6%. Schmauch et al[28] also created a CNN which detects and characterizes benign and malignant focal liver lesions on 2-D ultrasound images from 367 patients from various institutions. When applied to a new dataset of 177 patients, the model achieved a weighted mean AUC of 0.891. Recently, Brehar et al[29] conducted a study comparing CNN’s performance for HCC detection on ultrasound images against conventional machine learning alg
In addition to ultrasound images, cross-sectional imaging from CT or MRI studies serve as an extremely abundant and promising source of data for DL. In 2018, Yasaka et al[31] used CT image sets of liver masses from 460 patients to train a CNN that can classify liver lesions into five categories of: (1) HCC; (2) Other malignant tumors; (3) Indeterminate masses; (4) Hemangiomas; and (5) Cysts with a median AUC of 0.92. Shi et al[32] showed that incorporation of a CNN enabled identification of HCC using a three-phase CT imaging protocol with a diagnostic accuracy similar to that of a four-phase protocol, which would allow patients to receive lower doses of radiation. Segmentation of HCC, liver parenchyma, and other organs on CT scan is very important for determination of tumor extent and treatment planning, but manual contouring of the images is highly time-consuming and subject to inter-observer variability. The 2017 International Conference On Medical Image Computing Computer Assisted Intervention called for a Liver Tumor Segmentation Benchmark (LITS) challenge, encouraging researchers to develop automatic segmentation algorithms to segment liver lesions using 200 CT scans (training: 130; testing: 70) provided by clinical sites around the world. Several teams participating in the challenge have developed DL algorithms with promising performances for HCC segmentation using CT images[33-37]. Beyond the LITS challenge, there are ongoing research efforts to improve segmentation using different architectures of DL networks[38-42].
Hamm et al[43] used MRI images from 494 patients to train a CNN which can classify hepatic lesions into six different categories. When applied to random cases in the test set, the CNN outperformed expert radiologists (90% sensitivity and 98% specificity vs 82.5% sensitivity and 96.5% specificity) and especially for HCC detection (90% sensitivity vs 60%-70% sensitivity). The same group conducted additional studies to make their CNN interpretable by generating highlighted feature maps corresponding to liver lesions[44]. Wu et al[45] built a CNN using multiphase MRI images and achieved an AUC of 0.95 for distinguishing Liver Imaging Reporting and Data System (LI-RADS) grade 3 from LI-RADS 4 and 5 lesions for HCC diagnosis. Zhen et al[46] also trained a CNN model combining unenhanced MRI images and clinical variables from 1210 patients with liver tumors, which demonstrated diagnostic performances on par with three experienced radiologists using enhanced MRI images.
In addition to serving as accurate and efficient tools for diagnosis of HCC, DL models utilizing radiology data can also be used for prognostication, treatment planning, and assessing tumor response to therapy. Vascular invasion is a key prognostic element in patients with HCC. Recent studies developed CNN models with promising ability to detect microvascular invasion on MRI images of HCC patients undergoing surgical resection[47-49]. An et al[50] used an unsupervised CNN-based deformable image registration technique to assess the relationship between ablative margins and local tumor progression in 141 patients with single HCC who underwent microwave ablation, and demonstrated that patients with ablative margins < 5 mm were at significantly higher risk of local tumor progression. Liu et al[51] developed a DL radiomics model to predict responses to trans-arterial chemoembolization (TACE) using ultrasound images of 130 HCC patients, which accurately predicted TACE response with an AUC of 0.93. The same group also assessed their ultrasound-based DL radiomics model to predict 2-year progression-free survival among 419 HCC patients and facilitate optimized treatment selection. Peng et al[52] trained a residual CNN model to predict response to TACE using CT images from 562 patients with intermediate-stage HCC undergoing TACE, which showed accuracies of 85.1% and 82.8% in two external validation cohorts. Another study developed a DL score for disease-specific survival by using CT images in a cohort of 243 patients with HCC treated with TACE, with a higher score predicting poor prognosis [hazard ratio (HR): 3.01; 95% cumulative incidence (CI): 2.02-4.50][53]. Finally, Zhang et al[54] built a DL-based model predicting overall survival using CT images from 201 patients with unresectable HCC treated with TACE and sorafenib, which achieved superior predictive performance compared to the clinical nomogram (C-index of 0.730 vs 0.679, P = 0.023).
Automated interpretation of histopathologic images from liver biopsy is another major area of medical imaging in patients with HCC where DL can be utilized. In addition to effectively replicating the human pathologists’ jobs of diagnosing and grading HCC, DL models can help identify and analyze additional complex imaging features and patterns which are related to specific mutations and disease prognosis. Lin et al[55] used images from multiphoton microscopy of 113 HCC patients to train a CNN with over 90% accuracy for determining HCC differentiation. Kiani et al[56] developed a CNN-based “Liver Cancer Assistant” which accurately differentiated hematoxylin and eosin (H&E) images of HCC and cholangiocarcinoma and helped improve the diagnostic performance of nine pathologists. Liao et al[57] used TCGA dataset for training a CNN that distinguished HCC from adjacent normal tissues with perfect performance (AUC: 1.00) and predicted the presence of specific somatic mutations with AUCs over 0.70. Wang et al[58] trained a CNN for automated segmentation and classification of individual nuclei at single-cell levels on H&E-stained tissue sections of HCC tumors from TCGA, and performed feature extraction to identify 246 quan
Lu et al[60] applied three pre-trained CNN models to extract imaging features from HCC histopathology and performed Cox proportional hazards analysis to predict overall survival and disease-free survival, and observed significant correlations between the imaging features and established biological pathways. Saillard et al[61] used two DL algorithms based on whole-side digitized histological slides from 194 patients with HCC to predict the survival of patients treated by surgical resection. When tested on an independent validation set from TCGA, both DL models had a higher discriminatory power than a score combining all baseline variables associated with survival. Shi et al[62] built an interpretable DL framework using pathologic images from 1445 patients with HCC and developed a “tumor risk score” which showed prognostic performances independent of and superior to clinical staging systems and stratified patients into five groups of different prognosis. A recent study by Yamashita et al[63] developed a histopathology-based DL based system which stratified patients with risk scores for postsurgical recurrence of HCC.
There are several key issues to address before DL-based AI models can be universally implemented in real world clinical practice settings. Due to their complexity, DL models are traditionally considered to be “black-box” models, meaning humans cannot understand how the DL models make their predictions. Interpretability of the DL models are crucial for physicians to accept and trust them in everyday clinical practice, and for troubleshooting and improving the models for rare cases. This is being addressed by recent developments in various “explainable AI” techniques but currently there is no clear consensus on the best methodology. Another potential limitation is the generalizability of the individual DL algorithms. Concerns have been raised that AI algorithms developed at highly specialized academic medical centers using their own patients’ data may over-represent certain groups of patients and not accurately reflect the real-world population of patients seen at local community hospitals. Finally, AI models, like other prediction models, are often not publicly available, limiting external validation. Independent validation of the proposed model and comparison to old models are as important as deriving new models. Large-scale, prospective, multi-centered studies involving diverse populations with external validation will be necessary before DL algorithms can be widely accepted.
A currently under-explored, but highly promising and exciting area for the application of DL is the field of autonomous robotics. In a recent editorial, Gumbs et al[64] state that while the current form of robotic surgery seems like a form of minimally invasive surgery, the true power of robotic surgery exists in its potential to create autonomous actions. Recently, a DL-based surgical instrument tracking algorithm was able to closely track the instruments during robotic surgery and evaluate the surgeons’ performance, demonstrating that DL algorithms can learn the correct steps of robotic surgery[65]. With the help of DL and other AI technologies, it may be possible to imagine a future where fully autonomous robots perform resection of large, complex HCC in ways that no human surgeons can mimic. However, there are significant barriers before the idea of fully autonomous robotic surgery can become a reality, including the current technical limitations of autonomous surgical robotics, as well as the hesitation of patients and providers to fully trust autonomous robots to perform invasive operations. “Explainability” of the DL algorithms will be critical here, as humans would need to be able to understand and correct every single mistake that an autonomous robot makes during surgery. Therefore, for the foreseeable future, DL will most likely remain as a helpful, adjunctive tool to assist human surgeons.
This review has provided a comprehensive overview of various ways in which DL algorithms can be employed to assist medical providers and enhance the care of patients with HCC (Table 1). DL algorithms not only can efficiently and accurately replicate the same jobs performed by human physicians, but more importantly can help discover novel biologic pathways and disease subgroups with clinical sig
| Study | Cohort | Data source | Deep learning | Input | Output | Main findings |
| Predicting HCC risk using clinical variables | ||||||
| Ioannou et al[14] 2020 | 48151 HCV cirrhosis (T: 90%, V: 10%) | VHA database | RNN | Clinical variables | Risk of HCC development | RNN predicted HCC development with AUC of 0.759, and AUC of 0.806 among those who achieved SVR |
| Phan et al[15] 2020 | 6052 HBV and HCV (T: 70%, V: 30%) | Taiwanese NHIRD | CNN | Disease history data | Risk of HCC development | CNN achieved an accuracy of 0.980 and AUC of 0.886 for predicting HCC development among viral hepatitis patients |
| Nam et al[16] 2020 | T: 424 HBV cirrhosis; V: 316 HBV cirrhosis | 2 Korean centers | ResNet | Clinical variables | Risk of HCC development | DL model achieved an accuracy of 0.763 and AUC of 0.782 in the validation cohort and outperformed previous models |
| Nam et al[17] 2020 | T: 349 LT recipients; V: 214 LT recipients | 3 Korean LT centers | ResNet | Clinical variables | Recurrent HCC after LT | DL model significantly outperformed conventional models in prediction of post-T HCC recurrence with AUC of 0.75 |
| Multi-omics-based HCC diagnosis and prognostication | ||||||
| Xie et al[20] 2018 | T: 133 HCC/54 HV; V: 52 HCC/34 HV | 1 center in China | ANN | Gene expression | HCC detection | ANN using nine genes had an AUC of 0.943, 98% sensitivity, and 85% specificity for classifying HCC |
| Choi et al[21] 2018 | 135 HCC (10-fold CV) | TCGA | G2Vec | Gene expression | HCC prognosis | G2Vec showed significantly higher prediction accuracy for patient outcomes compared to existing gene selection tools |
| Chaudhary et al[22] 2018 | T: 360 HCC; V: 220, 221, 166, 40, 27 HCC | TCGA; 5 external datasets | Auto-encoder | RNA-seq, miRNA-seq, methylation | HCC prognosis | DL model distinguished groups with survival differences and identified mutations and pathways predicting aggressive tumor behavior |
| Radiology-based HCC diagnosis/prediction | ||||||
| Streba et al[25] 2012 | 112 FLL (10-fold CV) | 1 center in Romania | ANN | US images | FLL type | ANN had 87.12% testing accuracy, 93.2% sensitivity, and 89.7% specificity for classifying 5 classes of liver lesions |
| Hassan et al[26] 2017 | 110 FLL (10-fold CV) | 1 center in Egypt | Auto-encoder | US images | FLL type | The proposed system had 97.2% accuracy, 98% sensitivity, and 95.70% specificity for classifying liver lesions |
| Bharti et al[27] 2018 | 24 normal, 25 CLD, 25 cirrhosis, 20 HCC | 1 center in India | CNN | US images | Liver stages | CNN achieved 96.6% classification accuracy for differentiating normal liver, CLD, cirrhosis, and HCC |
| Schmauch et al[28] 2019 | T: 367 FLL; V: 177 FLL | Centers in France | ResNet | US images | FLL type | DL model reached mean AUC of 0.935 for focal liver lesion detection and 0.916 for focal liver lesion characterization |
| Brehar et al[29] 2020 | T: 200 HCC; V: 68 HCC | 1 center in Romania | CNN | US images | HCC detection | CNN achieved AUC of 0.95, accuracy of 0.91, 94.4% sensitivity and 88.4% specificity for HCC detection |
| Jin et al[30] 2021 | 434 HBV (3:1:1 split) | 1 center in China | DL radiomics | US images | Risk of HCC development | DL radiomics model predicted 5-yr HCC development risk with AUC of 0.900 in the test set |
| Yasaka et al[31] 2018 | T: 460 liver masses; V: 100 liver masses | 1 center in Japan | CNN | CT images | Liver mass type | CNN classified liver lesions into five categories with a median AUC of 0.92 |
| Shi et al[32] 2020 | 449 FLL; (T: 80%, V: 20%) | 1 center in China | CNN | CT images | FLL type | CNN applied to three-phase CT protocol images achieved AUC of 0.925 for differentiating HCC from other FLLs |
| Hamm et al[43] 2019 | T: 434 FLL; V: 60 FLL | 1 center in United States | CNN | MRI images | FLL type | CNN achieved 90% sensitivity and 98% specificity for classifying FLLs and AUC of 0.992 for HCC classification |
| Wang et al[44] 2019 | T: 434 FLL; V: 60 FLL | 1 center in United States | CNN | MRI images | FLL type | Interpretable DL system achieved 76.5% PPV and 82.9% sensitivity for identifying correct radiological features |
| Wu et al[45] 2020 | 89 liver tumors; (60: 20: 20) | 1 center in United States | CNN | MRI images | LI-RADS grading | CNN achieved AUC of 0.95, 90% accuracy, 100% sensitivity and 83.5% PPV for LI-RADS grading of liver tumors |
| Zhen et al[46] 2020 | T: 1210 liver tumors; V: 201 liver tumors | 1 center in China | CNN | MRI images | Liver tumor type | CNN combined with clinical data showed AUC of 0.985 for classifying HCC with 91.9% agreement with pathology |
| Radiology-based HCC prognostication, treatment planning, and response to treatment | ||||||
| Zhang et al[47] 2021 | T: 158 HCC; V: 79 HCC | 1 center in China | CNN | MRI images | MVI in HCC | CNN achieved AUC of 0.72, 55% sensitivity, and 81% specificity for preoperative MVI in HCC patients |
| Wang et al[48] 2020 | T: 60 HCC; V: 40 HCC | 1 center in China | CNN | MRI images | MVI in HCC | Fusion of deep features from MRI images yielded AUC of 0.79 for MVI prediction in HCC patients |
| Jiang et al[49] 2021 | 405 HCC; (T: 80%, V: 20%) | 1 center in China | CNN | CT images | MVI in HCC | CNN achieved AUC of 0.906 for prediction of MVI. Mean survival was significantly better in the group without MVI |
| An et al[50] 2020 | 141 single HCC resect MWA | 1 center in China | CNN | MRI images | Ablative margin | Deep learning model accurately estimated ablative margins and risk of local tumor progression |
| Liu et al[51] 2020 | T: 89 HCC resect TACE; V: 41 HCC rec. TACE | 1 center in China | CNN | Ultrasound images | Response to TACE | Deep learning radiomics model predicted tumor response to TACE with AUC of 0.93 |
| Peng et al[52] 2020 | T: 562 HCC resect TACE; V:227 HCC rec. TACE | 3 centers in China | CNN | CT images | Response to TACE | Deep learning model had accuracies of 85.1% and 82.8% for predicting TACE response in 2 validation cohorts |
| Liu et al[53] 2020 | 243 HCC resect TACE (6:1:3 split) | 1 center in China | CNN | CT images | Post-TACE survival | Higher DL score was an independent prognostic factor and predicted overall survival with AUCs of 0.85-0.90 |
| Zhang et al[54] 2020 | 201 HCC resect TACE + sorafenib (T: 120, V: 81) | 3 centers in China | CNN | CT images | OS on TACE + sorafenib | Deep learning signature achieved C-index of 0.714 for predicting OS in HCC patients receiving TACE + sorafenib |
| Histopathology-based HCC diagnosis, subtyping, and outcome predictions | ||||||
| Lin et al[55]2019 | 113 HCC | 1 center in China | CNN | Histopath images | HCC differentiation | CNN achieved an accuracy of 0.941 for determining HCC differentiation on multiphoton microscopy |
| Kiani et al[56] 2020 | 70 WSI (35 HCC, 35 CC) | TCGA | CNN | Histopath images | HCC vs CC | CNN-based “Liver Cancer Assistant” accurately differentiated HCC vs cholangiocarcinoma |
| Liao et al[57] 2020 | T: 491 HCC; V: 455 HCC | TCGA; 1 center in China | CNN | Histopath images | HCC detection, mutations | CNN distinguished HCC from adjacent tissues with AUC of 1.00 and predicted specific mutations with AUC over 0.70 |
| Wang et al[58] 2020 | T: 99 HCC; V: 205 HCC | TCGA | CNN | Histopath images | Histological HCC subtype | Unsupervised clustering identified 3 histological subtypes complementing molecular pathways and prognostic value |
| Chen et al[59] 2020 | T: 402 HCC/89 normal; V: 67 HCC/34 normal | GDC portal; 1 center in China | CNN | Histopath images | HCC grade mutations | CNN achieved 89.6% accuracy for tumor differentiation stage and predicted presence of specific gene mutations |
| Lu et al[60] 2020 | 421 HCC/105 normal (6-fold CV) | GDC portal | CNN | Histopath images | HCC prognosis | Pre-trained CNN predicted OS using pathology images and identified HCC subgroups with different prognosis |
| Saillard et al[61] 2020 | T: 194 HCC; V: 328 HCC | 1 French center TCGA | CNN | Histopath images | Survival after HCC resection | CNN models using pathology images predicted survival with C-index 0.75-0.78 and outperformed conventional models |
| Shi et al[62] 2021 | T: 1125 HCC; V: 320 HCC | 1 center in China; TCGA | CNN | Histopath images | HCC outcomes | Deep learning-based “tumor risk score” was superior to clinical staging and stratified 5 groups of different prognosis |
| Yamashita et al[63] 2021 | T: 36 WSI; V: 30 WSI | 1 center in United States; TCGA | CNN | Histopath images | Post-surgical recurrence | CNN risk scores outperformed TNM system for predicting recurrence and identified high-and low-risk subgroups |
Despite some important limitations to overcome, application of state-of-the-art AI technologies such as DL for the care of patients with HCC is no longer a futuristic idea but is rapidly becoming a reality. Most of the studies covered in this review were published within the past two years, and the number of studies utilizing DL continues to increase exponentially. We anticipate that DL algorithms will soon take a major role in the diagnosis, prognostication, and treatment of patients with HCC.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: Cedars-Sinai Medical Center, Cedars-Sinai Medical Center.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Gumbs A, Shafqat S S-Editor: Wang JJ L-Editor: Webster JR P-Editor: Wang JJ
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55615] [Article Influence: 7945.0] [Reference Citation Analysis (131)] |
| 2. | Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2184] [Cited by in RCA: 2862] [Article Influence: 477.0] [Reference Citation Analysis (17)] |
| 3. | Stepanova M, De Avila L, Afendy M, Younossi I, Pham H, Cable R, Younossi ZM. Direct and Indirect Economic Burden of Chronic Liver Disease in the United States. Clin Gastroenterol Hepatol. 2017;15:759-766.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 118] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 4. | Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1343] [Cited by in RCA: 3777] [Article Influence: 944.3] [Reference Citation Analysis (3)] |
| 5. | Yang JD, Heimbach JK. New advances in the diagnosis and management of hepatocellular carcinoma. BMJ. 2020;371:m3544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 240] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 6. | Ahn JC, Connell A, Simonetto DA, Hughes C, Shah VH. Application of Artificial Intelligence for the Diagnosis and Treatment of Liver Diseases. Hepatology. 2021;73:2546-2563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 93] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 7. | Haenlein M, Kaplan A. A Brief History of Artificial Intelligence: On the Past, Present, and Future of Artificial Intelligence. Calif Manage Rev. 2019;. [DOI] [Full Text] |
| 8. | Koza JR, Bennett FH, Andre D, Keane MA. Automated Design of Both the Topology and Sizing of Analog Electrical Circuits Using Genetic Programming. In: Gero JS, Sudweeks F. Artificial Intelligence in Design ’96. Dordrecht: Springer Netherlands, 1996: 151-170. |
| 9. | Cunningham P, Cord M, Delany SJ. Supervised Learning. In: Cord M, Cunningham P. Machine Learning Techniques for Multimedia: Case Studies on Organization and Retrieval. Berlin, Heidelberg: Springer Berlin Heidelberg, 2008: 21-49. |
| 10. | Ghahramani Z, Unsupervised Learning. In: Bousquet O, von Luxburg U, Rätsch G. Advanced Lectures on Machine Learning: ML Summer Schools 2003, Canberra, Australia, February 2 - 14, 2003, Tübingen, Germany, August 4 - 16, 2003, Revised Lectures. Berlin, Heidelberg: Springer Berlin Heidelberg, 2004: 72-112. |
| 11. | Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S, Dean J. A guide to deep learning in healthcare. Nat Med. 2019;25:24-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1123] [Cited by in RCA: 1472] [Article Influence: 245.3] [Reference Citation Analysis (0)] |
| 12. | Puttagunta M, Ravi S. Medical image analysis based on deep learning approach. Multimed Tools Appl. 2021;1-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 64] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 13. | Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, Roberts LR, Heimbach JK. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68:723-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2121] [Cited by in RCA: 3208] [Article Influence: 458.3] [Reference Citation Analysis (1)] |
| 14. | Ioannou GN, Tang W, Beste LA, Tincopa MA, Su GL, Van T, Tapper EB, Singal AG, Zhu J, Waljee AK. Assessment of a Deep Learning Model to Predict Hepatocellular Carcinoma in Patients With Hepatitis C Cirrhosis. JAMA Netw Open. 2020;3:e2015626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 87] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 15. | Phan DV, Chan CL, Li AA, Chien TY, Nguyen VC. Liver cancer prediction in a viral hepatitis cohort: A deep learning approach. Int J Cancer. 2020;147:2871-2878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 16. | Nam JY, Sinn DH, Bae J, Jang ES, Kim JW, Jeong SH. Deep learning model for prediction of hepatocellular carcinoma in patients with HBV-related cirrhosis on antiviral therapy. JHEP Rep. 2020;2:100175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (1)] |
| 17. | Nam JY, Lee JH, Bae J, Chang Y, Cho Y, Sinn DH, Kim BH, Kim SH, Yi NJ, Lee KW, Kim JM, Park JW, Kim YJ, Yoon JH, Joh JW, Suh KS. Novel Model to Predict HCC Recurrence after Liver Transplantation Obtained Using Deep Learning: A Multicenter Study. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 18. | Behne T, Copur MS. Biomarkers for hepatocellular carcinoma. Int J Hepatol. 2012;2012:859076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, Waljee AK, Singal AG. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients With Cirrhosis: A Meta-analysis. Gastroenterology. 2018;154:1706-1718.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 638] [Cited by in RCA: 798] [Article Influence: 114.0] [Reference Citation Analysis (0)] |
| 20. | Xie H, Xue YQ, Liu P, Zhang PJ, Tian ST, Yang Z, Guo Z, Wang HM. Multi-parameter gene expression profiling of peripheral blood for early detection of hepatocellular carcinoma. World J Gastroenterol. 2018;24:371-378. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 6] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 21. | Choi J, Oh I, Seo S, Ahn J. G2Vec: Distributed gene representations for identification of cancer prognostic genes. Sci Rep. 2018;8:13729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 22. | Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res. 2018;24:1248-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 579] [Article Influence: 82.7] [Reference Citation Analysis (0)] |
| 23. | Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, Kadoury S, Tang A. Deep Learning: A Primer for Radiologists. Radiographics. 2017;37:2113-2131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 511] [Cited by in RCA: 686] [Article Influence: 98.0] [Reference Citation Analysis (0)] |
| 24. | Azer SA. Deep learning with convolutional neural networks for identification of liver masses and hepatocellular carcinoma: A systematic review. World J Gastrointest Oncol. 2019;11:1218-1230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 70] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (4)] |
| 25. | Streba CT, Ionescu M, Gheonea DI, Sandulescu L, Ciurea T, Saftoiu A, Vere CC, Rogoveanu I. Contrast-enhanced ultrasonography parameters in neural network diagnosis of liver tumors. World J Gastroenterol. 2012;18:4427-4434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 53] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 26. | Hassan TM, Elmogy M, Sallam E-S. Diagnosis of Focal Liver Diseases Based on Deep Learning Technique for Ultrasound Images. Arab J Sci Eng. 2017;42:3127-3140. [RCA] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (1)] |
| 27. | Bharti P, Mittal D, Ananthasivan R. Preliminary Study of Chronic Liver Classification on Ultrasound Images Using an Ensemble Model. Ultrason Imaging. 2018;40:357-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 28. | Schmauch B, Herent P, Jehanno P, Dehaene O, Saillard C, Aubé C, Luciani A, Lassau N, Jégou S. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging. 2019;100:227-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 29. | Brehar R, Mitrea DA, Vancea F, Marita T, Nedevschi S, Lupsor-Platon M, Rotaru M, Badea RI. Comparison of Deep-Learning and Conventional Machine-Learning Methods for the Automatic Recognition of the Hepatocellular Carcinoma Areas from Ultrasound Images. Sensors (Basel). 2020;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 30. | Jin J, Yao Z, Zhang T, Zeng J, Wu L, Wu M, Wang J, Wang Y, Yu J, Zheng R. Deep learning radiomics model accurately predicts hepatocellular carcinoma occurrence in chronic hepatitis B patients: a five-year follow-up. Am J Cancer Res. 2021;11:576-589. [PubMed] |
| 31. | Yasaka K, Akai H, Abe O, Kiryu S. Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology. 2018;286:887-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 386] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 32. | Shi W, Kuang S, Cao S, Hu B, Xie S, Chen S, Chen Y, Gao D, Zhu Y, Zhang H, Liu H, Ye M, Sirlin CB, Wang J. Deep learning assisted differentiation of hepatocellular carcinoma from focal liver lesions: choice of four-phase and three-phase CT imaging protocol. Abdom Radiol (NY). 2020;45:2688-2697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 33. | Alirr OI. Deep learning and level set approach for liver and tumor segmentation from CT scans. J Appl Clin Med Phys. 2020;21:200-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 34. | Chlebus G, Schenk A, Moltz JH, van Ginneken B, Hahn HK, Meine H. Automatic liver tumor segmentation in CT with fully convolutional neural networks and object-based postprocessing. Sci Rep. 2018;8:15497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 116] [Article Influence: 16.6] [Reference Citation Analysis (1)] |
| 35. | Deng Z, Guo Q, Zhu Z. Dynamic Regulation of Level Set Parameters Using 3D Convolutional Neural Network for Liver Tumor Segmentation. J Healthc Eng. 2019;2019:4321645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 36. | Wardhana G, Naghibi H, Sirmacek B, Abayazid M. Toward reliable automatic liver and tumor segmentation using convolutional neural network based on 2.5D models. Int J Comput Assist Radiol Surg. 2021;16:41-51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 37. | Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation From CT Volumes. IEEE Trans Med Imaging. 2018;37:2663-2674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1078] [Cited by in RCA: 861] [Article Influence: 123.0] [Reference Citation Analysis (0)] |
| 38. | Ahn SH, Yeo AU, Kim KH, Kim C, Goh Y, Cho S, Lee SB, Lim YK, Kim H, Shin D, Kim T, Kim TH, Youn SH, Oh ES, Jeong JH. Comparative clinical evaluation of atlas and deep-learning-based auto-segmentation of organ structures in liver cancer. Radiat Oncol. 2019;14:213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 39. | Almotairi S, Kareem G, Aouf M, Almutairi B, Salem MA. Liver Tumor Segmentation in CT Scans Using Modified SegNet. Sensors (Basel). 2020;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 40. | Ayalew YA, Fante KA, Mohammed MA. Modified U-Net for liver cancer segmentation from computed tomography images with a new class balancing method. BMC Biomed Eng. 2021;3:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 41. | Budak Ü, Guo Y, Tanyildizi E, Şengür A. Cascaded deep convolutional encoder-decoder neural networks for efficient liver tumor segmentation. Med Hypotheses. 2020;134:109431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 42. | Chen Y, Wang K, Liao X, Qian Y, Wang Q, Yuan Z, Heng PA. Channel-Unet: A Spatial Channel-Wise Convolutional Neural Network for Liver and Tumors Segmentation. Front Genet. 2019;10:1110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 43. | Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Duncan JS, Weinreb JC, Chapiro J, Letzen B. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. 2019;29:3338-3347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 203] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 44. | Wang CJ, Hamm CA, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Weinreb JC, Duncan JS, Chapiro J, Letzen B. Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur Radiol. 2019;29:3348-3357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 45. | Wu Y, White GM, Cornelius T, Gowdar I, Ansari MH, Supanich MP, Deng J. Deep learning LI-RADS grading system based on contrast enhanced multiphase MRI for differentiation between LR-3 and LR-4/LR-5 liver tumors. Ann Transl Med. 2020;8:701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 46. | Zhen SH, Cheng M, Tao YB, Wang YF, Juengpanich S, Jiang ZY, Jiang YK, Yan YY, Lu W, Lue JM, Qian JH, Wu ZY, Sun JH, Lin H, Cai XJ. Deep Learning for Accurate Diagnosis of Liver Tumor Based on Magnetic Resonance Imaging and Clinical Data. Front Oncol. 2020;10:680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 47. | Zhang Y, Lv X, Qiu J, Zhang B, Zhang L, Fang J, Li M, Chen L, Wang F, Liu S, Zhang S. Deep Learning With 3D Convolutional Neural Network for Noninvasive Prediction of Microvascular Invasion in Hepatocellular Carcinoma. J Magn Reson Imaging. 2021;54:134-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 48. | Wang G, Jian W, Cen X, Zhang L, Guo H, Liu Z, Liang C, Zhou W. Prediction of Microvascular Invasion of Hepatocellular Carcinoma Based on Preoperative Diffusion-Weighted MR Using Deep Learning. Acad Radiol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 49. | Jiang YQ, Cao SE, Cao S, Chen JN, Wang GY, Shi WQ, Deng YN, Cheng N, Ma K, Zeng KN, Yan XJ, Yang HZ, Huan WJ, Tang WM, Zheng Y, Shao CK, Wang J, Yang Y, Chen GH. Preoperative identification of microvascular invasion in hepatocellular carcinoma by XGBoost and deep learning. J Cancer Res Clin Oncol. 2021;147:821-833. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 98] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 50. | An C, Jiang Y, Huang Z, Gu Y, Zhang T, Ma L, Huang J. Assessment of Ablative Margin After Microwave Ablation for Hepatocellular Carcinoma Using Deep Learning-Based Deformable Image Registration. Front Oncol. 2020;10:573316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 51. | Liu F, Liu D, Wang K, Xie X, Su L, Kuang M, Huang G, Peng B, Wang Y, Lin M, Tian J. Deep Learning Radiomics Based on Contrast-Enhanced Ultrasound Might Optimize Curative Treatments for Very-Early or Early-Stage Hepatocellular Carcinoma Patients. Liver Cancer. 2020;9:397-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 101] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 52. | Peng J, Kang S, Ning Z, Deng H, Shen J, Xu Y, Zhang J, Zhao W, Li X, Gong W, Huang J, Liu L. Residual convolutional neural network for predicting response of transarterial chemoembolization in hepatocellular carcinoma from CT imaging. Eur Radiol. 2020;30:413-424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 120] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 53. | Liu QP, Xu X, Zhu FP, Zhang YD, Liu XS. Prediction of prognostic risk factors in hepatocellular carcinoma with transarterial chemoembolization using multi-modal multi-task deep learning. EClinicalMedicine. 2020;23:100379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 54. | Zhang L, Xia W, Yan ZP, Sun JH, Zhong BY, Hou ZH, Yang MJ, Zhou GH, Wang WS, Zhao XY, Jian JM, Huang P, Zhang R, Zhang S, Zhang JY, Li Z, Zhu XL, Gao X, Ni CF. Deep Learning Predicts Overall Survival of Patients With Unresectable Hepatocellular Carcinoma Treated by Transarterial Chemoembolization Plus Sorafenib. Front Oncol. 2020;10:593292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 55. | Lin H, Wei C, Wang G, Chen H, Lin L, Ni M, Chen J, Zhuo S. Automated classification of hepatocellular carcinoma differentiation using multiphoton microscopy and deep learning. J Biophotonics. 2019;12:e201800435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 56. | Kiani A, Uyumazturk B, Rajpurkar P, Wang A, Gao R, Jones E, Yu Y, Langlotz CP, Ball RL, Montine TJ, Martin BA, Berry GJ, Ozawa MG, Hazard FK, Brown RA, Chen SB, Wood M, Allard LS, Ylagan L, Ng AY, Shen J. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digit Med. 2020;3:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 157] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 57. | Liao H, Long Y, Han R, Wang W, Xu L, Liao M, Zhang Z, Wu Z, Shang X, Li X, Peng J, Yuan K, Zeng Y. Deep learning-based classification and mutation prediction from histopathological images of hepatocellular carcinoma. Clin Transl Med. 2020;10:e102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 58. | Wang H, Jiang Y, Li B, Cui Y, Li D, Li R. Single-Cell Spatial Analysis of Tumor and Immune Microenvironment on Whole-Slide Image Reveals Hepatocellular Carcinoma Subtypes. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 59. | Chen M, Zhang B, Topatana W, Cao J, Zhu H, Juengpanich S, Mao Q, Yu H, Cai X. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis Oncol. 2020;4:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 123] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 60. | Lu L, Daigle BJ Jr. Prognostic analysis of histopathological images using pre-trained convolutional neural networks: application to hepatocellular carcinoma. PeerJ. 2020;8:e8668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 61. | Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, Pronier E, Laurent A, Amaddeo G, Regnault H, Sommacale D, Ziol M, Pawlotsky JM, Mulé S, Luciani A, Wainrib G, Clozel T, Courtiol P, Calderaro J. Predicting Survival After Hepatocellular Carcinoma Resection Using Deep Learning on Histological Slides. Hepatology. 2020;72:2000-2013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 179] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 62. | Shi JY, Wang X, Ding GY, Dong Z, Han J, Guan Z, Ma LJ, Zheng Y, Zhang L, Yu GZ, Wang XY, Ding ZB, Ke AW, Yang H, Wang L, Ai L, Cao Y, Zhou J, Fan J, Liu X, Gao Q. Exploring prognostic indicators in the pathological images of hepatocellular carcinoma based on deep learning. Gut. 2021;70:951-961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 105] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 63. | Yamashita R, Long J, Saleem A, Rubin DL, Shen J. Deep learning predicts postsurgical recurrence of hepatocellular carcinoma from digital histopathologic images. Sci Rep. 2021;11:2047. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 64. | Gumbs AA, Perretta S, d’Allemagne B, Chouillard E. What is Artificial Intelligence Surgery? Art Int Surg. 2021;1:1-10. [DOI] [Full Text] |
| 65. | Lee D, Yu HW, Kwon H, Kong HJ, Lee KE, Kim HC. Evaluation of Surgical Skills during Robotic Surgery by Deep Learning-Based Multiple Surgical Instrument Tracking in Training and Actual Operations. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |