Published online Oct 27, 2021. doi: 10.4254/wjh.v13.i10.1378
Peer-review started: February 27, 2021
First decision: April 18, 2021
Revised: May 4, 2021
Accepted: August 27, 2021
Article in press: August 27, 2021
Published online: October 27, 2021
Processing time: 236 Days and 18.6 Hours
Liver-secreted hepcidin is the systemic master switch of iron homeostasis and decreased levels of hepcidin are considered to cause iron overload not only in hereditary hemochromatosis but also in hemolytic anemia and chronic liver diseases. The regulation of hepcidin is complex and its response to iron is still not completely understood.
To study the direct effect of iron on various established hepcidin signaling pathways in hepatoma cells or primary hepatocytes.
Hepcidin mRNA expression was studied by quantitative real-time (qRT)-PCR in the presence of various forms of iron including ferric ammonium citrate (FAC) in hepatoma cells (Huh7), murine primary hepatocytes and an established co-culture model of phorbol myristate acetate-differentiated THP-1 monocytes and Huh7 cells. To analyze hepcidin signaling, the response to bone morphogenetic protein 6 (BMP6), interleukin (IL)-6, IL-1β, hypoxia and lipopolysaccharide (LPS) were studied. Hepcidin and small mothers against decapentaplegic 6 (SMAD6) mRNA levels were assessed by qRT-PCR and the expression of phosphorylated signal transducer and activator of transcription 3 (phospho-STAT3), STAT3, phospho-SMAD1/5/8 and SMAD1 proteins were analyzed by western blot.
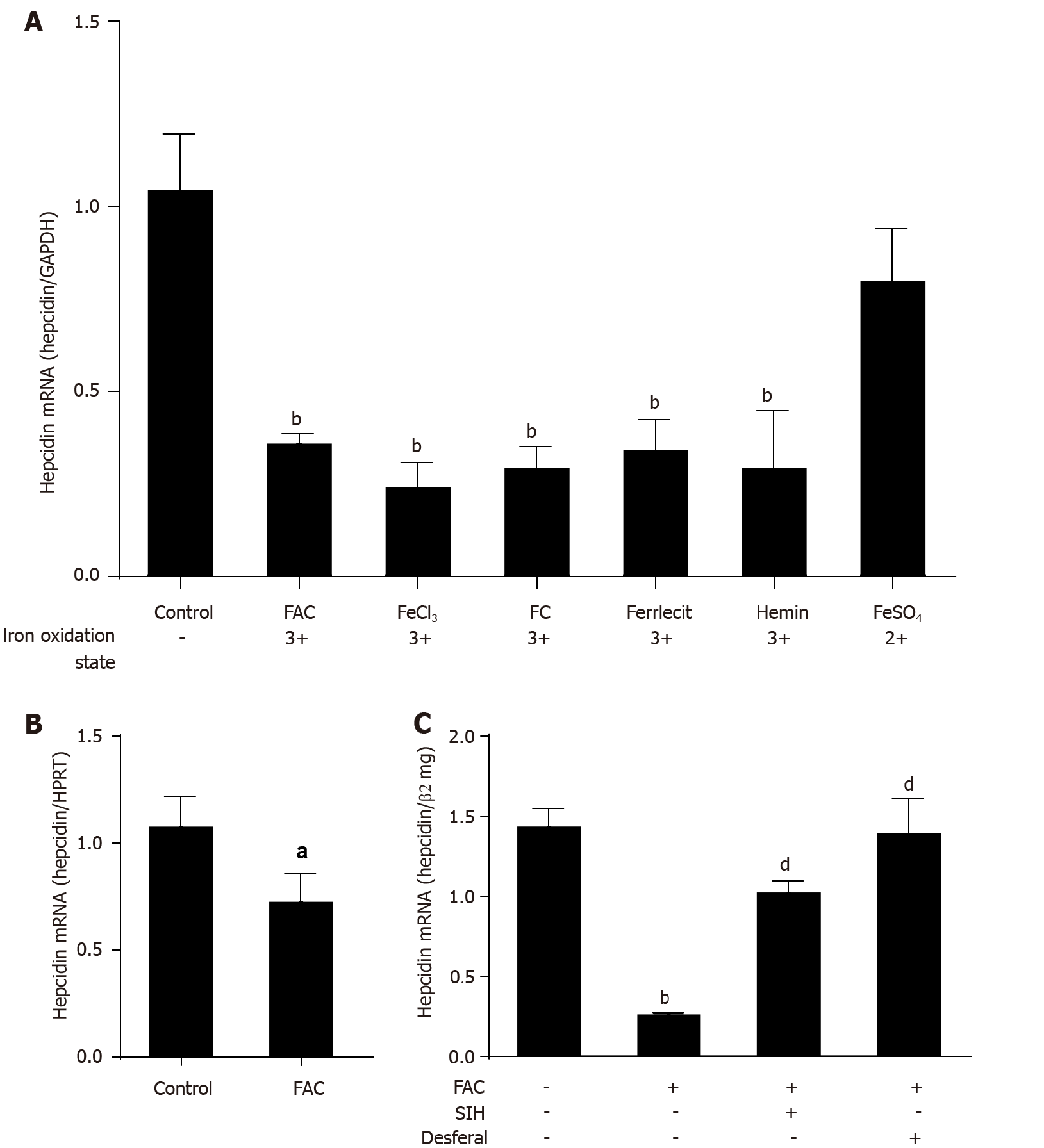
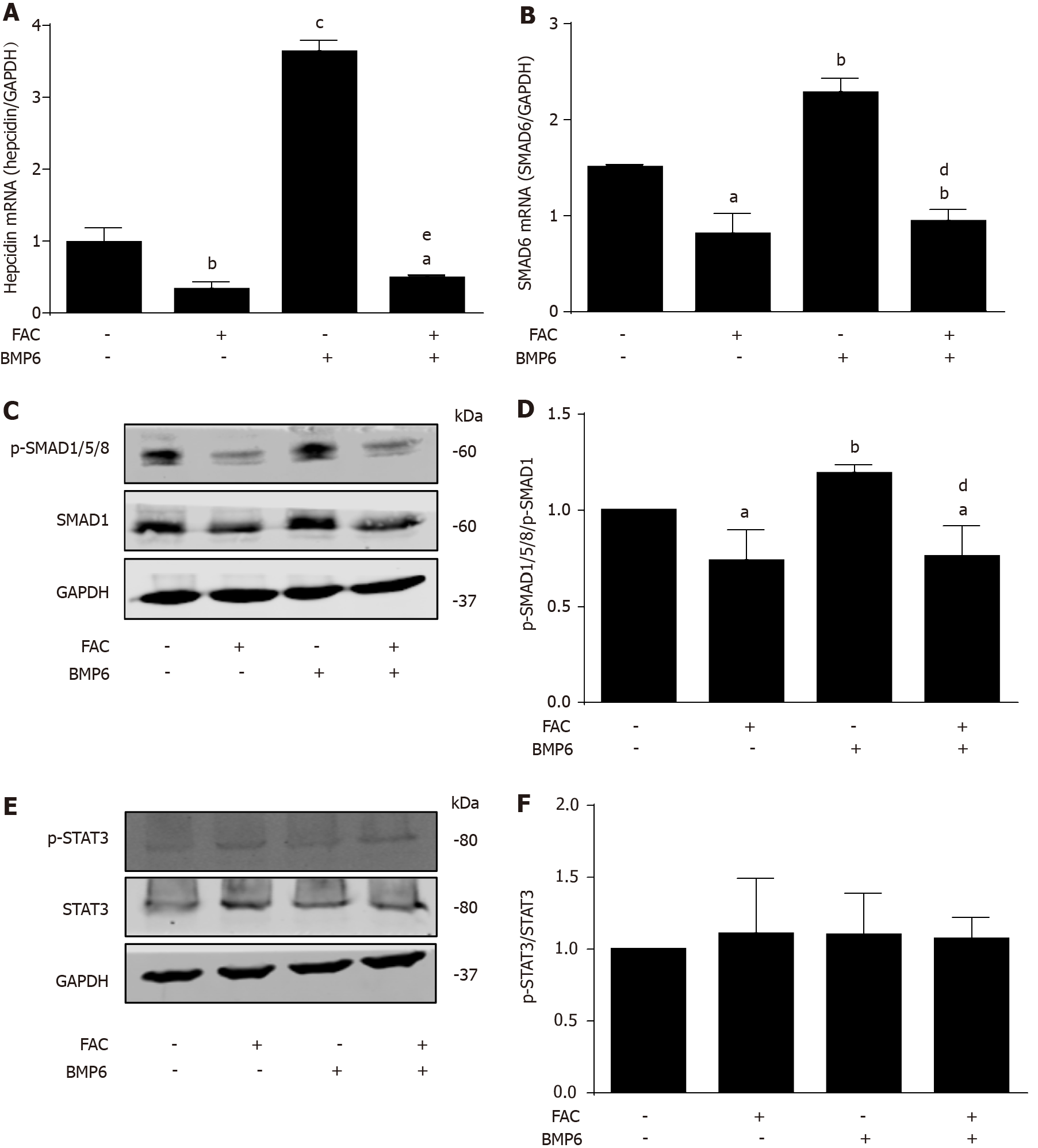
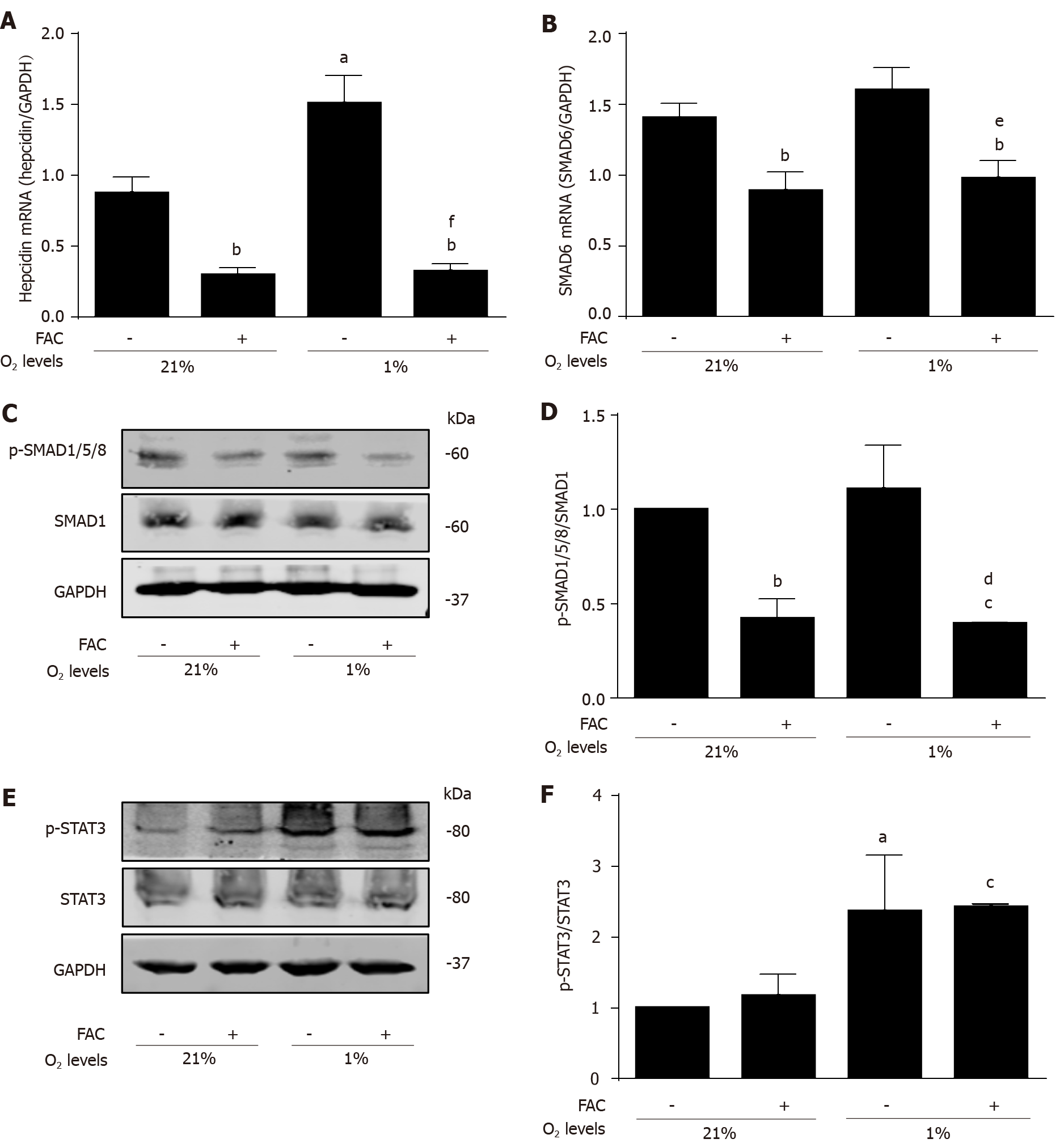
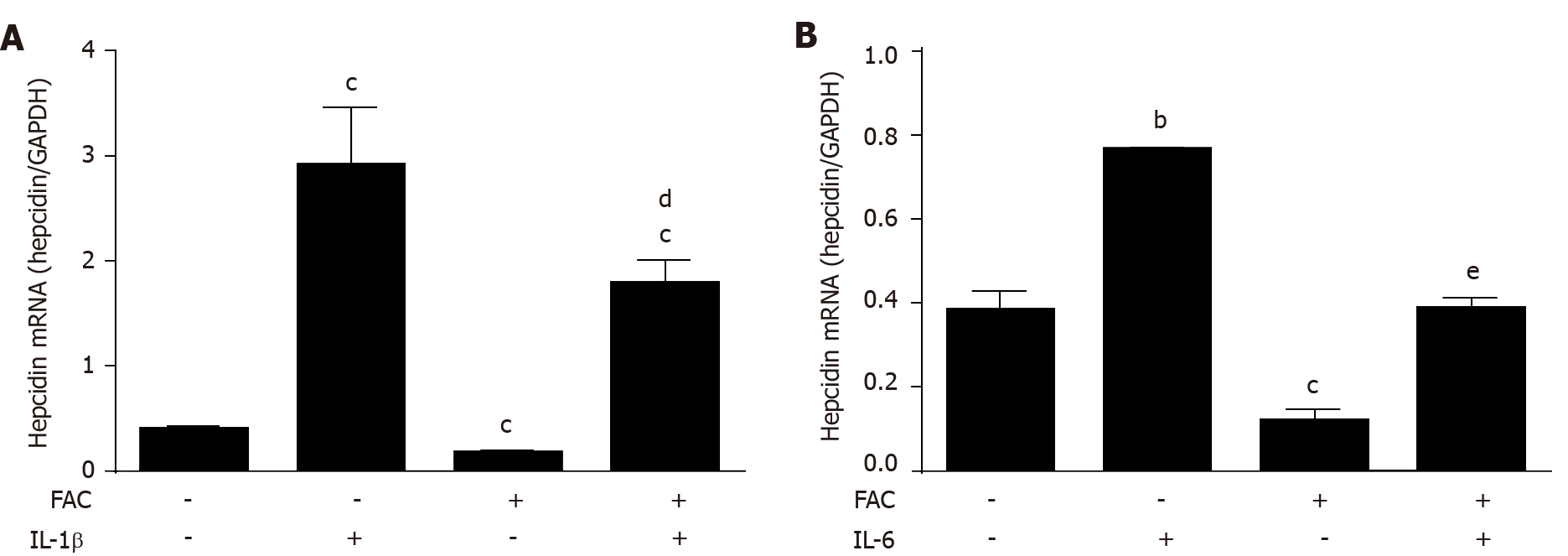
All iron III forms including FAC efficiently blocked hepcidin mRNA expression at non-toxic dosages in Huh7 cells or primary hepatocytes in a time and dose-dependent manner (P < 0.001; P < 0.05). Hepcidin blockage could be efficiently blunted by iron chelators salicylaldehyde isonicotinoyl hydrazone (SIH) and Desferal (P < 0.001). FAC also inhibited BMP6, hypoxia, IL-1β and IL-6-mediated hepcidin induction (P < 0.001; P < 0.001; P < 0.05; P < 0.001), and FAC also inhibited LPS-mediated hepatic hepcidin induction in co-culture model (P < 0.001). Moreover, FAC reduced SMAD6 mRNA and p-SMAD1/5/8 protein expression at basal or upon stimulation by BMP6 (P < 0.05; P < 0.01), and FAC also reduced SMAD6 and p-SMAD1/5/8 expression under hypoxia (P < 0.01; P < 0.05). However, FAC has no significant effect on p-STAT3 protein expression at basal or upon stimulation by various stimuli. Notably, in the presence of the BMP/SMAD signaling pathway inhibitor LDN193189 Hydrochloride (LDN), FAC was unable to further decrease hepcidin, SMAD6 and p-SMAD1/5/8 expression compared with LDN alone.
Iron directly blocks hepatocellular hepcidin signaling through the BMP/SMAD pathway but independent of STAT3. This mechanism may contribute to continued iron overload in many pathophysiological conditions ultimately causing a vicious cycle of continued hepcidin suppression.
Core Tip: Hepcidin is paradoxically and strongly suppressed during hemolytic iron overload. Although various upstream regulators of hepcidin have been discovered, the direct iron sensing mechanisms by hepcidin remain obscure. This study investigated the direct effect of iron on hepcidin signaling and for the first time to show that iron directly blocks hepcidin transcription via bone morphogenetic protein/small mothers against decapentaplegic but not the STAT3 signaling in various established in vitro models of hepcidin signaling.
- Citation: Yu LN, Wang SJ, Chen C, Rausch V, Elshaarawy O, Mueller S. Direct modulation of hepatocyte hepcidin signaling by iron. World J Hepatol 2021; 13(10): 1378-1393
- URL: https://www.wjgnet.com/1948-5182/full/v13/i10/1378.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i10.1378
Excess iron causes cancer and severe tissue damage and chronic iron overload is not only driving the rather rare hereditary iron overload diseases but also secondary iron overload diseases due to hemolysis or common chronic liver diseases such as alcoholic liver disease or hepatitis C[1]. In most of these diseases, suppression of hepcidin, the systemic master switch of iron homeostasis in mammals, has been identified to play a key role. Hepcidin is primarily expressed in hepatocytes as a precursor pro-peptide and to a lesser extent in macrophages or cardiomyocytes[2-4]. It is regulated at the transcription side, and its mRNA levels correspond well with concentrations of the peptide[5]. By binding to and degrading the iron exporter ferroportin 1 (Fpn1) which is localized at the basolateral membranes of duodenal enterocytes, macrophages and hepatocytes[6], circulating hepcidin efficiently blocks iron absorption, iron recycling and iron storage[7,8]. Consequently, its overexpression leads to hypoferremia and anemia[9], while the reduction of hepcidin levels causes iron overload[10,11].
The regulation of hepcidin is complex and the direct mechanisms of iron sensing are still not completely understood. Bone morphogenetic protein 6 (BMP6) released from endothelial cells (ECs) can efficiently induce hepcidin transcription via the SMAD pathway[12]. BMP6 binds to the BMP receptor on the liver cell membrane and its co-receptor hemojuvelin to promote the phosphorylation of the receptor-associated proteins small mothers against decapentaplegic (SMAD) 1/5/8. The latter interacts with SMAD4 to form the SMAD complex, translocates into the nucleus and binds to the hepcidin promoter[13]. In addition, inflammation mediators (e.g., IL-6, IL-1β, hypoxia or ROS/H2O2) can also induce hepcidin transcription by promoting the phosphorylation of STAT3 to initiate STAT3-mediated hepcidin signaling[14]. Cytokines namely IL-6 and microbial molecules such as lipopolysaccharide (LPS) represent an important evolutionary conserved mechanism during infection/inflammation to strongly induce hepatic hepcidin secretion leading to a rapid decrease of serum iron, which is thought to function as anti-bacterial defense mechanism[15]. More recently, the central redox signaling molecule H2O2 has been also identified as a potent inducer of hepcidin[16] with hypoxia further enhancing hepcidin-expression via the STAT3 signaling pathway[17]. Further data suggest that intracellular oxidases such as NOX4 may play an important upstream role in controlling hepcidin via the STAT3 pathway[17].
C/EBPα, BMP6, SMAD 1, 5, 8 and 4, TMPRSS6, IL-6, CREBH, CHOP and TLR4), an overall and conclusive regulatory network regarding the control of iron is not yet fully understood. This includes the experimental and clinical finding that hepcidin responds differentially to iron overload in vitro and in vivo[18-20]. Although recent data suggest important intercellular crosstalks e.g., between hepatocytes and endothelial cells or macrophages[14,21-23], the direct iron sensing mechanisms by hepcidin remain obscure. It has been reported that TfR1, ERFE or GDF15 overexpression contributes to iron overload by suppressing hepcidin in vivo[24-28]. However, there are examples that the seemingly paradox direct negative impact of iron on hepcidin, identified in vitro[19], may have direct clinical implications. For instance, in the most common human liver disease, alcoholic liver disease[29], hepatic iron overload is one of the key factors that drive the diseases and determine survival[30] with alcohol directly suppressing hepcidin[31]. In thalassemia, hepcidin is also strongly suppressed during hemolysis. While repetitive blood transfusions have been long thought to cause iron overload[32], a recently established thalassemia mouse model could demonstrate that hepatic iron overload occurs without additional blood supply through suppressed hepcidin levels[33].
These considerations prompted us to study the direct effect of iron in an in vitro setting on various established hepcidin signaling pathways including the BMP/SMAD signaling pathway and STAT3-mediated hepcidin signaling via cytokines, hypoxia, and LPS using a recently established macrophage-hepatocyte co-culture model[14]. Our data show that iron inhibits primarily the BMP/SMAD pathway but does not affect the STAT3 pathway. In conclusion, direct exposure of hepatocytes to pathophy
Huh7 cells from the Japanese Cancer Research Resources Bank (JCRB, Tokyo, Japan) were grown under standard conditions using Dulbecco's modified Eagle medium (Sigma-Aldrich, Taufkirchen, Germany), 25 mmol/L glucose and 10% fetal calf serum under 210 mL/L O2 (21% O2) and 50 mL/L CO2 (5% CO2)[16]. Murine primary hepatocytes kindly provided by Dr. Sai Wang (University of Heidelberg, Germany) were grown under standard conditions using Williams’ medium (Sigma-Aldrich, Taufkirchen, Germany), 10% fetal bovine serum, 1% P/S (Penicillin and Strep
PMA, LPS, LDN, FAC, FeCl3, FC, FeSO4, Hemin, Desferal, human recombinant IL-6 were all purchased from Sigma-Aldrich. Ferrlecit (sodium ferric gluconate) was obtained from a commercial pharmacy in its retail packaging. Human recombinant IL-1β was purchased from Enzo Lifesciences (Lörrach, Germany) and human recombinant BMP6 was purchased from R&D, Germany. SIH was a gift of Dr. P. Ponka (McGill University, Montreal, Canada).
THP-1 monocytes were differentiated to macrophages and co-cultured as described recently[14]. Briefly, THP-1 cells were seeded for differentiation with PMA (100 ng/mL) at a density of 0.25 × 105 cells/well in 12-well plates. After 48 h of differentiation, Huh7 cells were seeded on the top of macrophages at a density of 0.7 × 105 cells/well and incubated overnight for attachment. The co-culture was conditioned to LPS (0.5 μg/mL) and/or FAC (50 μmol/L) under 21% O2 and 5% CO2 for 24 h. Aiming at studying the effects of macrophage-conditioned medium, differentiated THP-1 macrophages were conditioned to LPS and/or to FAC for 24 h. Huh7 cells were exposed to the macrophage-conditioned medium for 24 h. In the co-culture experiments, a pathophysiological hepatocytes-to-macrophages ratio of 4 to 1 was used as described previously[14].
Huh7 cells were seeded at a cell density of 0.7 x 105 cell/well in 12-well plates. Huh7 cells were treated with or without FAC. Hypoxia was induced as described recently using a hypoxia chamber[14]. Briefly, cell culture plates were placed in the hypoxia chamber and flushed with a gas mixture of 1% O2, 5% CO2 and 940 mL/L N2 (94% N2) for 3 min and incubated at 37 °C for 24 h[16].
Total RNA was isolated with Trifast (Peqlab biotechnology GmbH, Erlangen, Germany) according to the manufacturer specifications. Reverse transcription and quantitative real-time PCR (qRT-PCR) reactions were performed as previously described[16]. Primers and probes were designed using the Probefinder software (Roche, Mannheim, Germany) and the sequences are shown in Table 1. Primarily, levels of hepcidin mRNA were assessed since they correspond well to the levels of the propeptide. The levels of secreted peptide are only used in clinical studies where liver biopsies are not available[5].
| Gene | Primer sequence |
| human β2-mg | forward: 5’-tga ctt tgt cac agc cca aga ta-3’ |
| reverse: 5’-aat cca aat gcg gca tct tc-3’ | |
| probe: FAM-tga tgc tgc tta cat gtc tcg atc cca-TAM | |
| human GAPDH | forward: 5′-gaa ggt gaa ggt cgg agt-3’ |
| reverse: 5′-gaa gat ggt gat ggg att tc-3’ | |
| probe: FAM-caa gct tcc cgt tct cag cc-TAM | |
| human hepcidin | forward 5′-cag gac aga gct gga gcc a-3′ |
| reverse: 5′-gca gca cat ccc aca ctt tg-3′ | |
| probe: FAM-ctg ctg cgg ctg ctg tca tcg a-TAM | |
| human SMAD6 | forward: 5′-tgc aac ccc tac cac ttc a-3′ |
| reverse: 5′-cga gga gac agc cga gag t-3′ | |
| probe UPL # 10 (Roche) | |
| mouse HPRT | forward: 5′-ggt cca ttc cta tga ctg tag att tt-3′ |
| reverse: 5′-caa tca aga cgt tct ttc cag tt-3′ | |
| probe UPL # 22 (Roche) |
Cells were washed in ice-cold 1xPBS and harvested in RIPA buffer plus 1 × Complete® protease inhibitor with EDTA (Roche Applied Sciences, Penzberg, Germany) on ice. Western Blotting was performed as described previously[16]. Following the transfer, the proteins immobilized on nitrocellulose membranes were incubated overnight with the antibodies anti-pSTAT3, anti-STAT3 (1:1000 dilution; Cell Signaling Technology, Frankfurt am Main, Germany); anti-pSMAD1/5/8, anti-SMAD1 (1:1000 dilution; Cell Signaling Technology, Frankfurt am Main, Germany) or anti-GAPDH (1:2000 dilution; Cell Signaling Technology, Frankfurt am Main, Germany). After incubation with the IRDye-conjugated 680 anti-mouse or 800 anti-rabbit antibodies (1:10000 dilutions; LI-COR, Inc., Lincoln, NE, United States), the membranes were scanned using an infrared imaging system (Odyssey CLx; LI-COR, Inc., Lincoln, NE, United States).
All the data were expressed as mean ± SD. Significant differences (P < 0.05) between means of data sets were assessed by one-way ANOVA with Tukey's test or two-way ANOVA with Sidak's test using GraphPad Prism 6 software.
Although iron injection in vivo causes strong induction of hepcidin[34,35], direct exposure of isolated hepatoma cells or murine primary hepatocytes to various forms of iron causes an efficient suppression of hepcidin mRNA expression (Figure 1A and B; P < 0.001 and P < 0.05 vs control). The inhibiting effect of iron was observed over a wide concentration range (Supplementary Figure 1) and could be efficiently blocked by two iron chelators (SIH and Desferal) (Figure 1C; P < 0.001 vs FAC group). While this “paradox” response towards iron may be explained by the absence of co-factors or other neighboring cells in vitro, the direct inhibition of hepcidin by iron may have important pathophysiological implications for hepatic iron overload in the context of chronic liver diseases or due to hemolysis. We further demonstrate that the suppression of hepcidin mRNA expression is not due to toxic or subtoxic effects as even five times higher FAC concentration did not affect growth or cell division (see Supplementary Figure 2A). Moreover, a significant suppression of hepcidin mRNA expression by FAC was observed at 6 h and continued over the observed time interval of 24 h (Supplementary Figure 2B; P < 0.001 vs control). In summary, in vitro exposure of hepatocytes to high levels of iron suppresses hepcidin, which may have important pathophysiological implications by initiating a vicious iron overloading cycle. Further experiments were carried out with FAC as a standard model for iron exposure.
We next studied the influence of iron (FAC) on BMP6-mediated hepcidin signaling, one of the major pathways in basal and iron-responsive expression of hepcidin. As shown in Figure 2A, recombinant BMP6 efficiently increased hepcidin mRNA levels by almost four times (P < 0.001 vs control). However, the presence of iron FAC not only blocked basal hepcidin expression under control conditions but completely inhibited BMP6-mediated hepcidin induction (Figure 2A; P < 0.001 vs BMP6 group). In fact, even in the presence of BMP6, FAC inhibited hepcidin mRNA levels by ca. 50% (Figure 2A; P < 0.05 vs control). Notably, BMP6 was unable to induce SMAD6 mRNA and p-SMAD1/5/8 protein expression under FAC conditions (Figure 2B, C and D; P < 0.01 vs BMP6 group), while no effect on p-STAT3 protein expression was seen (Figure 2E and F). In conclusion, in vitro, external iron has a profound inhibitory effect of basal hepcidin expression and completely abolished BMP6-mediated hepcidin signaling through SMAD but not the STAT3 pathway.
Recently, hypoxia and hydrogen peroxide have been identified as important modulators of hepcidin expression predominantly through the STAT3 pathway and involving oxidase enzymes of the NOX family[16,17]. To avoid direct interactions between iron and e.g., peroxide, we therefore next focused on hypoxia to study the role of FAC in a STAT3-mediated hepcidin signaling. In confirmation of previous experiments[14], Figure 3A demonstrates that hypoxia is able to significantly increase hepcidin mRNA levels (P < 0.05 vs normoxia control). However, hypoxia was unable to induce hepcidin mRNA expression under FAC conditions (Figure 3A; P < 0.01 vs normoxia control and P < 0.001 vs hypoxia control). Expectedly, hypoxia did not have any significant effect on SMAD6 mRNA and p-SMAD1/5/8 protein expression (Figure 3B, C and D), but efficiently upregulated p-STAT3 protein expression as shown previously (Figure 3E and F; P < 0.05 vs normoxia control). In contrast, FAC still decreased SMAD6 mRNA and p-SMAD1/5/8 protein expression under hypoxia (Figure 3B, C and D; P < 0.01 and P < 0.05 vs hypoxia control), but had no effect on p-STAT3 protein expression even under hypoxia (Figure 3E and F). These results demonstrate that FAC also and primarily affects hepcidin even in a typical STAT3-signaling setting through basal modulation of the SMAD pathway.
Cytokines such as IL-6 and IL-1β are important upstream regulators of hepcidin playing an important role in the so-called anemia of chronic disease response[36]. For instance, they are primarily responsible for the general hypoferremia observed during infections[37,38]. To study the effect of iron on cytokine signaling, hepatoma cells were exposed to FAC and/or IL-1β or IL-6 in vitro for 24 h and hepcidin mRNA was assessed by qRT-PCR. As shown in Figure 4A and B, both cytokines efficiently increased hepcidin mRNA levels while FAC blocked IL-1β-mediated induction by about 50% and IL-6-mediated induction completely (P < 0.05 vs IL-1β group and P < 0.001 vs IL-6 group). FAC not only decreased the basal but also the SMAD6 mRNA and p-SMAD1/5/8 protein expression induced by IL-1β (see Supplemen
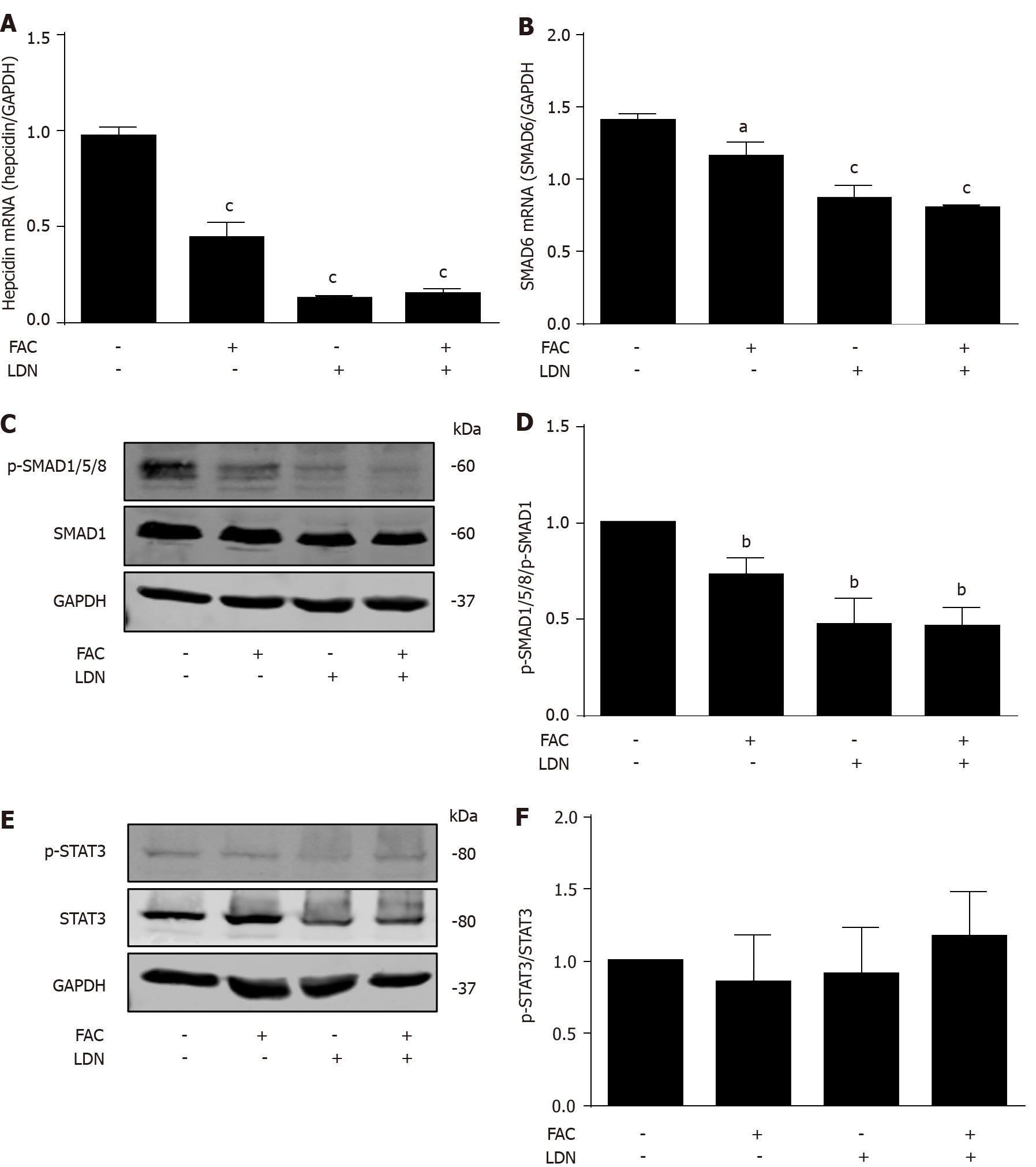
We next studied the role of BMP/SMAD signaling in the modulation of hepatocellular hepcidin by FAC using a BMP/SMAD signaling inhibitor LDN193189 (LDN)[39]. LDN suppressed the basal hepcidin mRNA expression (Figure 5A; P < 0.001 vs control), while FAC in combination with LDN could not further suppress hepcidin mRNA expression compared with LDN alone (Figure 5A). FAC in combination with LDN could not further suppress SMAD6 mRNA and p-SMAD1/5/8 protein expression compared with LDN alone (Figure 5B, C and D). Neither FAC nor LDN had a significant effect on p-STAT3 protein expression (Figure 5E and F). In conclusion, these data suggest that the BMP/SMAD signaling is necessary for FAC to inhibit hepcidin expression.
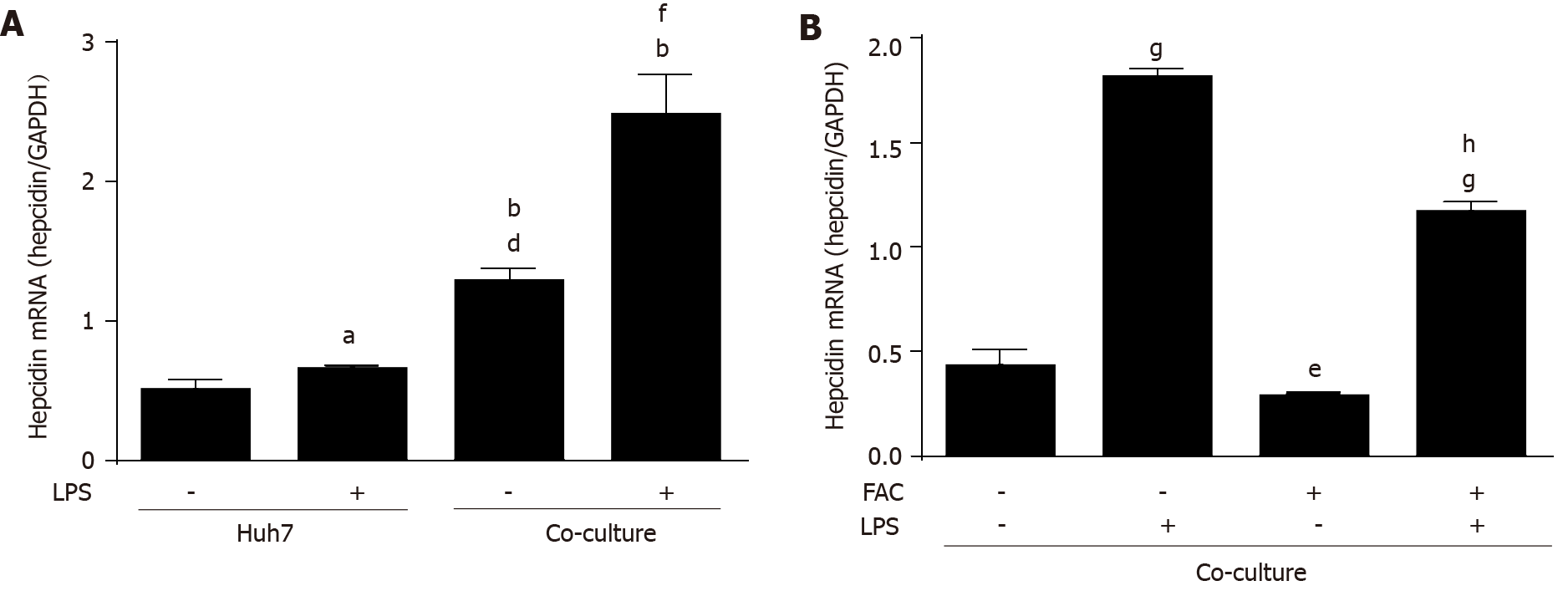
We finally studied the effect of FAC on a more complex and recently established co-culture model of macrophages and hepatocytes to mimic an inflammatory bacterial response by LPS under crosstalk conditions of both cell lines. Human THP-1 monocytes were differentiated into macrophages using PMA as described recently[40]. We examined the effect of LPS on hepatocellular hepcidin mRNA expression in the presence or absence of macrophages. A co-culture model of macrophages and hepatocytes was established according to the cell ratio of 4 to 1 of hepatocytes to macrophages in order to mimic pathophysiological cell ratios in the liver microenvironment[14]. In a normal experimental setting, THP-1 monocytes were differentiated with PMA for 24 h, washed with PBS, and then cultured in fresh medium for another 24h followed by co-cultivation for another 24h with huh7 cells. Huh7 cells were treated by LPS for 24h, and Huh7 cells were co-cultured with THP-1 macrophages in the presence of LPS or exposed to LPS-conditioned macrophage medium for 24 h. LPS slightly induced hepcidin mRNA expression in Huh7 cell monoculture. Co-culture with macrophages induced hepcidin mRNA expression (Figure 6A; P < 0.001 vs Huh7 control), which was further enhanced by LPS (Figure 6A; P < 0.001 vs co-culture control) in line with recent studies[14,41]. Notably, the effects of macrophages on hepcidin mRNA expression are even stronger than direct LPS-stimulation (Figure 6A; P < 0.001 vs Huh7 LPS group). FAC also significantly decreased hepatic hepcidin mRNA expression in our co-culture model (see Figure 6B; P < 0.05 vs control), and the presence of FAC also significantly attenuated the LPS-mediated expression of hepatic hepcidin mRNA in our co-culture model (see Figure 6B; P < 0.001 vs LPS group). As demonstrated in Supplementary Figure 5A, FAC decreased the LPS-induced SMAD6 mRNA and p-SMAD1/5/8 protein expression (P < 0.05 vs LPS group). Moreover, LPS induced p-STAT3 protein expression (see Supplementary Figure 5B; P < 0.05 vs control), while FAC had no significant effect on p-STAT3 (see Supple
We here show that iron suppresses hepatocellular hepcidin signaling directly under in vitro conditions. By exploring several established in vitro models of hepcidin signaling, we further demonstrate that this direct inhibitory effect of iron on hepcidin transcription unanimously affects the BMP-SMAD pathway but not the STAT3 pathway. Since iron-mediated blockage of hepcidin mRNA expression is also observed in primary hepatocytes at higher iron dosages and can be prevented by iron chelators, we suggest that this mechanism could contribute to hepcidin suppression in various iron overload diseases including hemolytic iron overload.
Although not widely gained attention, it has already been known for many years that hepatocellular hepcidin rapidly loses its responsiveness to iron under cultured conditions[19,41]. While this could be due to the loss of serum factors, the “in vivo liver microenvironment”, altered oxygen conditions or loss of metabolic demand ex vivo, the absence of an essential intercellular crosstalk could be another explanation. Namely with the identification of the BMP6-SMAD pathway, the role of endothelial released BMP6 has been identified as a major upstream event of the hepcidin response[23,26]. Indeed, and also shown here, exposure of cultured hepatocytes to recombinant BMP6 is able to efficiently recover the hepcidin response.
On the other hand, such paradox responses of hepcidin towards iron levels have been also well documented in patients with severe thalassemia. These patients show pronounced hemolytic anemia and require repeated blood transfusion[32]. Patients with severe disease typically show progressive liver damage and cirrhosis due to serious iron toxicity[42]. The recent establishment of a murine thalassemia model clearly demonstrates that hepatic iron overload occurs also in the absence of additional blood supply under continued hemolysis-mediated suppression of hepcidin[33].
The mechanisms behind this hepcidin suppression in hemolytic diseases are still controversially discussed. Erythropoietin (EPO) has been proposed as an important factor although the underlying mechanisms are not completely understood and cannot be recapitulated by direct exposure of hepatocytes to EPO[43]. The recent identification of bone marrow-derived erythroferrone (ERFE) and Growth Differentiation Factor-15 (GDF15) in response to EPO stimulation suggests that these factors at least partly contribute to hepcidin suppression during hemolysis[28,44-46]. However, our data on the direct inhibiting effect of iron on hepcidin signaling in vitro suggest that iron per se could also contribute to hepcidin suppression.
Chronic liver diseases represent another important model of chronic iron overload and ca. 50% of chronic liver diseases show hepatic iron overload with an inadequate hepcidin response[30]. While primary liver damage either through alcohol damage or viral replication could account for the total loss of hepcidin response[47-49], iron itself could also play a regulatory role. In our various in vitro models of hepcidin signaling, we here demonstrate that iron efficiently blocks hepcidin response primarily through the SMAD pathway. Although this seems rather counteractive towards the iron-mediated BMP-hepcidin response, this experiment deserves serious consideration especially during pathophysiological conditions such as severe hemolysis or damage to the liver sinus-endothelial layer. It may explain why continued hepatic iron overload would initiate a vicious cycle of hepcidin suppression and further iron uptake through the duodenal brush border[50]. It would also implicate that besides pharmacological approaches to re-introduce hepcidin or increase hepcidin peptide levels (e.g., mini hepcidins), removal of iron remains the cornerstone of the treatment. Not only would it remove the primary toxic agent iron but it would interrupt the suppressing effect of hepcidin on iron. It may also stimulate a mechanistic discussion on the therapeutic usage of iron chelators vs phlebotomy.
Although our data clearly show an exclusive effect of in vitro iron on the SMAD signaling cascade, the direct molecular mechanisms still remain elusive. Notably, hepcidin signaling was inhibited by iron in all explored models including the co-culture model with macrophages. Even in primary STAT3-mediated processes such as cytokines, hypoxia or LPS, iron efficiently blocked hepcidin transcription underlining the important role of the SMAD pathway for basal hepcidin expression. In line with this is the observation that efficient SMAD blockage by the SMAD inhibitor LDN could not be further enhanced by iron. Second, experiments with membrane permeable or non-permeable iron chelators (SIH or Desferal) show that iron chelators efficiently counteract the inhibitory effect of iron on hepcidin. Although do not provide definite answers to the underlying mechanisms of the iron-mediated hepcidin inhibition, the almost immediate effect restricted to the SMAD pathway and the fact that only oxidized forms of iron are effective suggests to us that iron may directly act through the BMP receptor or associated molecules such as TfR1 or TfR2[30].
On a final note, we were surprised not to see any interaction of iron with the STAT3 pathway. Since STAT3 is responsive to peroxide and iron and H2O2 are known for decades to chemically interfere via the Fenton chemistry[30], it would have been no surprise to see direct effects on hepcidin transcription. However, it remains open whether compensating mechanisms exist to counteract decreased peroxide levels e.g. by upregulating oxidases etc.
In summary, to our knowledge, this work is the first to show that iron directly blocks hepcidin transcription, at baseline or upon stimulation by different stimuli, through the BMP/SMAD but not STAT3 signaling in vitro. A summarizing scheme is shown in Figure 7. We think that in addition to potential hepcidin suppressing factors such as GDF15 or ERFE, iron could directly block hepcidin transcription under conditions of either excess iron or a liver endothelial fenestration with larger access to the hepatocellular membrane. Specifically under pathological conditions such as severe hemolysis or chronic iron overload as observed in alcoholic liver disease, this novel mechanism may contribute to further iron overload and initiate a vicious cycle. To interrupt this cycle, the removal of iron should be the most efficient therapeutic goal. It will not be an easy task to validate this concept in in vivo models since iron levels in the direct environment of hepatocytes are not easy to quantitate.
In conclusion, iron including FAC per se, directly blocks hepcidin transcription and the inhibitory effect could be observed over a large concentration range involving all forms of iron-III, which was not caused by toxicity or inhibition of cell growth. FAC has a profound inhibitory effect on hepcidin expression at baseline or upon stimulation by stimuli in various cell models, which was controlled through the BMP/SMAD pathway but independent of STAT3. We suggest that this mechanism may contribute to continued iron overload in many pathophysiological conditions ultimately causing a vicious cycle of continued hepcidin suppression. Anyway, this study provides a new idea for in-depth exploration of iron overload diseases and provides an experimental basis for the underlying therapeutic goal.
Excess iron causes cancer and severe tissue damage and chronic iron overload is not only driving the rather rare hereditary iron overload diseases but also secondary iron overload diseases due to hemolysis or common chronic liver diseases such as alcoholic liver disease or hepatitis C. In most of these diseases, suppression of hepcidin, the systemic master switch of iron homeostasis in mammals, has been identified to play a key role. Hepcidin is primarily expressed in hepatocytes as a precursor pro-peptide and to a lesser extent in macrophages or cardiomyocytes. Elevated hepcidin causes hypoferremia and anemia by efficiently blocking iron absorption, iron recycling and iron storage by binding to and degrading the major iron export pump ferroportin 1.
The direct iron sensing mechanisms by hepcidin remain obscure and seemingly paradox response of hepcidin have been observed in various clinical scenarios. Thus, direct intravenous injection of iron causes rapid induction of hepcidin, iron release in the context of hemolytic diseases such as thalassemia efficiently block hepcidin expression and cause further detrimental iron accumulation. Moreover, it still remains largely unexplained why hepatocellular hepcidin is downregulated under in vitro conditions. These observations prompted us to study in detail the direct effect of iron in cultured hepatocytes.
The authors here aimed to study the direct effect of iron on various established hepcidin signaling pathways including the bone morphogenetic protein (BMP)/small mothers against decapentaplegic (SMAD) signaling pathway and signal transducer and activator of transcription 3 (STAT3)-mediated hepcidin signaling via cytokines, hypoxia, and lipopolysaccharide (LPS) using a recently established macrophage-hepatocyte co-culture model.
Hepcidin mRNA expression in presence of various forms of iron was studied, using hepatoma cells (Huh7), murine primary hepatocyte and a co-culture model of phorbol myristate acetate-differentiated THP-1 monocytes and hepatoma cells. The response to BMP6, interleukin (IL)-6, IL-1β, hypoxia and LPS were studied in order to analyze hepcidin signaling. Hepcidin and SMAD6 mRNA levels were assessed and the expression of phospho-STAT3, STAT3, phospho-SMAD1/5/8 and SMAD1 proteins were analyzed.
All iron III forms including ferric ammonium citrate efficiently blocked hepcidin mRNA expression at non-toxic dosages in hepatoma cells or primary hepatocytes. Using iron chelators, the blockage of hepcidin by iron could be efficiently blunted. Iron also had a profound inhibitory effect of basal hepcidin expression and completely abolished BMP6-mediated hepcidin signaling through SMAD but not the STAT3 pathway. Iron also and primarily affected hepcidin even in a typical STAT3-signaling setting through basal modulation of the SMAD pathway and iron significantly attenuated hepcidin response to cytokines, which is SMAD dependent but does not involve STAT3. In the co-culture model, iron inhibited LPS-mediated hepcidin induction.
In conclusion, iron directly blocks hepatocellular hepcidin transcription involving all forms of iron III and the effect was not caused by toxicity or reduced cell growth. Iron also inhibits hepcidin upregulation in various models of hepcidin stimulation primarily through the BMP/SMAD pathway but independent of STAT3 signaling. We propose that his mechanism may contribute to continued iron overload at least under pathophysiological conditions of iron release ultimately causing a vicious cycle of continued hepcidin suppression and further iron overload.
This study provides a new concept for better understanding the seemingly paradox response of hepcidin in in vivo and in vitro settings. Moreover, understanding the direct inhibitory effects of iron on hepcidin signaling at the hepatocellular side could help to identify novel molecular targets for future therapies.
We thank Dr. Wang S for kindly providing murine primary hepatocytes and Dr. Ponka P for kindly providing SIH. We thank the financial support of the China Scholarship Council (CSC) for Yu LN, Wang SJ, and Chen C.
Manuscript source: Invited manuscript
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: Germany
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Rychtrmoc D S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Wu RR
| 1. | Abergel A, Ruivard M, Bonny C. [Iron overload and chronic liver diseases]. Rev Prat. 2006;56:2130-2134. [PubMed] |
| 2. | Sow FB, Florence WC, Satoskar AR, Schlesinger LS, Zwilling BS, Lafuse WP. Expression and localization of hepcidin in macrophages: a role in host defense against tuberculosis. J Leukoc Biol. 2007;82:934-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 156] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 3. | Papanikolaou G, Pantopoulos K. Systemic iron homeostasis and erythropoiesis. IUBMB Life. 2017;69:399-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 4. | Lakhal-Littleton S. Cardiomyocyte hepcidin: From intracellular iron homeostasis to physiological function. Vitam Horm. 2019;110:189-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 5. | Détivaud L, Nemeth E, Boudjema K, Turlin B, Troadec MB, Leroyer P, Ropert M, Jacquelinet S, Courselaud B, Ganz T, Brissot P, Loréal O. Hepcidin levels in humans are correlated with hepatic iron stores, hemoglobin levels, and hepatic function. Blood. 2005;106:746-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 132] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 6. | Ganz T. Systemic iron homeostasis. Physiol Rev. 2013;93:1721-1741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 623] [Cited by in RCA: 773] [Article Influence: 64.4] [Reference Citation Analysis (0)] |
| 7. | Ward DM, Kaplan J. Ferroportin-mediated iron transport: expression and regulation. Biochim Biophys Acta. 2012;1823:1426-1433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 272] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 8. | Ross SL, Tran L, Winters A, Lee KJ, Plewa C, Foltz I, King C, Miranda LP, Allen J, Beckman H, Cooke KS, Moody G, Sasu BJ, Nemeth E, Ganz T, Molineux G, Arvedson TL. Molecular mechanism of hepcidin-mediated ferroportin internalization requires ferroportin lysines, not tyrosines or JAK-STAT. Cell Metab. 2012;15:905-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 9. | Nicolas G, Bennoun M, Porteu A, Mativet S, Beaumont C, Grandchamp B, Sirito M, Sawadogo M, Kahn A, Vaulont S. Severe iron deficiency anemia in transgenic mice expressing liver hepcidin. Proc Natl Acad Sci U S A. 2002;99:4596-4601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 618] [Cited by in RCA: 612] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 10. | Nicolas G, Bennoun M, Devaux I, Beaumont C, Grandchamp B, Kahn A, Vaulont S. Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc Natl Acad Sci U S A. 2001;98:8780-8785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 917] [Cited by in RCA: 908] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 11. | Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, Loukopoulos D, Camaschella C. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 634] [Cited by in RCA: 585] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 12. | Andriopoulos B Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, Knutson MD, Pietrangelo A, Vukicevic S, Lin HY, Babitt JL. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41:482-487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 633] [Cited by in RCA: 592] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 13. | Core AB, Canali S, Babitt JL. Hemojuvelin and bone morphogenetic protein (BMP) signaling in iron homeostasis. Front Pharmacol. 2014;5:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 14. | Silva I, Peccerella T, Mueller S, Rausch V. IL-1 beta-mediated macrophage-hepatocyte crosstalk upregulates hepcidin under physiological low oxygen levels. Redox Biol. 2019;24:101209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Weiss G, Goodnough LT. Anemia of chronic disease. N Engl J Med. 2005;352:1011-1023. [PubMed] |
| 16. | Millonig G, Ganzleben I, Peccerella T, Casanovas G, Brodziak-Jarosz L, Breitkopf-Heinlein K, Dick TP, Seitz HK, Muckenthaler MU, Mueller S. Sustained submicromolar H2O2 Levels induce hepcidin via signal transducer and activator of transcription 3 (STAT3). J Biol Chem. 2012;287:37472-37482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 17. | Silva I, Rausch V, Peccerella T, Millonig G, Seitz HK, Mueller S. Hypoxia enhances H2O2-mediated upregulation of hepcidin: Evidence for NOX4-mediated iron regulation. Redox Biol. 2018;16:1-10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 18. | Ramos E, Kautz L, Rodriguez R, Hansen M, Gabayan V, Ginzburg Y, Roth MP, Nemeth E, Ganz T. Evidence for distinct pathways of hepcidin regulation by acute and chronic iron loading in mice. Hepatology. 2011;53:1333-1341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 186] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 19. | Nemeth E, Valore EV, Territo M, Schiller G, Lichtenstein A, Ganz T. Hepcidin, a putative mediator of anemia of inflammation, is a type II acute-phase protein. Blood. 2003;101:2461-2463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 962] [Cited by in RCA: 995] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 20. | Trombini P, Paolini V, Pelucchi S, Mariani R, Nemeth E, Ganz T, Piperno A. Hepcidin response to acute iron intake and chronic iron loading in dysmetabolic iron overload syndrome. Liver Int. 2011;31:994-1000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Takayama G, Taniguchi A, Okano T. Identification of differentially expressed genes in hepatocyte/endothelial cell co-culture system. Tissue Eng. 2007;13:159-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 22. | Canali S, Zumbrennen-Bullough KB, Core AB, Wang CY, Nairz M, Bouley R, Swirski FK, Babitt JL. Endothelial cells produce bone morphogenetic protein 6 required for iron homeostasis in mice. Blood. 2017;129:405-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 165] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 23. | Silvestri L, Nai A, Dulja A, Pagani A. Hepcidin and the BMP-SMAD pathway: An unexpected liaison. Vitam Horm. 2019;110:71-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 24. | Li H, Choesang T, Bao W, Chen H, Feola M, Garcia-Santos D, Li J, Sun S, Follenzi A, Pham P, Liu J, Zhang J, Ponka P, An X, Mohandas N, Fleming RE, Rivella S, Li G, Ginzburg YZ. Decreasing TfR1 expression reverses anemia and hepcidin suppression in β-thalassemic mice. Blood. 2017;129:1514-1526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 25. | El-Gamal RAE, Abdel-Messih IY, Habashy DM, Zaiema SEG, Pessar SA. Erythroferrone, the new iron regulator: evaluation of its levels in Egyptian patients with beta thalassemia. Ann Hematol. 2020;99:31-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 26. | Arezes J, Foy N, McHugh K, Sawant A, Quinkert D, Terraube V, Brinth A, Tam M, LaVallie ER, Taylor S, Armitage AE, Pasricha SR, Cunningham O, Lambert M, Draper SJ, Jasuja R, Drakesmith H. Erythroferrone inhibits the induction of hepcidin by BMP6. Blood. 2018;132:1473-1477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 205] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 27. | Kim A, Nemeth E. New insights into iron regulation and erythropoiesis. Curr Opin Hematol. 2015;22:199-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 120] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 28. | Tanno T, Bhanu NV, Oneal PA, Goh SH, Staker P, Lee YT, Moroney JW, Reed CH, Luban NL, Wang RH, Eling TE, Childs R, Ganz T, Leitman SF, Fucharoen S, Miller JL. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin. Nat Med. 2007;13:1096-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 600] [Cited by in RCA: 609] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 29. | World Health Organization. Global status report on alcohol and health. Available from https://www.who.int/substance_abuse/publications/alcohol_2014/en. |
| 30. | Mueller S, Rausch V. The role of iron in alcohol-mediated hepatocarcinogenesis. Adv Exp Med Biol. 2015;815:89-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 31. | Harrison-Findik DD, Klein E, Evans J, Gollan J. Regulation of liver hepcidin expression by alcohol in vivo does not involve Kupffer cell activation or TNF-alpha signaling. Am J Physiol Gastrointest Liver Physiol. 2009;296:G112-G118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 32. | Wahidiyat PA, Iskandar SD, Sekarsari D. Evaluation of Iron Overload Between Age Groups Using Magnetic Resonance Imaging and Its Correlation with Iron Profile in Transfusion-dependent Thalassemia. Acta Med Indones. 2018;50:230-236. [PubMed] |
| 33. | Liu J, Liu W, Liu Y, Miao Y, Guo Y, Song H, Wang F, Zhou H, Ganz T, Yan B, Liu S. New thiazolidinones reduce iron overload in mouse models of hereditary hemochromatosis and β-thalassemia. Haematologica. 2019;104:1768-1781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 34. | Burden RJ, Pollock N, Whyte GP, Richards T, Moore B, Busbridge M, Srai SK, Otto J, Pedlar CR. Effect of Intravenous Iron on Aerobic Capacity and Iron Metabolism in Elite Athletes. Med Sci Sports Exerc. 2015;47:1399-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 35. | Wang JW, Xu LH, Yao X, Fang JP. [Expression Changes of Hepcidin and Ferroportin 1 in Murine Model of Iron Overload]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2017;25:936-940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 36. | Matsumura I, Kanakura Y. [Pathogenesis of anemia of chronic disease]. Nihon Rinsho. 2008;66:535-539. [PubMed] |
| 37. | Moran-Lev H, Weisman Y, Cohen S, Deutsch V, Cipok M, Bondar E, Lubetzky R, Mandel D. The interrelationship between hepcidin, vitamin D, and anemia in children with acute infectious disease. Pediatr Res. 2018;84:62-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 38. | Queiroz DM, Rocha AM, Melo FF, Rocha GA, Teixeira KN, Carvalho SD, Bittencourt PF, Castro LP, Crabtree JE. Increased gastric IL-1β concentration and iron deficiency parameters in H. pylori infected children. PLoS One. 2013;8:e57420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 39. | Cuny GD, Yu PB, Laha JK, Xing X, Liu JF, Lai CS, Deng DY, Sachidanandan C, Bloch KD, Peterson RT. Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg Med Chem Lett. 2008;18:4388-4392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 307] [Cited by in RCA: 285] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 40. | Matak P, Chaston TB, Chung B, Srai SK, McKie AT, Sharp PA. Activated macrophages induce hepcidin expression in HuH7 hepatoma cells. Haematologica. 2009;94:773-780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 41. | Montosi G, Corradini E, Garuti C, Barelli S, Recalcati S, Cairo G, Valli L, Pignatti E, Vecchi C, Ferrara F, Pietrangelo A. Kupffer cells and macrophages are not required for hepatic hepcidin activation during iron overload. Hepatology. 2005;41:545-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 56] [Reference Citation Analysis (0)] |
| 42. | Hershko C. Pathogenesis and management of iron toxicity in thalassemia. Ann N Y Acad Sci. 2010;1202:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 104] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 43. | Pasricha SR, McHugh K, Drakesmith H. Regulation of Hepcidin by Erythropoiesis: The Story So Far. Annu Rev Nutr. 2016;36:417-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 44. | Ganz T. Erythropoietic regulators of iron metabolism. Free Radic Biol Med. 2019;133:69-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 45. | Sasaki Y, Noguchi-Sasaki M, Yasuno H, Yorozu K, Shimonaka Y. Erythropoietin stimulation decreases hepcidin expression through hematopoietic activity on bone marrow cells in mice. Int J Hematol. 2012;96:692-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 46. | Kautz L, Jung G, Valore EV, Rivella S, Nemeth E, Ganz T. Identification of erythroferrone as an erythroid regulator of iron metabolism. Nat Genet. 2014;46:678-684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 817] [Cited by in RCA: 822] [Article Influence: 74.7] [Reference Citation Analysis (0)] |
| 47. | Varghese J, James JV, Sagi S, Chakraborty S, Sukumaran A, Ramakrishna B, Jacob M. Decreased hepatic iron in response to alcohol may contribute to alcohol-induced suppression of hepcidin. Br J Nutr. 2016;115:1978-1986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 48. | Fujita N, Sugimoto R, Takeo M, Urawa N, Mifuji R, Tanaka H, Kobayashi Y, Iwasa M, Watanabe S, Adachi Y, Kaito M. Hepcidin expression in the liver: relatively low level in patients with chronic hepatitis C. Mol Med. 2007;13:97-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 156] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 49. | Gao YH, Wang JY, Liu PY, Sun J, Wang XM, Wu RH, He XT, Tu ZK, Wang CG, Xu HQ, Niu JQ. Iron metabolism disorders in patients with hepatitis B-related liver diseases. World J Clin Cases. 2018;6:600-610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 20] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (1)] |
| 50. | Bergamaschi G, Di Sabatino A, Pasini A, Ubezio C, Costanzo F, Grataroli D, Masotti M, Alvisi C, Corazza GR. Intestinal expression of genes implicated in iron absorption and their regulation by hepcidin. Clin Nutr. 2017;36:1427-1433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |