Published online Dec 27, 2021. doi: 10.4254/wjh.v13.i12.1875
Peer-review started: May 7, 2021
First decision: June 4, 2021
Revised: June 15, 2021
Accepted: November 15, 2021
Article in press: November 15, 2021
Published online: December 27, 2021
Processing time: 233 Days and 16.4 Hours
Hepatitis B virus (HBV) (sub)genotypes A1, D3 and E circulate in sub-Saharan Africa, the region with one of the highest incidences of HBV-associated hepatocellular carcinoma globally. Although genotype E was identified more than 20 years ago, and is the most widespread genotype in Africa, it has not been extensively studied. The current knowledge status and gaps in its origin and evolution, natural history of infection, disease progression, response to antiviral therapy and vaccination are discussed. Genotype E is an African genotype, with unique molecular characteristics that is found mainly in Western and Central Africa and rarely outside Africa except in individuals of African descent. The low prevalence of this genotype in the African descendant populations in the New World, phylogeographic analyses, the low genetic diversity and evidence of remnants of genotype E in ancient HBV samples suggests the relatively recent re-introduction into the population. There is scarcity of information on the clinical and virological characteristics of genotype E-infected patients, disease progression and outcomes and efficacy of anti-HBV drugs. Individuals infected with genotype E have been characterised with high hepatitis B e antigen-positivity and high viral load with a lower end of treatment response to interferon-alpha. A minority of genotype E-infected participants have been included in studies in which treatment response was monitored. Of concern is that current guidelines do not consider patients infected with genotype E. Thus, there is an urgent need for further large-scale investigations into genotype E, the neglected genotype of HBV.
Core Tip: Although genotype E was identified more than 20 years ago, and is the most widespread genotype in Africa, it has not been extensively studied. The current knowledge status and gaps in its origin and evolution, natural history of infection, disease progression, response to antiviral therapy and vaccination discussed in this review highlight the urgent need for further more in-depth and large-scale investigations into genotype E, the neglected genotype of hepatitis B virus.
- Citation: Ingasia LAO, Wose Kinge C, Kramvis A. Genotype E: The neglected genotype of hepatitis B virus. World J Hepatol 2021; 13(12): 1875-1891
- URL: https://www.wjgnet.com/1948-5182/full/v13/i12/1875.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i12.1875
Hepatitis B virus (HBV), a common cause of liver disease, is the prototype member of the family Hepadnaviridae. Despite the availability of vaccines, HBV infection remains a public health concern causing high morbidity and mortality rates, as a result of the serious clinical consequences of cirrhosis and hepatocellular carcinoma (HCC)[1]. It is estimated that a third of the world’s population is or has been infected with HBV at some point in their lives[1]. As a result of its unusual mechanism of replication by reverse transcription through an RNA intermediate, and lack of proof reading ability of its viral polymerase[2], HBV displays sequence heterogeneity, which leads to the existence of at least 9 genotypes. Four genotypes, A to D, were recognized initially, with genotypes E to I being recognized subsequently[3]. A putative 10th genotype J, has been proposed[4]. All genotypes, except E and G, are further subdivided into subgenotypes. Most HBV genotypes and, in some cases subgenotypes have a distinct geographical distribution. HBV genotypes A and D have global distributions while genotypes B and C are predominantly found in East and Southeast Asia. Genotype E is found in West and Central Africa, genotypes F and H are found among various population groups, including indigenous peoples in Central and South America[5,6], while genotype G is found in the Americas and Europe[6]. Genotype I was reported in Vietnam and Laos[6], with the most recent putative genotype J identified in a Japanese patient living in Borneo island[4].
Together with south-east Asia, Africa is one of the two regions in the world where HBV remains endemic. West Africa is the only major region in the world where HBV is still hyperendemic[5] — [> 8% of hepatitis B surface antigen (HBsAg) chronic carriers in the general population] and there is a correspondingly high incidence of HCC[7]. Genotype E was first described in 1992 from a HBsAg-positive Cameroonian blood donor[8]. It predominates in sub-Saharan Africa (SSA) accounting for 97% of individual infections and 17.6% of all HBV infections globally[9-11]. It is found almost exclusively throughout the vast expanses of the Western and Central Africa crescent including Angola, Liberia, Senegal[12,13], Ivory Coast[14], the Gambia, Nigeria[15], Mali, Burkina Faso, Togo, Guinea, Benin, Democratic Republic of Congo, Cameroon[16] and Namibia. The prevalence of genotype E decreases in proportions towards Eastern Africa, where, with the exception of Madagascar (genotype E), mainly genotype A has been found[5,9,11].
Genotype E has been found only in Africa, with some rare exceptions on other continents mainly in persons with a link to Africa[17,18]. Nonetheless, two cases, where no link to Africa could be established, have been documented, one in India[19] and another in Colombia[20]. Genotype A, on the other hand, circulates on every continent, including Africa, where it has the highest genetic diversity of 4% over the complete genome compared to 3% outside Africa[21]. Despite its high genetic diversity in Africa, genotype A is rarely found in West Africa. The dispersal routes of genotype A have previously been described to coincide with the slave trade leading to the dispersal of this genotype to the Americas and the Indian subcontinent[19,21-23]. Despite the forced migrations of slaves from West Africa to the New world[3,17], only sporadic cases of genotype E have been reported in the Americas[17,24], Northern Europe[25] including Belgium[26] and the Netherlands[27]. This may suggest that genotype E was not in circulation before and during the slave trade (9th to 19th century) and has only been introduced into the West African population after the end of the slave trade in the late 1800s[23].
The conspicuously low genetic diversity of genotype E ranging between 1.2% and 1.95%[11,16,23,28,29] further supports a short natural history in Africa[16] and relatively recent introduction into the general population[16,30]. Various times from the most recent common ancestor (tMRCA) of genotype E have been calculated using Bayesian inference, with a median tMRCA of 130 years[30] whereas in Nigeria, a more recent tMRCA was estimated to be year 1948 [95% higher posterior density (HPD): 1924-1966] (73 years), with an increase in the genotype E-infected population over the last approximately 40 years to 50 years[31]. A recent study focusing on ancient HBV estimated a median MRCA to be year 1016 (95% HPD: 712-1358)[32]. These times differ from the estimated tMRCA of 6000 years[33]. Differences in the calculation of the nucleotide substitution rate of HBV are responsible for the variance of the estimated age of genotype E. Our recent study describing the phylogeography of full genomes of genotype E showed localized transmission, and limited movements within West and Central Africa. The study showed West Africa to be the most probable origin of the genotype E epidemic, with strains dispersing to the European region from there, whereas the strains dispersed to the Americas originated in Central Africa[29].
Studies on HBV-infected mummies from the 16th century revealed a very close relationship between the ancient and modern HBV genomes dating 400-500 years[34,35]. Furthermore, studies conducted by Krause-Kyora et al[36] reported ancient HBV sequences in the Neolithic age, while studies by Mühlemann et al[32] reported archeological ancient HBV and predicted recombination breakpoints in the polymerase gene leading to the formation of genotype A with similar recombination events involved in the creation of genotypes E and G[32,36-38] in the Bronze age[32]. Concurring with Mühlemann et al[32]’s study, Krause showed recombination events over time and similarity between the earliest ancient HBV sequences of the Neolithic era and modern HBV genotypes E and G[36]. By comparing the sequences from the above two studies, Datta et al[39] was able to confirm the previous findings of the presence of remnants of genotype E in ancient sequences from the Neolithic and Bronze age[32,36,39].
At first glance, the widespread prevalence and extensive geographic distribution of genotype E[17,28,29] may be difficult to reconcile with the long natural history of genotype A in Africa. However, isolation of genotype E in indigenous isolated tribes of Africa; Pygmies[37] and Khoi San (Kramvis unpublished data), believed to be direct descendants of earliest human lineages[6,37,40], and the recent discovery of the ancient HBV sequences in the Neolithic and Bronze era from skeletal remains of humans with remnants of genotype E[32,36,39], may support the theory that genotype E pre-existed but has been re-introduced into the population thus replacing genotype A. Similarly, the presence of recombinant sequences similar to extant genotypes D (subgenotype D6) and E, which are presently endemic in certain regions of Africa[6], together with the co-existence of genotypes E/A/D in SSA, including Sudan and Cameroon, also support the aforesaid possibility[37,41,42]. Possible mechanisms of introduction and routes of transmission include mass vaccination programmes carried out in Western Africa and a high frequency of hepatitis B e antigen (HBeAg)-positivity in mothers infected with genotype E [mother to child transmission (MTCT)][43,44] leading to chronicity due to HBe/HBcAg-specific T helper cell tolerance in utero[44]. In constrast to genotype E, the two subgenotypes of A, A1 and A3, circulating in Africa, are characterized by early loss of HBeAg seroconversion and a high frequency of HBeAg-negativity[10].
Genotype E, closely related to human strains, has also been isolated from captive and wild born chimpanzees originating from West and Central Africa[12,41,45]. The direction of transmission was not established[17] although, it was suggested that the practice of injecting human serum into chimpanzees after their capture in Africa was the most probable explanation[41,42,46]. Thus, chimpanzees may be a possible source of separate primate to human transmission events of HBV in West Africa[41,42,46]. Moreover, a closer relationship between the Neolithic and the African non-human primate strains compared to other human strains suggests African origin of extinct HBV genotypes and reciprocal cross-species transmission in the past[38,47] supporting preceding suppositions[48].
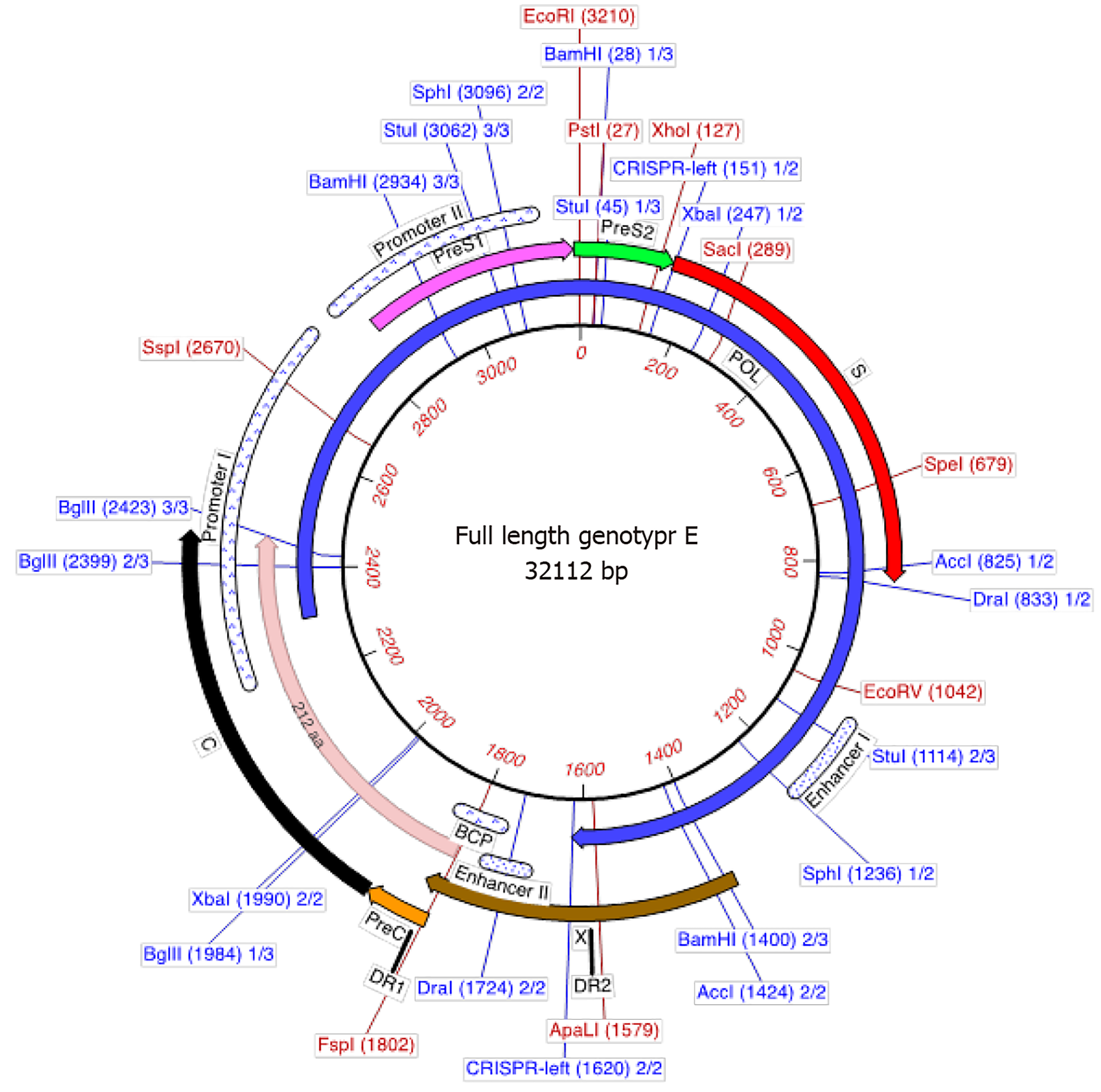
Genotype E is the most prevalent genotype of HBV in Africa estimated to have infected close to 20% of chronic HBV carriers globally. However, due to limited studies and the lack of surveillance data in Africa, this estimate may be higher[17]. Genotype E is the second shortest genotype after D with a complete genome length of 3212 bp (Figure 1). It has a unique three-nucleotide deletion in the preS1 that can differentiate it from other genotypes (Figure 1) and a signature pattern of amino acids in the preS1. In addition, genotype E has a putative additional start codon in the preS1, which may lead to an elongated middle hepatitis B surface protein (317 amino acids in length instead of 281 amino acids)[11]. This elongated middle HBsAg has not been detected to date. The amino acids of the preS1, preS2 and S genes are well conserved, with signature motifs Leu3SerTrpThrValProLeuGluTrp11 in the preS1 specific to genotype E[11]. Additional signature amino acids are also found at Thr18, Arg38, His44, Thr52, Met83, Lys85 and Thr108 in the preS1. All genotype E strains have a His at amino acid position 15 of the preS1 but no known unique signature motifs in the pre-S2 region. Arg122, Lys160 and Leu127 residues are a characteristic of the S gene in this genotype and encodes for a unique serological subtype ayw4[11,12]. Although the reactivity to different diagnostic assays has been determined for genotypes A to D[49], it has not been tested for genotype E. The L209V substitution in the HBsAg was described as a unique feature among all genotype E sequences deposited in GenBank to date[50]. The spacer region of the polymerase (POL) has eight amino acids unique to genotype E: Met64, Glu16, His21, Arg52, Asp55, Lys88, Asn110 and His111. Within the reverse transcriptase, Met164 is the only unique amino acid substitution in this genotype[11]. This introduces a start codon that theoretically could be translated into a protein of 344 amino acids. Although genotype E has the T1858 mutation in the precore (preC) region it does not frequently develop the G1896A mutation[44,51], which has been shown to stabilize the encapsidation signal (ε) converting the wobble to a stable Watson-Crick T-A pair[52]. This introduces a stop codon in the HBeAg precursor leading to no expression of the mature HBeAg[10,44,51]. As a result of its unique molecular structure, genotype E has a restriction map that differentiates it from other genotypes of HBV (Figure 1).
Variants can play a critical role in HBV epidemics. From the limited studies on genotype E, a number of variants and mutants that can hypothetically affect detection, vaccination response and pathogenicity of HBV, have been described. Within the ‛a’ determinant of HBsAg, the vaccine and immune escape mutations R48T, P120T and G145R have been reported in genotype E HBV isolated from infected individuals[3,53]. The preS2 F22L mutation, associated with cirrhosis, and a risk factor for the development of HCC, was found in genotype E isolates from Sudanese HCC patients[54].
Variants can also be generated through recombination[38] within an individual co-infected, with more than one genotype, resulting in drug resistant or diverse HBV strains. Recombinants can only occur when the various genotypes co-circulate in a population. Genotype E presents high chances of recombination, with A/E and D/E recombinants found in Ghana, A/E recombinant has been reported in Cameroon[37], Guinea, Burkina Faso and Nigeria[31] while D/E recombinant has been found in Gabon, Sudan, South Africa, Niger and Guinea[55,56].
Table 1, summarizes the different recombination events of genotype E with either D or A, mostly reported within Africa with different breakpoints within the HBV genome[37,54-61].
| Parental genotype | Region | Genome position (from the EcoRI site) | Country |
| D/E | preS1 | Niger, Ghana, Gabon, and Sudan[53-58] | |
| D/E | preC/C | Ireland[59] and South Africa[60] | |
| D/E | Pol | 978, 1230 | Sudan[56] |
| X | 1643 | ||
| C/Pol overlapping region | 2384 | ||
| Pol | 2756 | ||
| preS1/Pol overlapping region | 3000 | ||
| D/E | X/preC overlapping region | 1649, 1932 | Niger[58] |
| C/Pol overlapping region | 2392, 2385 | ||
| Pol | 2831, 2836 | ||
| Pol/preS1 overlapping region | 3075, 3083 | ||
| D/E | X | 1651 | Ghana[57] |
| C/Pol overlapping region | 2406 | ||
| Pol | 2823 | ||
| Pol/preS1 overlapping region | 3081 | ||
| E/D | preS | 85-505 | Niger[58] |
| S-Pol overlapping region | 796-1306 | ||
| A/E | C | Ghana[57] | |
| A/E | Pol | 874-1062 | Cameroon[37] |
| X | |||
| E/A | Pol | 908-1026 | |
| X-C | |||
| A/E | preC/C | Guinea[57] and France[61] | |
| E/A | X |
The F22L mutation and various deletions in the preS2 and the 1753V and 1762T/1764A mutations in the basic core promoter (BCP), are mostly found in HBV strains isolated from HCC patients[62] than in those from non-HCC controls[54,63]. Deletions in the core region have been reported in HBsAg-positive genotype E asymptomatic blood donors in Guinea. Another study conducted by Yousif et al[54] found preS2 deletion mutations in HBV from patients infected with either genotypes D or E in Sudan. The preS deletions in genotype E were found in the HBV isolated from HCC patients, while genotype D deletion mutants were detected in non-HCC patients[54]. The significance of this difference remains to be determined. On the other hand subgenotype A1, which is mostly found in SSA[5], has been shown to have a higher carcinogenic potential compared to other (sub)genotypes[64]. A meta-analysis study associated the preS deletion mutants with a 3.77-fold increased risk of HCC[65]. Furthermore, a prospective study revealed the predictive value of a combination of the preS and BCP mutants in the development of HCC and pro-oncogenic role of mutated envelope proteins through their intracellular accumulation[66]. These mutations may be used as biomarkers for screening high-risk individuals in resource limited regions such as SSA, who may potentially develop HCC[67].
The prevalence of chronic HBV infection varies widely according to geographic area and is closely linked with the predominant routes of HBV transmission. In regions of Africa, where genotype E prevails, transmission can occur horizontally or vertically in utero, intrapartum or via breast-feeding[68] from mother to child[69]. However, about 50% of the infection in children cannot be accounted for by MTCT and in many endemic regions, prior to the introduction of neonatal vaccination, the prevalence peaked among children aged between 7 years to 14 years[70]. In the pre-vaccine era, most chronic carriers were infected horizontally in SSA and only 10% were infected through MTCT compared to 40% in Asia[71,72]. Horizontal transmission can occur early in life mainly from HBeAg-positive family members/household contacts, playmates or by unsafe medical interventions. Very few studies have been carried out in terms of identifying routes of transmission for genotype E. In the Gambia, MTCT is responsible for 16% of chronic infections and increases the risk of persistent viral replication and severe liver disease[73]. Strong evidence from a phylogenetic analysis showed intrafamilial transmission of HBV[73]. A study conducted in Ghana also concluded that the HBV is predominantly transmitted through horizontal transmission in childhood with intrafamilial, rather than interfamilial environment being the primary place of transmission[74]. However, a study conducted in Nigeria in two semi-isolated rural communities suggested that HBV transmission between siblings was not the major route of transmission with a complex pattern of transmission among the residents of the two communities[31]. So it appears that other factors may be at play in the transmission of genotype E in various communities. As has been shown in Burkina Faso, co-infection with human immunodeficiency virus (HIV), which leads to an increase in HBV viral load and frequency of HBeAg-positivity, can increase the risk of HBV transmission by as much as 2.5-fold[75,76]. Traditional cultural practices such as scarification and tattooing have been shown to be responsible for the transmission of HBV[77].
Genotypes and subgenotypes can influence the natural history of infection. Comparing different (sub)genotypes is often difficult because the (sub)genotypes do not circulate in the same populations. The majority of the studies have compared genotypes B and C as well as A and D and have shown different clinical manifestations and the serious outcomes of disease [cirrhosis (LC) and HCC][78-81]. The natural history of infection in individuals infected with genotype E has not been extensively studied, and has mostly been derived from anecdotal evidence. Genotype E has clinically been characterized, with high viral loads and the patients infected with this genotype are more likely to be HBeAg-positive than the patients infected with genotype D[5,10,53,54,56]. A higher HBeAg-positivity of this genotype has been shown to confer tolerance, with a milder clinical manifestation[10]. This could be the reason for the higher prevalence of genotype E in Sudanese blood donors, whereas genotype D is more prevalent in those patients with liver disease[28,54,56]. In addition, infection with genotype E has previously been linked to higher chronicity rates than other genotypes[10,54,56].
Table 2, which was compiled from limited data comparing genotype E to D in Sudan (Yousif et al[53,54]) and studies in the Gambia (Shimakawa et al[72]), sum
| Genotypes | ||||||||
| E | A | B | C | D | F | G | H | |
| HBV DNA level | Increased | Decreased | Decreased | Increased | Not studied | Not studied | Not studied | Not studied |
| Frequency of precore G1896A mutation | Increased1 | Decreased | Increased | Decreased | Increased | Not studied | Not studied | Not studied |
| Frequency of basic core promoter T1762A/A1764G mutation | Not studied | Increased | Decreased | Increased | Decreased | Not studied | Not studied | Not studied |
| Frequency of preS deletion mutation | Not studied | Increased | Decreased | Increased | Not studied | Not studied | Not studied | Not studied |
| Tendency of chronicity | ||||||||
| High | + | + | ||||||
| Low | + | + | ||||||
| Not studied | + | + | + | + | ||||
| HBeAg positivity | ||||||||
| High | + | + | ||||||
| Low | + | + | + | |||||
| Not studied | + | + | + | |||||
| HBeAg seroconversion | ||||||||
| Early | + | + | ||||||
| Late | + | + | ||||||
| Not studied | + | + | + | + | ||||
| HBsAg seroconversion | ||||||||
| More | + | + | ||||||
| Less | + | + | ||||||
| Not studied | + | + | + | + | ||||
In their study, Yousif et al[54] observed that genotype E infected liver disease patients and blood donors[56] had a higher frequency of HBeAg-positivity and higher viral loads compared to patients infected with genotype D (Table 2)[53,54]. Both genotype D and E have the 1858T, and thus can develop the G1896A mutation, however, what is puzzling is that G1896A is positively associated with genotype D and negatively associated with genotype E[51].
This lack of association may be the reason for the high frequency of HBeAg-positivity in individuals infected with genotype E compared to genotype D. A study focusing on chronic hepatitis B (CHB) and HCC in Burkina Faso showed patients infected with genotype E had lower viral loads, lower frequency of HBeAg-positivity and higher prevalence of cirrhosis than those infected with genotype C or C/E recombinants. With the majority of HCC, infected with genotype E (78%), HCC-associated risk factors were old age, male with high HBV viral load when comparing CHB in HCC patients to non-HCC patients[83]. Another longitudinal study conducted in Gambia showed that a majority of the genotyped CHB carriers were infected with genotype E[72]. Although the mean viral load and alanine aminotransferase levels were higher in carriers with HBsAg-positive mothers, a majority (47%) had undetectable viral loads with 22% of all chronic HBV infections having viral loads ranging between 50 and 200 IU/mL. HBV viral load has been used to predict progression from cirrhosis to HCC[84]. From this study, the rate at which the HBV DNA cleared was faster when compared to age progression making it difficult to predict HCC[72]. What should be noted from this study is that, the samples that were assayed for viral loads were from a different time frame (2012-2013), while the genotyped samples were from 2003. Successful genotyping would require viral loads high enough to allow amplification of the DNA and thus higher viral loads may be a factor that biases genotyping making it hard to draw any conclusion on the infecting genotype for the chronic carriers who had undetectable or low HBV DNA.
African regions in which genotype E is endemic are characterized by a higher incidence of HCC[85] and epidemiological studies have suggested the carcinogenic potential of genotype E[86]. Although the mechanisms underlying this oncogenic potential have not yet been clarified for genotype E, they could be related to immune escape phenomena[87], as well as to other possible cofounders that may be involved, such as HIV co-infection, dietary iron overload or aflatoxin consumption[85,88,89].
Globally, an estimated 10% of the 37 million HIV infected individuals are co-infected with HBV[90]. HBV/HIV co-infection in SSA accounts for 36% (2-4 million) with the highest rates reported in West- and Southern Africa[90]. Epidemiological and virological characteristics of HIV-infected individuals in West Africa showed an average of 13% prevalence of HBsAg-positivity, ranging between 1.1% in blood donors and 35.7% in pregnant women attending antenatal care[76,91-93], while 4.75% of HBV-HIV infected individuals were HBeAg-positive with the prevalence ranging between 3.2% and 7.2% in adults and anti-retroviral (ART) naïve adults, respectively[94,95]. An average HBV exposure rate of 74% (64%-81.7%) in ART naïve and adults initiating ART[90,94,96,97] has been documented. A high rate of morbidity has been reported in HBV/HIV co-infected individuals, while the progression of CHB to HCC is more rapid in genotype E HIV-positive individuals than in those with HBV alone[98]. In a study of Senegalese children, 47% who were HBV genotype E-HIV co-infected had elevated levels of drug resistance mutations (L180M, M204V/I, and S202N) to both HIV and HBV, significant levels of HBsAg escape mutations, HBV DNA persistence and HIV virologic failure[99]. This suggests that the use of the Tenofovir Disoproxil Fumarate regimen in the management of HBV, HIV and HBV-HIV co-infection is ideal in the SSA setting.
Occult HBV infection (OBI) is defined as the presence of replication-competent HBV DNA (i.e., episomal HBV covalently closed circular DNA) in the liver and/or HBV DNA in the blood of people who test negative for HBsAg by currently available assays)[100]. OBI is frequent in HIV-infected individuals and has been described in individuals infected with genotype E, with a prevalence 10% and 15% in HIV-positive patients from the Ivory Coast and Sudan, respectively[97,101].
Biomarkers are very important in assessing risk factors for the development of serious clinical manifestations. As is evident from the above observations the same risk biomarkers may not be applicable to all (sub)genotypes and cannot be extrapolated from studies on other genotypes. Therefore, it is important that biomarkers are studied exclusively in genotype E.
Current antiviral therapies, which include nucleos(t)ide analogues (NA) and interferon-alpha (IFN-α) reduce but do not eliminate the risk of liver cancer. As curative therapies are developed, it will be important to monitor patients for progression to liver cancer, even if they have been cured of CHB infection. HBV genotype may influence the efficacy of the antiviral therapy but most studies that analyzed the role of HBV genotype in the treatment with NA mostly focused on genotypes A, B, C and D. Lamivudine (LAM) is the earliest used NA in the world and the association between HBV genotype and LAM has been demonstrated both in terms of response and the development of resistance mutations. Various response rates have been observed for various studies with genotype A being more likely to develop resistance mutations[102,103]. Studies have shown that HBeAg-positive patients infected with genotype B have a higher response rate to IFN-α than those infected with genotype C, while patients infected with genotype A have a higher response rate to IFN-α than those infected with genotype D[104].
There is a scarcity of information on the clinical and virological characteristics of genotype E-infected patients as well as on the efficacy of anti-HBV drugs[86]. However, a few studies have described genotype E’s response to treatment[86,105-108] in a variety of scenarios: Treatment-naïve CHB patients initiating treatment with NA [entecavir (ETV) or tenofovir][86], HBV-HIV co-infected patients[109], rescued after LAM failure[110], adefovir phase III clinical trials[111]; a follow-up study of HBsAg decline in ETV-responding patients[107] and response to IFN[106,112]. As is evident from the above list, only one study looked at tenofovir the drug recommended by the World Health Organization (WHO), American Association for the Study of Liver Diseases, and the European Association for the Study of the Liver for antiviral therapy.
The phase III clinical trial of adefovir dipivoxil conducted by Westland et al[111] included a total of 6 genotype E patients and reported antiviral efficacy in patients on a 48-wk therapy regardless of the HBV genotype. Studies by Boglione et al[107] and Cuenca-Gómez et al[86] focused on genotype E treatment-naïve, CHB patients of SSA origin, on ETV or tenofovir antiviral therapy. A higher rate of HBsAg loss in patients infected with genotype E compared to genotypes A or D was observed. In addition, a high response rate to NA was reported with undetectable viral load and loss of HBeAg in a median time of 31.8 mo with no cases of HCC[86].
Two different treatment regimens were compared in CHB patients infected with genotype E, who had migrated to Italy. In the one arm, CHB patients with low viral loads, where given pegIFN for 24 wk, whereas in the second arm, CHB patients with high viral loads were treated sequentially with ETV for 12 wk and thereafter pegIFN for 24 wk. Those treated with monotherapy did not respond as well as those on dual therapy[106]. In a follow-up study, genotype E CHB patients were treated with pegIFN for varying lengths of time 48-, 72- and 96-wk. Prolonged treatment was beneficial and recommended for individuals infected with genotype E[106,108]. Thus, from these limited studies it is evident that genotype E infected individuals are unresponsive to conventional pegIFN treatment. However, in concurring with the Boglione et al[107] and Cuenca-Gómez et al[86] studies, a retrospective study conducted in Europe by Erhardt et al[105], focusing on HBV genotypes E-H the response to IFN-α or NAs (LAM, adefovir, ETV) therapy concluded that genotype E infected patients treated with IFN-α had lower end of treatment response but overall sustained virological response, while the patients on NAs had viral suppression within 48 wk[105]. It should be noted that the conclusion was reached with only 5 treatment-naïve genotype E mono-infected patients[103].
Taken together, the current international treatment guidelines do not consider patients with genotype E CHB. Thus, better management strategies for HBV infected patients are recommended taking into account the genotype in question. In order to deliver proper medical care, improve knowledge on the response to treatment, and the development of resistance of relatively under-studied genotypes like E, it is critical to issue proper and specific recommendations that could differ from those issued for other genotypes. Moreover, all gathered information on response to treatment of genotype E in Africa is useful, especially considering that the development of immune escape mutations[87] can have an epidemiological impact in other parts of the world with the dispersal of these strains via increased migration from Africa. As new finite cure strategies are developed it is important that the clinical trials include CHB patients infected with genotype E.
The risk of developing chronic infection is about 90% following perinatal infection up to 6 mo but decreases to about 20%-60% between the ages of 6 mo to 5 years[68,73]. Thus, prevention of HBV infection by vaccination is very important and is most successful when it targets infants, and when prevention begins with administration of the first dose of HBV vaccine soon after birth. The HBV vaccine is about 80%-100% effective in managing HBV infection or clinical hepatitis following completion of the dose. However, inoculation will not help those chronically infected[1]. The two commonly used efficacious vaccines are either plasma-derived vaccines prepared from purified HBsAg obtained from chronic HBV patients or recombinant vaccines from synthesized HBsAg[113]. As of 2020, more than 190 WHO member states immunized infants against HBV as part of their routine vaccination schedule, and 84% of children received HBV vaccines[1]. Even with the vaccine roll out, the burden of HBV infections in SSA remains of concern attributed to the delay in the implementation, lack of birth doses and low coverage of the vaccine programme[114-117]. The high HBeAg positivity in mothers infected with genotype E is a risk factor for MTCT[118] (one in ten infants vaccinated at birth) suggesting that vertical/perinatal infection is still present in African countries[119-122]. Antenatal HBV screening is hardly performed in SSA (0%-20%)[123], with only 33% of countries having official guidelines[124]. HBV was first classified on the basis of the amino acid substitution on the HBsAg at positions 122, 127, 134 and 160. The serological subtypes contain the common ‛a’ determinant and one of each of the mutually exclusive determinants d/y and w/r[125]. Additional serological specificities, originally designated as subdeterminants of ‛a’ and subsequently as subdeterminants of w, have allowed the identification of ten serological subtypes ayw1, ayw2, ayw3, ayw4, ayr, adw2, adw3 adw4, adrq- and adrq+[6,8,126]. The humoral immune response following vaccination with HBV vaccines is largely directed against the common ‛a’ determinant, with a lesser response directed against the d/y and r/w subdeterminant epitopes[113,127].
All currently available genetically engineered HBV vaccines are produced with the subgenotype A2, serotype adw, which differs from the genotype E subtype ayw4. Available data show that current HBV-A2 vaccines are highly effective at preventing infections and clinical disease caused by all known HBV genotypes[128]. However, a study conducted on blood donors in the United States[129] questioned the ability of subgenotype A2-derived HBV vaccines to protect against non-A2 HBV (sub)geno
The emergence of HBV escape mutants may occur under medically induced immune pressure (in association with vaccine or hepatitis B immune globulin) or naturally induced immune pressure (as a result of CHB)[130]. These HBV mutants may carry multiple amino acid substitutions around- and within the HBsAg ‛a’ determinant, which can affect the binding of neutralizing antibodies (anti-HBsAg), with some of the former remaining undetectable by certain diagnostic tests, thus implying a potential risk in transfusion events[130]. The emergence of S escape mutants, raised concerns about the efficacy of the current vaccine on the African continent. To this day, very few studies have focused on the genotype E response to vaccination, although vaccination began over four decades ago.
In conclusion, genotype E has unique molecular and epidemiological characteristics. The natural history of genotype E has not been studied and very little is known about the virological breakthrough as a result of vaccination. Only a few studies that focused on the treatment of a limited number of genotype E infected patients exist, making it difficult to reach any firm conclusions. In addition, most of these studies have been conducted outside of Africa on a small number of individuals that had migrated from Africa, with only a minority of studies carried out on the African continent. Consequently, it is important that African CHB patients infected with genotype E are included in clinical trials focusing on new antiviral therapy, biomarkers and other possible preventive methods. There are multiple reasons for this. Western Africa, where genotype E prevails, is the only region in the world where HBV continues to be hyperendemic. Although West Africa has a relatively long time span of vaccination against HBV, which began in the Gambia in the early 1980s, the infection is still being maintained in the community. There is a correspondingly high incidence of HBV-associated HCC, ranked fourth worldwide and in SSA, the second leading cancer for men and the third for women, with average age-standardised incidence rates of 18.9 and 8.0 per 100000 persons/year, respectively[85]. In this region, HCC presents in younger age groups and has a median survival rate of approximately 3-4 mo. Genotype E is being dispersed from high to low endemicity regions of the world as a result of migration and this may lead to changes in the natural history of HBV infection in countries of destination, where different genotypes predominate.
Toward achieving the WHO target for the worldwide elimination of viral hepatitis as a public health burden by 2030 there is an urgent need for more in-depth and large-scale investigations into genotype E, which has been under-represented in studies, resulting in the paucity of data on this neglected genotype.
We would like to thank all our colleagues in the Hepatitis Virus Diversity Research Unit for the support, the Swedish International Development Cooporation Agency (SIDA) in partnership with the Organization for Women in Science for the Developing World (OWSD) for the PhD fellowship, Poliomyelitis Research Foundation (PRF) and the Loreal-UNESCO For Women in Science awarded to Ms. Ingasia LAO.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: European Association for the Study of Liver Disease; Academy of Sciences of South Africa; International Coalition for the Elimination of HBV (ICE_HBV).
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: South Africa
Peer-review report’s scientific quality classification
Peer-review model: Single blind
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chemin IA S-Editor: Gao CC L-Editor: Webster JR P-Editor: Gao CC
| 1. | World Health Organization. Hepatitis B Fact Sheet 2021. [cited 7 April 2021]. In: World Health Organization [Internet]. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b. |
| 2. | Steinhauer DA, Holland JJ. Direct method for quantitation of extreme polymerase error frequencies at selected single base sites in viral RNA. J Virol. 1986;57:219-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 125] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 3. | Norder H, Couroucé AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 630] [Cited by in RCA: 649] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 4. | Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, Nakayoshi T, Wakuta M, Miyakawa Y, Mizokami M. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J Virol. 2009;83:10538-10547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 336] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 5. | Kramvis A, Kew MC. Epidemiology of hepatitis B virus in Africa, its genotypes and clinical associations of genotypes. Hepatol Res. 2007;37:S9-S19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 230] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 6. | Kramvis A. Genotypes and genetic variability of hepatitis B virus. Intervirology. 2014;57:141-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 323] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 7. | Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 4902] [Article Influence: 700.3] [Reference Citation Analysis (1)] |
| 8. | Norder H, Hammas B, Löfdahl S, Couroucé AM, Magnius LO. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J Gen Virol. 1992;73 (Pt 5):1201-1208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 263] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 9. | Velkov S, Ott JJ, Protzer U, Michler T. The Global Hepatitis B Virus Genotype Distribution Approximated from Available Genotyping Data. Genes (Basel). 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 108] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 10. | Kramvis A. Molecular characteristics and clinical relevance of African genotypes and subgenotypes of hepatitis B virus. S Afr Med J. 2018;108:17-21. [PubMed] |
| 11. | Kramvis A, Restorp K, Norder H, Botha JF, Magnius LO, Kew MC. Full genome analysis of hepatitis B virus genotype E strains from South-Western Africa and Madagascar reveals low genetic variability. J Med Virol. 2005;77:47-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 56] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Norder H, Couroucé AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 556] [Cited by in RCA: 587] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 13. | Vray M, Debonne JM, Sire JM, Tran N, Chevalier B, Plantier JC, Fall F, Vernet G, Simon F, Mb PS. Molecular epidemiology of hepatitis B virus in Dakar, Sénégal. J Med Virol. 2006;78:329-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 14. | Suzuki S, Sugauchi F, Orito E, Kato H, Usuda S, Siransy L, Arita I, Sakamoto Y, Yoshihara N, El-Gohary A, Ueda R, Mizokami M. Distribution of hepatitis B virus (HBV) genotypes among HBV carriers in the Cote d'Ivoire: complete genome sequence and phylogenetic relatedness of HBV genotype E. J Med Virol. 2003;69:459-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 58] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 15. | Odemuyiwa SO, Mulders MN, Oyedele OI, Ola SO, Odaibo GN, Olaleye DO, Muller CP. Phylogenetic analysis of new hepatitis B virus isolates from Nigeria supports endemicity of genotype E in West Africa. J Med Virol. 2001;65:463-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 59] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 16. | Mulders MN, Venard V, Njayou M, Edorh AP, Bola Oyefolu AO, Kehinde MO, Muyembe Tamfum JJ, Nebie YK, Maiga I, Ammerlaan W, Fack F, Omilabu SA, Le Faou A, Muller CP. Low genetic diversity despite hyperendemicity of hepatitis B virus genotype E throughout West Africa. J Infect Dis. 2004;190:400-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 141] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 17. | Andernach IE, Hübschen JM, Muller CP. Hepatitis B virus: the genotype E puzzle. Rev Med Virol. 2009;19:231-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 18. | Bannister EG, Yuen L, Littlejohn M, Edwards R, Sozzi V, Colledge D, Li X, Locarnini S, Hardikar W, Revill PA. Molecular characterization of hepatitis B virus (HBV) in African children living in Australia identifies genotypes and variants associated with poor clinical outcome. J Gen Virol. 2018;99:1103-1114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 19. | Singh J, Dickens C, Pahal V, Kumar R, Chaudhary R, Kramvis A, Kew MC. First report of genotype e of hepatitis B virus in an Indian population. Intervirology. 2009;52:235-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Alvarado Mora MV, Romano CM, Gomes-Gouvêa MS, Gutierrez MF, Carrilho FJ, Pinho JR. Molecular epidemiology and genetic diversity of hepatitis B virus genotype E in an isolated Afro-Colombian community. J Gen Virol. 2010;91:501-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 21. | Kramvis A, Paraskevis D. Subgenotype A1 of HBV--tracing human migrations in and out of Africa. Antivir Ther. 2013;18:513-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 22. | Hassan MA, Kim WR, Li R, Smith CI, Fried MW, Sterling RK, Ghany MG, Wahed AS, Ganova-Raeva LM, Roberts LR, Lok ASF; Hepatitis B Research Network. Characteristics of US-Born Versus Foreign-Born Americans of African Descent With Chronic Hepatitis B. Am J Epidemiol. 2017;186:356-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Andernach IE, Nolte C, Pape JW, Muller CP. Slave trade and hepatitis B virus genotypes and subgenotypes in Haiti and Africa. Emerg Infect Dis. 2009;15:1222-1228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 98] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 24. | Chu CJ, Keeffe EB, Han SH, Perrillo RP, Min AD, Soldevila-Pico C, Carey W, Brown RS Jr, Luketic VA, Terrault N, Lok AS. Hepatitis B virus genotypes in the United States: results of a nationwide study. Gastroenterology. 2003;125:444-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 229] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 25. | Kidd-Ljunggren K, Couroucé AM, Oberg M, Kidd AH. Genetic conservation within subtypes in the hepatitis B virus pre-S2 region. J Gen Virol. 1994;75 ( Pt 6):1485-1490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 26. | Liu HF, Sokal E, Goubau P. Wide variety of genotypes and geographic origins of hepatitis B virus in Belgian children. J Pediatr Gastroenterol Nutr. 2001;32:274-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 27. | van Steenbergen JE, Niesters HG, Op de Coul EL, van Doornum GJ, Osterhaus AD, Leentvaar-Kuijpers A, Coutinho RA, van den Hoek JA. Molecular epidemiology of hepatitis B virus in Amsterdam 1992-1997. J Med Virol. 2002;66:159-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 28. | Hübschen JM, Andernach IE, Muller CP. Hepatitis B virus genotype E variability in Africa. J Clin Virol. 2008;43:376-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 29. | Ingasia LAO, Kostaki EG, Paraskevis D, Kramvis A. Global and regional dispersal patterns of hepatitis B virus genotype E from and in Africa: A full-genome molecular analysis. PLoS One. 2020;15:e0240375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 30. | Andernach IE, Hunewald OE, Muller CP. Bayesian inference of the evolution of HBV/E. PLoS One. 2013;8:e81690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 31. | Forbi JC, Vaughan G, Purdy MA, Campo DS, Xia GL, Ganova-Raeva LM, Ramachandran S, Thai H, Khudyakov YE. Epidemic history and evolutionary dynamics of hepatitis B virus infection in two remote communities in rural Nigeria. PLoS One. 2010;5:e11615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 32. | Mühlemann B, Jones TC, Damgaard PB, Allentoft ME, Shevnina I, Logvin A, Usmanova E, Panyushkina IP, Boldgiv B, Bazartseren T, Tashbaeva K, Merz V, Lau N, Smrčka V, Voyakin D, Kitov E, Epimakhov A, Pokutta D, Vicze M, Price TD, Moiseyev V, Hansen AJ, Orlando L, Rasmussen S, Sikora M, Vinner L, Osterhaus ADME, Smith DJ, Glebe D, Fouchier RAM, Drosten C, Sjögren KG, Kristiansen K, Willerslev E. Ancient hepatitis B viruses from the Bronze Age to the Medieval period. Nature. 2018;557:418-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 131] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 33. | Paraskevis D, Magiorkinis G, Magiorkinis E, Ho SY, Belshaw R, Allain JP, Hatzakis A. Dating the origin and dispersal of hepatitis B virus infection in humans and primates. Hepatology. 2013;57:908-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 117] [Article Influence: 9.8] [Reference Citation Analysis (2)] |
| 34. | Kahila Bar-Gal G, Kim MJ, Klein A, Shin DH, Oh CS, Kim JW, Kim TH, Kim SB, Grant PR, Pappo O, Spigelman M, Shouval D. Tracing hepatitis B virus to the 16th century in a Korean mummy. Hepatology. 2012;56:1671-1680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 35. | Patterson Ross Z, Klunk J, Fornaciari G, Giuffra V, Duchêne S, Duggan AT, Poinar D, Douglas MW, Eden JS, Holmes EC, Poinar HN. Correction: The paradox of HBV evolution as revealed from a 16th century mummy. PLoS Pathog. 2018;14:e1006887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 36. | Krause-Kyora B, Susat J, Key FM, Kühnert D, Bosse E, Immel A, Rinne C, Kornell SC, Yepes D, Franzenburg S, Heyne HO, Meier T, Lösch S, Meller H, Friederich S, Nicklisch N, Alt KW, Schreiber S, Tholey A, Herbig A, Nebel A, Krause J. Neolithic and medieval virus genomes reveal complex evolution of hepatitis B. Elife. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 37. | Kurbanov F, Tanaka Y, Fujiwara K, Sugauchi F, Mbanya D, Zekeng L, Ndembi N, Ngansop C, Kaptue L, Miura T, Ido E, Hayami M, Ichimura H, Mizokami M. A new subtype (subgenotype) Ac (A3) of hepatitis B virus and recombination between genotypes A and E in Cameroon. J Gen Virol. 2005;86:2047-2056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 134] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 38. | Simmonds P, Midgley S. Recombination in the genesis and evolution of hepatitis B virus genotypes. J Virol. 2005;79:15467-15476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 189] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 39. | Datta S. Excavating new facts from ancient Hepatitis B virus sequences. Virology. 2020;549:89-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 40. | Foupouapouognigni Y, Mba SA, Betsem à Betsem E, Rousset D, Froment A, Gessain A, Njouom R. Hepatitis B and C virus infections in the three Pygmy groups in Cameroon. J Clin Microbiol. 2011;49:737-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 41. | Locarnini S, Littlejohn M, Aziz MN, Yuen L. Possible origins and evolution of the hepatitis B virus (HBV). Semin Cancer Biol. 2013;23:561-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 71] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 42. | Locarnini SA, Littlejohn M, Yuen LKW. Origins and Evolution of the Primate Hepatitis B Virus. Front Microbiol. 2021;12:653684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 43. | Milich D, Liang TJ. Exploring the biological basis of hepatitis B e antigen in hepatitis B virus infection. Hepatology. 2003;38:1075-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 297] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 44. | Kramvis A, Kostaki EG, Hatzakis A, Paraskevis D. Immunomodulatory Function of HBeAg Related to Short-Sighted Evolution, Transmissibility, and Clinical Manifestation of Hepatitis B Virus. Front Microbiol. 2018;9:2521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 45. | Takahashi K, Brotman B, Usuda S, Mishiro S, Prince AM. Full-genome sequence analyses of hepatitis B virus (HBV) strains recovered from chimpanzees infected in the wild: implications for an origin of HBV. Virology. 2000;267:58-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 79] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 46. | Hu X, Margolis HS, Purcell RH, Ebert J, Robertson BH. Identification of hepatitis B virus indigenous to chimpanzees. Proc Natl Acad Sci U S A. 2000;97:1661-1664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 77] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 47. | Souza BF, Drexler JF, Lima RS, Rosário Mde O, Netto EM. Theories about evolutionary origins of human hepatitis B virus in primates and humans. Braz J Infect Dis. 2014;18:535-543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 48. | Littlejohn M, Locarnini S, Yuen L. Origins and Evolution of Hepatitis B Virus and Hepatitis D Virus. Cold Spring Harb Perspect Med. 2016;6:a021360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 49. | Hassemer M, Finkernagel M, Peiffer KH, Glebe D, Akhras S, Reuter A, Scheiblauer H, Sommer L, Chudy M, Nübling CM, Hildt E. Comparative characterization of hepatitis B virus surface antigen derived from different hepatitis B virus genotypes. Virology. 2017;502:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (1)] |
| 50. | Mathet VL, Cuestas ML, Ruiz V, Minassian ML, Rivero C, Trinks J, Daleoso G, León LM, Sala A, Libellara B, Corach D, Oubiña JR. Detection of hepatitis B virus (HBV) genotype E carried--even in the presence of high titers of anti-HBs antibodies--by an Argentinean patient of African descent who had received vaccination against HBV. J Clin Microbiol. 2006;44:3435-3439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 51. | Kramvis A, Arakawa K, Yu MC, Nogueira R, Stram DO, Kew MC. Relationship of serological subtype, basic core promoter and precore mutations to genotypes/subgenotypes of hepatitis B virus. J Med Virol. 2008;80:27-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 162] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 52. | Lok AS, Akarca U, Greene S. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc Natl Acad Sci U S A. 1994;91:4077-4081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 280] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 53. | Yousif M, Mudawi H, Hussein W, Mukhtar M, Nemeri O, Glebe D, Kramvis A. Genotyping and virological characteristics of hepatitis B virus in HIV-infected individuals in Sudan. Int J Infect Dis. 2014;29:125-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 54. | Yousif M, Mudawi H, Bakhiet S, Glebe D, Kramvis A. Molecular characterization of hepatitis B virus in liver disease patients and asymptomatic carriers of the virus in Sudan. BMC Infect Dis. 2013;13:328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 55. | Araujo NM. Hepatitis B virus intergenotypic recombinants worldwide: An overview. Infect Genet Evol. 2015;36:500-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 56. | Mahgoub S, Candotti D, El Ekiaby M, Allain JP. Hepatitis B virus (HBV) infection and recombination between HBV genotypes D and E in asymptomatic blood donors from Khartoum, Sudan. J Clin Microbiol. 2011;49:298-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 57. | Garmiri P, Loua A, Haba N, Candotti D, Allain JP. Deletions and recombinations in the core region of hepatitis B virus genotype E strains from asymptomatic blood donors in Guinea, west Africa. J Gen Virol. 2009;90:2442-2451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 58. | Abdou Chekaraou M, Brichler S, Mansour W, Le Gal F, Garba A, Dény P, Gordien E. A novel hepatitis B virus (HBV) subgenotype D (D8) strain, resulting from recombination between genotypes D and E, is circulating in Niger along with HBV/E strains. J Gen Virol. 2010;91:1609-1620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 59. | Laoi BN, Crowley B. Molecular characterization of hepatitis B virus (HBV) isolates, including identification of a novel recombinant, in patients with acute HBV infection attending an Irish hospital. J Med Virol. 2008;80:1554-1564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 60. | Matlou MK, Gaelejwe LR, Musyoki AM, Rakgole JN, Selabe SG, Amponsah-Dacosta E. A novel hepatitis B virus recombinant genotype D4/E identified in a South African population. Heliyon. 2019;5:e01477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 61. | Martel N, Gomes SA, Chemin I, Trépo C, Kay A. Improved rolling circle amplification (RCA) of hepatitis B virus (HBV) relaxed-circular serum DNA (RC-DNA). J Virol Methods. 2013;193:653-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 62. | Mak D, Kramvis A. Molecular characterization of hepatitis B virus isolated from Black South African cancer patients, with and without hepatocellular carcinoma. Arch Virol. 2020;165:1815-1825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 63. | Cohen D, Ghosh S, Shimakawa Y, Ramou N, Garcia PS, Dubois A, Guillot C, Kakwata-Nkor Deluce N, Tilloy V, Durand G, Voegele C, Ndow G, d'Alessandro U, Brochier-Armanet C, Alain S, Le Calvez-Kelm F, Hall J, Zoulim F, Mendy M, Thursz M, Lemoine M, Chemin I. Hepatitis B virus preS2Δ38-55 variants: A newly identified risk factor for hepatocellular carcinoma. JHEP Rep. 2020;2:100144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 64. | Kew MC, Kramvis A, Yu MC, Arakawa K, Hodkinson J. Increased hepatocarcinogenic potential of hepatitis B virus genotype A in Bantu-speaking sub-saharan Africans. J Med Virol. 2005;75:513-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 119] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 65. | Lin CL, Kao JH. Clinical implications of hepatitis B virus variants. J Formos Med Assoc. 2010;109:321-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 66. | Chen CH, Hung CH, Lee CM, Hu TH, Wang JH, Wang JC, Lu SN, Changchien CS. Pre-S deletion and complex mutations of hepatitis B virus related to advanced liver disease in HBeAg-negative patients. Gastroenterology. 2007;133:1466-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 198] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 67. | Fang ZL, Sabin CA, Dong BQ, Ge LY, Wei SC, Chen QY, Fang KX, Yang JY, Wang XY, Harrison TJ. HBV A1762T, G1764A mutations are a valuable biomarker for identifying a subset of male HBsAg carriers at extremely high risk of hepatocellular carcinoma: a prospective study. Am J Gastroenterol. 2008;103:2254-2262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 97] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 68. | Beasley RP, Hwang LY, Lee GC, Lan CC, Roan CH, Huang FY, Chen CL. Prevention of perinatally transmitted hepatitis B virus infections with hepatitis B immune globulin and hepatitis B vaccine. Lancet. 1983;2:1099-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 526] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 69. | Botha JF, Ritchie MJ, Dusheiko GM, Mouton HW, Kew MC. Hepatitis B virus carrier state in black children in Ovamboland: role of perinatal and horizontal infection. Lancet. 1984;1:1210-1212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 121] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 70. | Szmuness W. Recent advances in the study of the epidemiology of hepatitis B. Am J Pathol. 1975;81:629-650. [PubMed] |
| 71. | Keane E, Funk AL, Shimakawa Y. Systematic review with meta-analysis: the risk of mother-to-child transmission of hepatitis B virus infection in sub-Saharan Africa. Aliment Pharmacol Ther. 2016;44:1005-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 101] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 72. | Shimakawa Y, Lemoine M, Njai HF, Bottomley C, Ndow G, Goldin RD, Jatta A, Jeng-Barry A, Wegmuller R, Moore SE, Baldeh I, Taal M, D'Alessandro U, Whittle H, Njie R, Thursz M, Mendy M. Natural history of chronic HBV infection in West Africa: a longitudinal population-based study from The Gambia. Gut. 2016;65:2007-2016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 130] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 73. | Dumpis U, Holmes EC, Mendy M, Hill A, Thursz M, Hall A, Whittle H, Karayiannis P. Transmission of hepatitis B virus infection in Gambian families revealed by phylogenetic analysis. J Hepatol. 2001;35:99-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 85] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 74. | Martinson FE, Weigle KA, Royce RA, Weber DJ, Suchindran CM, Lemon SM. Risk factors for horizontal transmission of hepatitis B virus in a rural district in Ghana. Am J Epidemiol. 1998;147:478-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 108] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 75. | Sangaré L, Sombié R, Combasséré AW, Kouanda A, Kania D, Zerbo O, Lankoandé J. [Antenatal transmission of hepatitis B virus in an area of HIV moderate prevalence, Burkina Faso]. Bull Soc Pathol Exot. 2009;102:226-229. [PubMed] |
| 76. | Ilboudo D, Simpore J, Ouermi D, Bisseye C, Sagna T, Odolini S, Buelli F, Pietra V, Pignatelli S, Gnoula C, Nikiema JB, Musumeci S. Towards the complete eradication of mother-to-child HIV/HBV coinfection at Saint Camille Medical Centre in Burkina Faso, Africa. Braz J Infect Dis. 2010;14:219-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 77. | Eke AC, Eke UA, Okafor CI, Ezebialu IU, Ogbuagu C. Prevalence, correlates and pattern of hepatitis B surface antigen in a low resource setting. Virol J. 2011;8:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 78. | Ogawa M, Hasegawa K, Naritomi T, Torii N, Hayashi N. Clinical features and viral sequences of various genotypes of hepatitis B virus compared among patients with acute hepatitis B. Hepatol Res. 2002;23:167-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 79. | Kobayashi M, Suzuki F, Arase Y, Akuta N, Suzuki Y, Hosaka T, Saitoh S, Kobayashi M, Tsubota A, Someya T, Ikeda K, Matsuda M, Sato J, Kumada H. Infection with hepatitis B virus genotype A in Tokyo, Japan during 1976 through 2001. J Gastroenterol. 2004;39:844-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 80. | Wai CT, Fontana RJ, Polson J, Hussain M, Shakil AO, Han SH, Davern TJ, Lee WM, Lok AS; US Acute Liver Failure Study Group. Clinical outcome and virological characteristics of hepatitis B-related acute liver failure in the United States. J Viral Hepat. 2005;12:192-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 100] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 81. | Zhang HW, Yin JH, Li YT, Li CZ, Ren H, Gu CY, Wu HY, Liang XS, Zhang P, Zhao JF, Tan XJ, Lu W, Schaefer S, Cao GW. Risk factors for acute hepatitis B and its progression to chronic hepatitis in Shanghai, China. Gut. 2008;57:1713-1720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 82. | Schaefer S. Hepatitis B virus: significance of genotypes. J Viral Hepat. 2005;12:111-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 166] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 83. | Wongjarupong N, Yonli AT, Nagalo BM, Djigma FW, Somda SK, Hassan MA, Mohamed EA, Sorgho AP, Compaore TR, Soubeiga ST, Kiendrebeogo I, Sanou M, Diarra B, Yang HI, Chen CJ, Ouattara AK, Zohoncon TM, Martinson JJ, Buetow K, Chamcheu JC, Antwi SO, Borad MJ, Simpore J, Roberts LR. Characteristics of Patients With Chronic Hepatitis B Virus Infection With Genotype E Predominance in Burkina Faso. Hepatol Commun. 2020;4:1781-1792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 84. | Ishikawa T, Ichida T, Yamagiwa S, Sugahara S, Uehara K, Okoshi S, Asakura H. High viral loads, serum alanine aminotransferase and gender are predictive factors for the development of hepatocellular carcinoma from viral compensated liver cirrhosis. J Gastroenterol Hepatol. 2001;16:1274-1281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 85. | Mak D, Kramvis A. Epidemiology and aetiology of hepatocellular carcinoma in Sub-Saharan Africa. Hepatoma Res. 2021;7:39. [DOI] [Full Text] |
| 86. | Cuenca-Gómez JÁ, Lozano-Serrano AB, Cabezas-Fernández MT, Soriano-Pérez MJ, Vázquez-Villegas J, Estévez-Escobar M, Cabeza-Barrera I, Salas-Coronas J. Chronic hepatitis B genotype E in African migrants: response to nucleos(t)ide treatment in real clinical practice. BMC Infect Dis. 2018;18:568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 87. | Malagnino V, Salpini R, Maffongelli G, Battisti A, Fabeni L, Piermatteo L, Colagrossi L, Fini V, Ricciardi A, Sarrecchia C, Perno CF, Andreoni M, Svicher V, Sarmati L. High rates of chronic HBV genotype E infection in a group of migrants in Italy from West Africa: Virological characteristics associated with poor immune clearance. PLoS One. 2018;13:e0195045. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 88. | Okeke E, Davwar PM, Roberts L, Sartorius K, Spearman W, Malu A, Duguru M. Epidemiology of Liver Cancer in Africa: Current and Future Trends. Semin Liver Dis. 2020;40:111-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 89. | Lemoine M, Thursz MR. Battlefield against hepatitis B infection and HCC in Africa. J Hepatol. 2017;66:645-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 74] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 90. | Matthews PC, Geretti AM, Goulder PJ, Klenerman P. Epidemiology and impact of HIV coinfection with hepatitis B and hepatitis C viruses in Sub-Saharan Africa. J Clin Virol. 2014;61:20-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 91. | Adesina O, Oladokun A, Akinyemi O, Adedokun B, Awolude O, Odaibo G, Olaleye D, Adewole I. Human immuno-deficiency virus and hepatitis B virus coinfection in pregnancy at the University College Hospital, Ibadan. Afr J Med Med Sci. 2010;39:305-310. [PubMed] |
| 92. | Anigilaje EA, Olutola A. Prevalence and Clinical and Immunoviralogical Profile of Human Immunodeficiency Virus-Hepatitis B Coinfection among Children in an Antiretroviral Therapy Programme in Benue State, Nigeria. ISRN Pediatr. 2013;2013:932697. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 93. | Tounkara A, Sarro YS, Kristensen S, Dao S, Diallo H, Diarra B, Noumsi TG, Guindo O. Seroprevalence of HIV/HBV coinfection in Malian blood donors. J Int Assoc Physicians AIDS Care (Chic). 2009;8:47-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 94. | Jobarteh M, Malfroy M, Peterson I, Jeng A, Sarge-Njie R, Alabi A, Peterson K, Cotten M, Hall A, Rowland-Jones S, Whittle H, Tedder R, Jaye A, Mendy M. Seroprevalence of hepatitis B and C virus in HIV-1 and HIV-2 infected Gambians. Virol J. 2010;7:230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 95. | Geretti AM, Patel M, Sarfo FS, Chadwick D, Verheyen J, Fraune M, Garcia A, Phillips RO. Detection of highly prevalent hepatitis B virus coinfection among HIV-seropositive persons in Ghana. J Clin Microbiol. 2010;48:3223-3230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 96. | Laurent C, Bourgeois A, Mpoudi-Ngolé E, Kouanfack C, Ciaffi L, Nkoué N, Mougnutou R, Calmy A, Koulla-Shiro S, Ducos J, Delaporte E. High rates of active hepatitis B and C co-infections in HIV-1 infected Cameroonian adults initiating antiretroviral therapy. HIV Med. 2010;11:85-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 97. | N'Dri-Yoman T, Anglaret X, Messou E, Attia A, Polneau S, Toni T, Chenal H, Seyler C, Gabillard D, Wakasugi N, Eholié S, Danel C. Occult HBV infection in untreated HIV-infected adults in Côte d'Ivoire. Antivir Ther. 2010;15:1029-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 98. | Thio CL, Seaberg EC, Skolasky R Jr, Phair J, Visscher B, Muñoz A, Thomas DL; Multicenter AIDS Cohort Study. HIV-1, hepatitis B virus, and risk of liver-related mortality in the Multicenter Cohort Study (MACS). Lancet. 2002;360:1921-1926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 760] [Cited by in RCA: 775] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 99. | Toyé RM, Lô G, Diop-Ndiaye H, Cissé AM, Ndiaye AJS, Kébé-Fall K, Dramé A, Cohen D, Pujol FH, Mboup S, Boye CS, Chemin I, Laborde-Balen G, Taverne B, Touré-Kane C. Prevalence and molecular characterization of hepatitis B virus infection in HIV-infected children in Senegal. Clin Res Hepatol Gastroenterol. 2021;45:101502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 100. | Raimondo G, Allain JP, Brunetto MR, Buendia MA, Chen DS, Colombo M, Craxì A, Donato F, Ferrari C, Gaeta GB, Gerlich WH, Levrero M, Locarnini S, Michalak T, Mondelli MU, Pawlotsky JM, Pollicino T, Prati D, Puoti M, Samuel D, Shouval D, Smedile A, Squadrito G, Trépo C, Villa E, Will H, Zanetti AR, Zoulim F. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J Hepatol. 2008;49:652-657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 598] [Cited by in RCA: 606] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 101. | Mudawi H, Hussein W, Mukhtar M, Yousif M, Nemeri O, Glebe D, Kramvis A. Overt and occult hepatitis B virus infection in adult Sudanese HIV patients. Int J Infect Dis. 2014;29:65-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 102. | Chen XL, Li M, Zhang XL. HBV genotype B/C and response to lamivudine therapy: a systematic review. Biomed Res Int. 2013;2013:672614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 103. | Zalewska M, Domagała M, Simon K, Gładysz A. [Hepatitis B virus genotypes and the response to lamivudine therapy]. Pol Arch Med Wewn. 2005;114:1190-1199. [PubMed] |
| 104. | Lin CL, Kao JH. The clinical implications of hepatitis B virus genotype: Recent advances. J Gastroenterol Hepatol. 2011;26 Suppl 1:123-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 279] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 105. | Erhardt A, Göbel T, Ludwig A, Lau GK, Marcellin P, van Bömmel F, Heinzel-Pleines U, Adams O, Häussinger D. Response to antiviral treatment in patients infected with hepatitis B virus genotypes E-H. J Med Virol. 2009;81:1716-1720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 106. | Boglione L, Cariti G, Ghisetti V, Burdino E, Di Perri G. Extended duration of treatment with peginterferon alfa-2a in patients with chronic hepatitis B, HBeAg-negative and E genotype: A retrospective analysis. J Med Virol. 2018;90:1047-1052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 107. | Boglione L, Cardellino CS, De Nicolò A, Cariti G, Di Perri G, D'Avolio A. Different HBsAg decline after 3 years of therapy with entecavir in patients affected by chronic hepatitis B HBeAg-negative and genotype A, D and E. J Med Virol. 2014;86:1845-1850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 108. | Boglione L, Cusato J, Cariti G, Di Perri G, D'Avolio A. The E genotype of hepatitis B: clinical and virological characteristics, and response to interferon. J Infect. 2014;69:81-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 109. | Boyd A, Moh R, Gabillard D, le Carrou J, Danel C, Anglaret X, Eholié SP, Maylin S, Delaugerre C, Zoulim F, Girard PM, Lacombe K; ANRS 12240 VarBVA study. Low risk of lamivudine-resistant HBV and hepatic flares in treated HIV-HBV-coinfected patients from Côte d'Ivoire. Antivir Ther. 2015;20:643-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 110. | De Francesco MA, Gargiulo F, Spinetti A, Zaltron S, Giagulli C, Caccuri F, Castelli F, Caruso A. Clinical course of chronic hepatitis B patients receiving nucleos(t)ide analogues after virological breakthrough during monotherapy with lamivudine. New Microbiol. 2015;38:29-37. [PubMed] |
| 111. | Westland C, Delaney W 4th, Yang H, Chen SS, Marcellin P, Hadziyannis S, Gish R, Fry J, Brosgart C, Gibbs C, Miller M, Xiong S. Hepatitis B virus genotypes and virologic response in 694 patients in phase III studies of adefovir dipivoxil1. Gastroenterology. 2003;125:107-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 171] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 112. | Boglione L, Cusato J, Cariti G, Di Perri G, D'Avolio A. Role of HBsAg decline in patients with chronic hepatitis B HBeAg-negative and E genotype treated with pegylated-interferon. Antiviral Res. 2016;136:32-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 113. | Shokrgozar MA, Shokri F. Subtype specificity of anti-HBs antibodies produced by human B-cell lines isolated from normal individuals vaccinated with recombinant hepatitis B vaccine. Vaccine. 2002;20:2215-2220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 114. | Spearman CW, Sonderup MW. Preventing hepatitis B and hepatocellular carcinoma in South Africa: The case for a birth-dose vaccine. S Afr Med J. 2014;104:610-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 115. | Kao JH. Hepatitis B vaccination and prevention of hepatocellular carcinoma. Best Pract Res Clin Gastroenterol. 2015;29:907-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 116. | Gerlich WH. Prophylactic vaccination against hepatitis B: achievements, challenges and perspectives. Med Microbiol Immunol. 2015;204:39-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 117. | Poorolajal J, Mahmoodi M, Majdzadeh R, Nasseri-Moghaddam S, Haghdoost A, Fotouhi A. Long-term protection provided by hepatitis B vaccine and need for booster dose: a meta-analysis. Vaccine. 2010;28:623-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 95] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 118. | Ismail AM, Raghavendran A, Sivakumar J, Radhakrishnan M, Rose W, Abraham P. Mother to child transmission of hepatitis B virus: a cause for concern. Indian J Med Microbiol. 2015;33 Suppl:140-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 119. | Graber-Stiehl I. The silent epidemic killing more people than HIV, malaria or TB. Nature. 2018;564:24-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 120. | Andersson MI, Rajbhandari R, Kew MC, Vento S, Preiser W, Hoepelman AI, Theron G, Cotton M, Cohn J, Glebe D, Lesi O, Thursz M, Peters M, Chung R, Wiysonge C. Mother-to-child transmission of hepatitis B virus in sub-Saharan Africa: time to act. Lancet Glob Health. 2015;3:e358-e359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 121. | Keeble S, Quested J, Barker D, Varadarajan A, Shankar AG. Immunization of babies born to HBsAg positive mothers: An audit on the delivery and completeness of follow up in Norfolk and Suffolk, United Kingdom. Hum Vaccin Immunother. 2015;11:1153-1156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 122. | Ma L, Alla NR, Li X, Mynbaev OA, Shi Z. Mother-to-child transmission of HBV: review of current clinical management and prevention strategies. Rev Med Virol. 2014;24:396-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 123. | Dionne-Odom J, Njei B, Tita ATN. Elimination of Vertical Transmission of Hepatitis B in Africa: A Review of Available Tools and New Opportunities. Clin Ther. 2018;40:1255-1267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 124. | Smith S, Harmanci H, Hutin Y, Hess S, Bulterys M, Peck R, Rewari B, Mozalevskis A, Shibeshi M, Mumba M, Le LV, Ishikawa N, Nolna D, Sereno L, Gore C, Goldberg DJ, Hutchinson S. Global progress on the elimination of viral hepatitis as a major public health threat: An analysis of WHO Member State responses 2017. JHEP Rep. 2019;1:81-89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 125. | Bancroft WH, Mundon FK, Russell PK. Detection of additional antigenic determinants of hepatitis B antigen. J Immunol. 1972;109:842-848. [PubMed] |
| 126. | Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69 ( Pt 10):2575-2583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 725] [Cited by in RCA: 769] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 127. | Hauser P, Voet P, Simoen E, Thomas HC, Pêtre J, De Wilde M, Stephenne J. Immunological properties of recombinant HBsAg produced in yeast. Postgrad Med J. 1987;63 Suppl 2:83-91. [PubMed] |
| 128. | Cassidy A, Mossman S, Olivieri A, De Ridder M, Leroux-Roels G. Hepatitis B vaccine effectiveness in the face of global HBV genotype diversity. Expert Rev Vaccines. 2011;10:1709-1715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 129. | Stramer SL, Wend U, Candotti D, Foster GA, Hollinger FB, Dodd RY, Allain JP, Gerlich W. Nucleic acid testing to detect HBV infection in blood donors. N Engl J Med. 2011;364:236-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 236] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 130. | Weber B. Genetic variability of the S gene of hepatitis B virus: clinical and diagnostic impact. J Clin Virol. 2005;32:102-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 149] [Article Influence: 7.5] [Reference Citation Analysis (0)] |