Published online Oct 27, 2021. doi: 10.4254/wjh.v13.i10.1234
Peer-review started: March 4, 2021
First decision: May 2, 2021
Revised: May 14, 2021
Accepted: September 10, 2021
Article in press: September 10, 2021
Published online: October 27, 2021
Processing time: 231 Days and 15.9 Hours
Hepatitis C virus (HCV) is responsible for no less than 71 million people chroni
Core Tip: Hepatitis C virus (HCV) remains a global health burden despite the successful introduction of direct-acting antiviral therapies. In order to achieve global control of HCV epidemic a vaccine is necessary. Its development has faced many hurdles, reason why it is still elusive. Herein, we describe all the challenges during HCV vaccine research, focusing on HCV immunology and emphasizing on current vaccine candidates, particularly nucleic acid-based as well as recombinant vector-based vaccines. We also highlight the impact of severe acute respiratory syndrome coronavirus-2 vaccine race on the renewed interest on HCV vaccine production. Finally, we present ideas on live-attenuated vaccine approaches against HCV.
- Citation: Echeverría N, Comas V, Aldunate F, Perbolianachis P, Moreno P, Cristina J. In the era of rapid mRNA-based vaccines: Why is there no effective hepatitis C virus vaccine yet? . World J Hepatol 2021; 13(10): 1234-1268
- URL: https://www.wjgnet.com/1948-5182/full/v13/i10/1234.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i10.1234
Hepatitis C virus (HCV), discovered in 1989[1], represents an important health burden. In 2015, the World Health Organization (WHO) estimated that there were at least 71 million people chronically infected with HCV, which represents a global prevalence of approximately 1%[2]. Additionally, around 400000 deaths occurred from infection complications.
Infections with HCV cause both acute as well as chronic liver disease in 60%-80% of the cases. Chronicity is associated with the development of cirrhosis (15%-30%) and hepatocellular carcinoma (HCC)[3]. Liver damage resulting from this infection makes it one of the most frequent indications for liver transplantation worldwide[4-8].
The problem of HCV infections worldwide has led the WHO to propose the elimination of viral hepatitis as a public health burden by 2030[2]. However, in order to achieve this goal, big scale interventions are needed, such as screening testing, effective treatment and hopefully vaccination, the latter still non-existing for HCV.
Access to widely available screening tests is uncommon and is hindered by economic reasons, particularly given the fact that new HCV infections are mainly asymptomatic[9]. This leads to an underestimation of the disease prevalence and does not contribute to the eradication goal. Concerning treatment, the development of interferon-free (IFN-free) regimens based in direct-acting antivirals (DAAs) has revolutionized HCV therapy. These antivirals have significantly increased response rates (up to 98%) and greatly reduced treatment duration to only 8-12 wk of oral treatment. DAAs have generated optimism on the global control front, and some consider that this pathogen can now be effectively controlled solely by means of antiviral therapy[10,11]. However, there are some limitations and obstacles to keep the virus in check, in particular, the cost and practical aspects of treatment access, which is uneven among different countries and leaves underdeveloped regions without treatment[11]. Additionally, resistance to DAAs emerged concomitantly with their development and implementation. Resistance-associated substitutions have been detected both before as well as during and after treatment with DAAs[12]. Another interesting aspect to consider is that eliminating HCV infection with DAAs does not eradicate the risk of developing liver cancer. Also, protective immunity is usually insufficient after natural or treatment-induced viral clearance, thus, the possibility of reinfection remains[13]. Together, these facts make HCV elimination in high-risk groups a very challenging task and the need for an effective prophylactic vaccine remains the greatest uncovered medical problem in the hepatitis C field[14]. Vacci
Proper immune responses are able to clear HCV acute infections, preventing the progression to chronicity (in 20%-40% of infected individuals). This fact suggests that vaccination could be a reasonable goal[19] provided we grasp a better understanding of immune responses against HCV in order to develop different vaccine candidates that allow for appropriate protection.
Global epidemic control will only be possible if the number of new HCV infections is reduced alongside with an increased number of cured patients[11,14]. However, a recent report showed that almost 60% of 91 surveyed countries had, in 2016, higher rates of infection than cures, making the goal of HCV elimination as a health burden by 2030, difficult to achieve[20].
For all the reasons previously mentioned, safe and effective prophylactic and/or therapeutic vaccines are necessary for the global control of HCV epidemic[11,21-24]. Indeed, no infectious disease has been controlled and eradicated with antimicrobial treatment, while it has in fact been possible by vaccination[10]. Furthermore, effective vaccination strategies widely available have been the only unfailing method to keep viral transmission at bay by providing herd immunity[25]. Modelling studies have indicated that, even with the introduction of new DAA treatments, only a quasi-eradication of HCV would be possible[26,27], highlighting the need for a vaccine against HCV.
Two extraordinary and unique situations that took place during this last year have fueled optimism on vaccine development against HCV. First, the Nobel Prize in Phy
This review focuses on different vaccine candidates designed to prevent or diminish HCV infection cases, and summarizes all the pitfalls encountered during vaccine development against this virus, including some key aspects of HCV immunology. We make special emphasis on nucleic acid-based vaccines as well as recombinant viral vectors and provide information on severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) vaccines as examples of approaches that might be important in HCV vaccine development.
Vaccine candidates with two different goals have been considered to control HCV epidemic: Prophylactic and therapeutic (primary and secondary prevention, respectively). The most widespread use of vaccination has always been to prevent a particular disease (prophylactic vaccination)[31] by building immunity in an indi
As we will later discuss in detail, the challenges for designing an effective prophylactic vaccine are vast (HCV variability and diversity, limited animal models and a complex immunological response). Many preventive vaccines against other viral patho
Therapeutic vaccines against HCV have great potential to aid in controlling chronic infections by increasing curing rates or reducing therapy duration[36]. In this new DAA era, sustained virological response (SVR) rates are extremely high (above 98%) and treatment duration has already been shortened compared to classic dual therapy (pegylated IFN-α plus ribavirin). However, there are difficult to treat patients (with active HCC or severe liver decompensation, those experiencing multiple DAA treatment failures, or those infected with HCV genotype 3)[37] for which this therapeutic approach would be beneficial. These vaccines would boost HCV-specific T cell responses and would help in three different ways: (1) Preventing viral relapse if therapeutic vaccines were to be administered in conjunction with DAA therapy; (2) Maximizing early viral clearance and thus increasing SVR rates by first employing a therapeutic vaccine followed by the antiviral treatment; and (3) Producing partial control of HCV infection just by means of therapeutic immunization and thus redu
In general, effective vaccine candidates should stimulate generation of nAbs and a proper cellular immune response. In order to design vaccines that elicit protective immunity against HCV, it is of utmost importance to consider the virus tropism (mainly hepatocytes), transmission route (parenteral transmission through contaminated blood) and pathogenesis[39].
A vaccine that induces immune responses similar to those produced by individuals which have successfully cleared the virus after an acute HCV infection, might prove valuable[19]. As we will discuss in the next section, vigorous responses of broadly cross-reactive CD4+, CD8+ T cells to conserved epitopes[40-42], as well as nAbs contri
Approximately 20%–40% of HCV-infected patients clear the virus spontaneously, while the rest develop a persistent infection that will result in severe fibrosis, cirrhosis and HCC[3,45]. Thus, it is essential to understand the immune protection induced during acute infections in patients that achieved spontaneous viral clearance in order to determine the immune parameters that a successful vaccine has to reach.
Multiple evidences in human and animal models have demonstrated the undoubted association of spontaneous viral clearance with a broad, sustained HCV-specific T cell-mediated immunity (CMI) to conserved HCV non-structural proteins[46,47] and nAb targeting conserved regions of viral envelope glycoproteins E1E2[48].
As will be detailed below, both arms of the immune response are primed during HCV infection, but the characteristics vary depending on whether an acute infection is spontaneously resolved or if it evolves to chronicity.
While HCV-specific CD8+ T cells are the main effector cells, the outcome of infection depends on eliciting efficient virus-specific CD4+ T cell responses[49]. These cells are the central regulators of adaptative immunity providing help for priming CD8+ T cell response as well as antibody response during viral infections. The breadth of the T cell response is a key determinant to spontaneously clear HCV. High numbers of CD4+ and CD8+ T cells targeting different epitopes were observed in individuals who resolved acute infections in comparison to those who evolve to chronicity[42,50,51]. These cells are multi-specifically targeting both structural and non-structural HCV proteins[46,52,53]. However, CD8+ T cells targeting non-structural proteins are immunodominant and associate with spontaneous clearance[54].
The strength of the CMI is also important for HCV infection outcome. Indeed, a robust HCV-specific CD8+ T cell response is associated with the resolution of acute HCV infection[55]. In an acute infection, cytotoxic T lymphocytes (CTLs) have cyto
Broad specific CD4+ T cells are detected during the acute phase regardless of the final outcome. However, these cells undergo an early decrease in frequency and breadth in persistent HCV infection compared to patients who clear the infection spontaneously[57]. Thus, spontaneous resolution is associated with a CD4+ T cell response significatively stronger in comparison to persistently, or chronically infected individuals[58,59].
In chronic infections, the limited functionality of specific CD4+ T cells due to the lack of proliferative capacity and cytokines production[59-61] leads to a dysregulated CD8+ T cell response which facilitates the emergence of escape viral variants[62]. Dysfunctional CD8+ T cells are unable to control the viral load and become exhausted because of the persistent exposure to HCV epitopes which have not mutated[63]. Thus, these exhausted T cells undergo a progressive loss of their cytotoxic activity, proliferative capacity and proinflammatory cytokines production[64,65]. However, it is of note, that the cytolytic activity, and in particular the Fas/FasL dependent function, are associated with HCV immunopathology. Fas expression is up-regulated in hepa
During acute HCV infection antibodies are produced and target epitopes in both structural and non-structural proteins, however, the envelope glycoproteins E1 and E2 are the main targets of the humoral immune response. Located at the N-terminal end of E2, the hypervariable region 1 (HVR1) is an immunodominant motif[67], which is the most variable region of the HCV genome[68]. Mutation in neutralizing epitopes allow the virus to escape from isolate-specific nAbs[69-71].
Early studies reported that nAbs developed against HCV target the HVR1 region of E2, however these nAbs were isolate-specific[67,69]. Thus, diverse studies have identified monoclonal antibodies (mAbs) that target conserved sites across multiple HCV genotypes located on either linear[72,73] or conformational[74,75] epitopes on E2 ectodomain.
Analyzing sera from different patients who were infected with the same HCV isolate showed that 43% of those who resolved their infections had nAbs against the main HVR1 variant, whereas these antibodies were present only in 13% of patients who evolved to chronicity[76]. Interestingly, plasma isolated from HCV-infected patients immediately prior to clearance has a better capacity to neutralize HCV strains from different genotypes compared to acute infection plasma from patients who subsequently evolve to persistence[77,78]. Furthermore, analysis from patients who cleared HCV infection showed detectable level of nAbs at earlier time points in comparison with acute infections that proceed to chronicity[79]. Chronic infections have been associated with a delayed cross-reactive nAbs response[43,77,78,80]. Although cross-reactive nAbs elicited during chronicity are not able to clear the infection, these have been associated with reduced liver fibrosis[81].
Despite the high genetic diversity of HCV, it was possible to isolate broadly neutra
The resolution of the initial HCV infection does not lead to sterilizing immunity so patients who previously controlled the primary HCV infection can be infected again[83]. However, differential rates of reinfection and/or chronicity have been reported among people who inject drugs (PWIDs) with the same risk of exposure, being reduced in people previously infected in comparison with people without previous infection[84]. Resolution is achieved in about 80% of HCV-reinfected patients[85].
Reinfection was characterized by a significant reduction in duration and magnitude of viremia compared with the primary infection and it was also shown to protect against persistence[85]. Moreover, clearance of reinfection was associated with an earlier and higher frequency of broadened T cells secreting IFN-γ as compared to primary infection[86-89] and an early induction of nAbs[85,90].
Long-lived memory HCV-specific CD4+ and CD8+ T cells are detected in the peripheral blood in humans following spontaneous resolution of the primary infection for up to 20 years[89,91]. CD4+ T cell depletion before reinfection leads to viral persistence even in the presence of functional CD8+ T cells which evidences the protective role of memory T cells upon re-exposure to HCV. While CD8+ T cells are the main effector cells in viral control, CD4+ T cells are essential for CD8+ T cell function and prevent viral escape within epitopes targeted by CD8+ T cells.
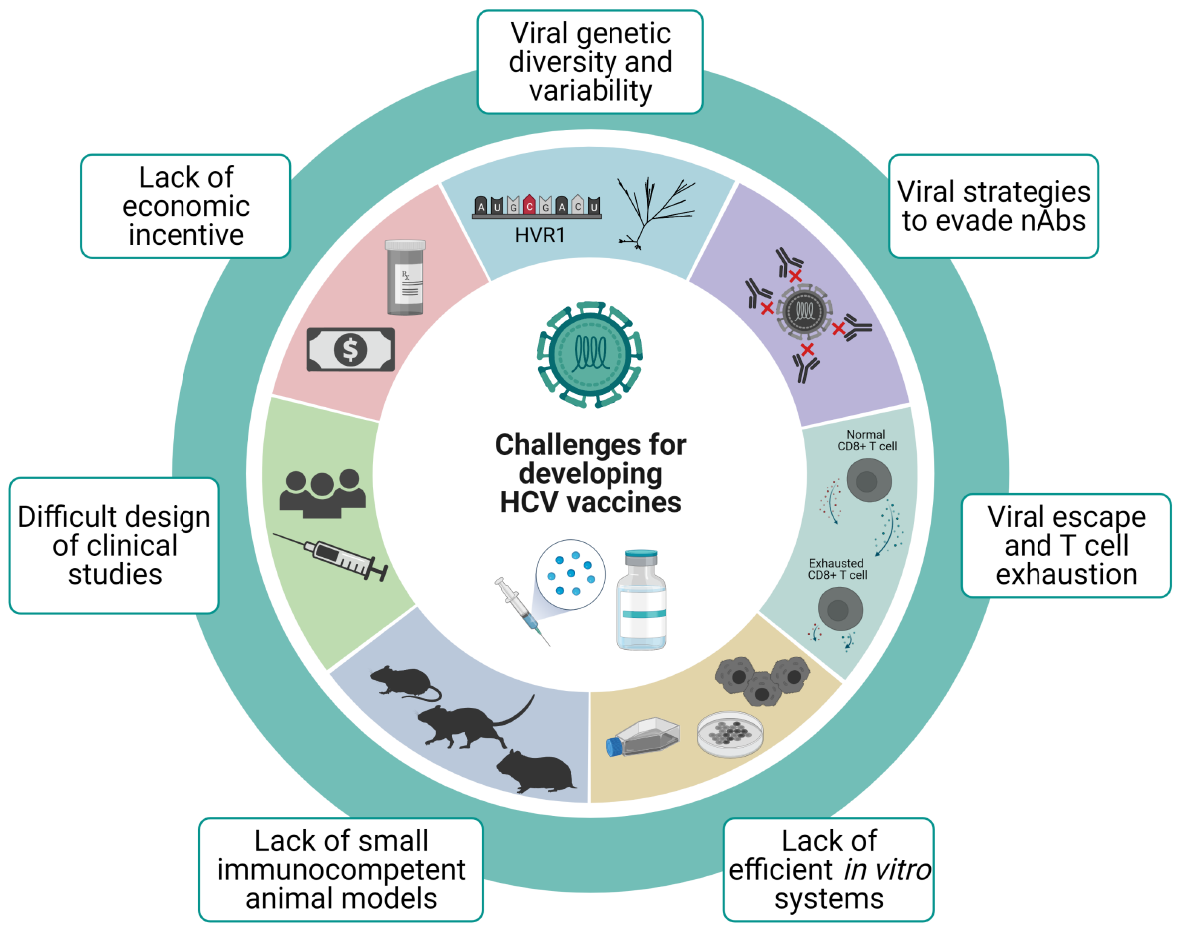
A number of difficulties have hindered the development of vaccines against HCV throughout the years (Figure 1). Despite all the knowledge acquired on the biology of this virus in recent years, a full understanding of key aspects of its pathogenesis and the host’s immune response remains elusive. Taking into account the correlate of protection, an effective vaccine needs to be able to prime both arms of the adaptative immune response. Thus, vaccination has to induce an early and sustained expansion of specific CD4+ and CD8+ T cell response. Alongside cellular immunity, cross-reactive nAbs need to be elicited to provide protection against different variants and geno
In this section we will go over the most important challenges on the design and validation of an effective vaccine against HCV.
Despite the fact that vaccines are great tools to prevent diseases, usually they are not as profitable as are drugs and other health services, and therefore investing in vaccine development is less appealing for the pharmaceutical industry[92]. Additionally, the development of vaccines with two different aims (prophylactic and therapeutic) would probably be expensive, and including prime/boost vaccination strategies may result impractical[19]. On another front, most newly infected individuals are PWID which mainly belong to populations with limited financial resources. This represents another discouraging aspect for companies interested in vaccine development[19].
From an economic perspective, though, there is well-reported evidence that vaccines are, in the long run, the most cost-effective public health measure after access to clean water[93,94]. A vaccine to fight HCV will, most likely, not be an exception.
HCV is an enveloped virus with a single-stranded positive RNA genome which has a single open reading frame (ORF) flanked by non-coding regions at both ends (5’ and 3’). For these features, it is classified as the prototype member of the Hepacivirus genus within the Flaviviridae family[95]. The ORF codes for a polyprotein of around 3000 amino acids which is co- and post-translationally processed into three structural (core, E1, E2) and seven non-structural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, NS5B)[96].
Mutation is a key mechanism contributing to HCV genetic diversity and it is mainly driven by the error prone NS5B RNA-dependent RNA-polymerase[97]. HCV has an approximate mutation rate of 10-5 mutations/nucleotide/replicative cycle[98,99], a characteristic which together with big population sizes, short generation times, and high replication rates generates the intra-host circulation of a complex population of closely related genome variants, usually termed as viral quasispecies[100,101]. Of utmost importance is the N-terminus of the envelope protein E2[67]. It contains the HVR1 region of about 30 amino acids which exhibits a huge variation among different isolates, and it is the most variable region of the entire HCV genome[68]. Even though most HCV-infected individuals develop nAbs against the virus, this high variability represents a problem as it allows the virus to escape immunologic surveillance and prevents the development of vaccines that induce cross-reactive nAbs[21]. Thus, a major challenge for the development of a broadly reactive vaccine for the control of HCV infection is identifying conserved neutralizing epitopes outside of HVR1.
Notably, mutations within HVR1 have also been associated with resistance to cross-neutralizing antibody response even if their epitopes are conserved, which highlights again the difficulties in achieving HCV neutralization as HCV could persist even in the presence of an antibody response to conserved epitopes[102,103]. This finding sugge
Mutation rates coupled with the selective pressure exerted by the host’s immune system has steered HCV diversification into 8 genotypes and 90 subtypes[104,105]. HCV strains from different genotypes differ by 30% in their nucleotide positions within the coding region, whereas subtypes exhibit 15% nucleotide variation[106]. Genotypes 1 and 3 are the most prevalent worldwide (accounting for 49.1% and 17.9% of diagnosed cases, respectively), and are most frequently found in developed countries[107].
The quasispecies dynamic as well as the resulting viral diversity confers HCV an amazing ability to adapt which in turn implies the possibility to escape from different therapeutic or preventive approaches such as antiviral drugs or vaccines[108-112]. Thus, T cell-based vaccines intended to induce broadly reactive immune responses by targeting more conserved regions/proteins of the virus are desirable if the aim is to protect against new infections and/or persistence[11,21].
Viral entry to host cells and viral interactions with different host factors could theoretically be blocked by nAbs targeting HCV envelope glycoproteins E1 and E2. However, the virus has evolved several mechanisms which affect the host´s ability to neutralize the virus. One of the mechanisms has been described extensively above (genetic diversity, particularly in HVR1 region), yet there are a number of other strategies employed by this virus to evade neutralization: (1) Glycosylation of structural proteins; (2) Cell-to-cell transmission; (3) Interfering antibodies; (4) Association with lipoproteins; (5) Antibody decoy; (6) Flexible conformational epitopes; and (7) Enhancing of viral entry.
Glycosylation of structural proteins: This feature reduces their immunogenicity as they are recognized as selfstructures. This is an important mechanism used by HCV to escape host humoral immune response. Glycans act by masking antigenic sites targeted by nAbs, interfering sterically with antibody neutralization[113]. Indeed, the deletion of N-glycans leads to an increase in E1E2 immunogenicity and can induce a more potent antibody response against HCV[114-116]. Glycan shift is another mechanism to induce neutralization resistance through glycosylation. Single point mutations which result in deleting a glyco
Cell-to-cell transmission: It is another mechanism for viral dissemination, which avoids the extracellular compartment and favors escaping host humoral immune responses[118,119].
Interfering antibodies: When non-nAb bind to sequences in the C-terminal region of HVR1, they disrupt the recognition of conserved epitopes by antibodies with neutra
Association with lipoproteins: HCV circulates in the blood in association with triglyceride-rich lipoproteins and low-density lipoproteins forming hybrid lipoviral particles, which are a hallmark of infectious HCV particles. Several host-derived factors play a role in evading antibody neutralization. Lipoproteins such as apolipoprotein E contribute to humoral immune escape by hiding relevant neutralization epitopes in E2 protein, preventing them to be exposed during HCV assembly and maturation, hence, abrogating antibody neutralization[123,124].
Antibody decoy: Interestingly, in vitro studies have reported that HCV-infected cells release E2-containing exosomes that act as antibody bait making HCV virions less susceptible to neutralization[125].
Flexible conformational epitopes: The capacity of some conserved neutralizing epito
Enhancing of viral entry: It has been shown that host mutations that alter the interaction of serum components like high-density lipoprotein with scavenger receptor BI enhance viral entry to the cell[127]. This, in turn, protects the virus against humoral response as the time window in which nAbs can bind and act is reduced[128,129]. Fofana et al[130] (2012) also showed that mutations in the E2 glycoprotein, conferred viral escape to humoral responses by altering the use of the T cell receptor CD81[130].
Despite these challenges, it has been possible to isolate broadly cross-neutralizing mAbs with the ability to block HCV infection of various genotypes and thus, protect against heterologous viral infection[75,131-134]. These findings suggest that a prophylactic vaccine against HCV may indeed be achievable.
The elucidation of the crystal structure of E2 has provided a better insight into different antigenic domains and regions that allow a rational vaccine design. A study showed that epitopes within E2, exhibiting moderate or conserved variability, were efficiently targeted by bNAbs[135,136]. Unfortunately, despite the relative conservation of some bNAbs epitopes, escape mutations have been identified[137,138].
Several studies have evidenced the key role of cellular immunity in the clearance of infection. An effective vaccine has to induce a rapid recall of the memory T cell respon
(1) Escape mutations within major histocompatibility (MHC) class I-restricted HCV epitopes represent the main mechanism used by HCV to evade CTL responses and thus it is associated with persistence. Unlike CD8+ epitopes, escape mutations within targeted CD4+ T cell epitopes are not common, suggesting that CD4+ T cells failure mechanisms cannot be completely explained by viral escape[139]. Escape mutations occur early in infection and they are rare during long-term chronic infection, possibly due to the lack of T cell-mediated selective pressure[140]. Interestingly, escape variants show an impaired replicative fitness[141,142] and this contributes to limiting the variability within some epitopes[143,144]. As a consequence, the ideal target for T cell-based vaccines are conserved epitopes less likely to mutate because of viral fitness cost[141,142]. Another effect of escape variants results in impaired recognition by T cells receptors and thus prevents CD8+ T cell recognition. Moreover, CD8+ T cells from infected patients with genotype 4 were not able to recognize epitopes from other genotypes[52]. This finding highlights the challenging task of choosing vaccine targets that protect against multiple HCV genotypes. Hence, identifying conserved epitopes recognizable by specific CD8+ T cells is a key point to develop efficient T cell-based vaccines.
(2) T cell exhaustion: While T cell-based vaccines likely provide protection against chronic virus infections, they also have the potential to generate immunopathology following subsequent virus infection. This is illustrated by the fact that during chronic infection an impaired HCV-specific CD8+ T cell response develops, known as T cell exhaustion. This phenotype is associated with the inability of the immune system to control viraemia during chronic infection. These exhausted T cells undergo a progre
Long-lived memory T cell response is only induced following spontaneous clearance and it can provide some protection. However, individuals who cannot maintain such long-lived memory T cell response due to T cell exhaustion are not protected upon re-exposure.
One of the major challenges for immunogenic T cell vaccines refers to the recovery of T cell immunity through vaccination in people with persistent HCV infection. Kelly et al[145] (2016) demonstrated that when an HCV T cell vaccine based on chimpanzee adenoviruses (ChAd3) are given to patients with chronic disease, the immune response is not able to restore T cell function[145]. Failure to respond to this vaccine approach may be the result of T cell exhaustion, as vaccination is stimulating memory responses that were induced early in infection but that ended up partially dysfunctional following viral exposure[145].
An essential step in vaccine research is the evaluation of antibodies generated as a result of natural infections or experimental immunizations, as well as the evaluation of vaccine candidates. For those purposes using different in vitro and animal models becomes a must[23].
As we will exemplify in a later section on vaccines against SARS-CoV-2, the generation of live-attenuated and/or inactivated whole virus vaccines has been possible against a number of different viruses (measles, mumps, rubella, rotavirus, hepatitis A virus, poliovirus, among others), however this strategy is not achievable to generate HCV vaccines. Since HCV was discovered[1], and only until recently, resear
As for in vitro models, propagating HCV in cultured cells remained limited for several years since inoculation of patient sera or plasma in different cell lines resulted in limited or no viral replication[147]. The first report of efficient replication came from working with HCV subgenomic replicons (where the structural region was replaced by a neomycin-encoding gene)[148]. However, the challenge was to generate an in vitro system that was able to produce infectious HCV particles at high titers that would allow further research[23]. The production of cell-culture derived viral particles (HCV
Further studies on HCVcc led to the discovery of more permissive cell clones derived from Huh7 cells (e.g., Huh7.5 and Huh7.5.1)[155,156] as well as to the generation of inter- and intragenotypic recombinant genomes that are able to recapitulate the complete HCV life cycle and produce high titers of infectious particles in vitro. These recombinants have been shown to be optimal in vitro models to study the neutralization ability both of mAbs as well as of sera from infected patients[82,157-160]. They have also been used to characterize antibody escape mutations[71,137,161]. Additionally, reporter and flag-tagged JFH-1-based genomes (J6/JFH1) have been generated[162-164] and used in vaccine development[165], the latter in particular to facilitate large-scale purification of viral particles[163]. However, the most important aim in this field would be to efficiently grow any virus derived from HCV infected patients, which unfortunately has not yet been achieved[153]. For now, we depend on the constructs described above as well as a few full-length consensus clones, which have been developed after a lot of research effort and had to be designed including numerous adaptive mutations[166-170], therefore, not quite resembling natural circulating isolates. In spite of the setbacks, all these constructs have the potential to be employed for producing inactivated whole-virus vaccines.
Another in vitro approach to assess the neutralizing ability of sera and mAbs, in addition to HCVcc, relies on the generation of HCV pseudoparticles (HCVpp). These are generated by cotransfecting HCV E1 and E2 genes together with a retroviral packing and reporter system[171]. Due to the struggles imposed by the generation of different HCVcc derived viral particles, HCVpps were actually developed earlier[172,173] but continue to be used in vaccine research nowadays[157,174-176].
Humans are the natural hosts of HCV, and in order to test the efficacy and safety of vaccine candidates in pre-clinical studies, in vivo animal models are needed. Foremost, in vivo studies on pathogenesis of HCV chronic infections have been problematic since HCV only infects humans and, under experimental conditions, also chimpanzees. The first and most successful immunocompetent animal model has indeed been the chimpanzee. However, ethical concerns and its inclusion on the United States Fish and Wildlife Service’s Endangered Species have led to a ban in its use for biomedical research[177]. Even before this prohibition, the continued use of these animals faced many issues such as high costs, small cohort sizes which made statistically significant results difficult to achieve, and the inability to genetically manipulate chimpanzees. Furthermore, it would require the need to have special and expensive facilities to breed and keep them under study[178].
Small animal models are frequently very useful tools to test potential vaccine candidates, but, since HCV does not infect rodents, a lot of effort has been devoted into developing strategies to adapt mice to evaluate HCV vaccines. This led to the use of chimeric humanized or transgenic mice with humanized livers[179] or expressing human CD81 and occludin[180], two cellular proteins that HCV uses as receptors for cell entry. However, mouse models are difficult to produce, and most are immunocompromised, which makes them inappropriate to study virus-host interactions and immune responses. Additionally, they do not exhibit cirrhosis or HCC[181]. In spite of this, genetically humanized fully immunocompetent inbred mice expressing human orthologs of HCV entry factors were developed[182], which have allowed the study of viral entry, yet not the full viral cycle. To address the latter, Chen et al[183] (2014), developed an immune-competent humanized mice model that is capable of developing persistent HCV infections and hepatopathological manifestations[183], yet the mice stock are outbred and genetically not well defined. More recently, Keng et al[184] (2016) were able to establish a new humanized mouse model including human hepatocytes as well as human immune system[184], which was able to recapitulate HCV infection and immunopathogenesis[181], although low levels of B cells were detected when compared to clinical settings.
For the difficulties in getting broad access to small immunocompetent mouse models, alternative experimental non-human primate models have been explored. However, no signs of infection were detected (for a detailed review see Ploss and Kapoor[178], 2020), with the exception of tree shrews (now classified in a separate order Scandentia, but previously designated as small squirrel-like primates) which can become symptomatic and even progress to chronicity[185]. Despite this encouraging finding, keeping these animals in captivity is a difficult task, and additionally they are genetically diverse for being an outbred species, which again poses issues to be widely used in HCV biomedical research[178].
Altogether, this shows us the difficulty we face when we need animals that can be employed for vaccine development but also to study HCV-associated pathogenesis. An alternative could be the use of substitutes and analogue viral models that can be propagated in mice lab strains and that appear to share basic immunological features with HCV. Recently, the discovery of non-primate hepaciviruses has raised interest since they can be used as analogues of HCV infection[23]. A rodent Hepacivirus discovered in Norway rats[186] has been shown to establish high-titer liver infections when inoculated in immunocompetent mice, and thus, provides insight into hepatic immune responses[187]. However, the main drawback of this model is the limited sequence homology to HCV[186]. On the other hand, equine hepacivirus (eqHV), formerly known as non-primate Hepacivirus, is the closest relative of HCV and both species share some important features such as the level of E1E2 glycosylation or the presence of miR-122 seed sites in their 5’ non-coding regions (2 sites in HCV and 1 site in eqHV)[188,189]. These approaches of using alternative and analogue viral models for vaccine development is extremely valuable, yet it is worth acknowledging that different mammalian immune systems might respond in different ways and this should be taken into consideration at the moment of interpreting data[23].
The design of clinical studies for HCV vaccine candidates poses its own hurdles. It must be considered that, in order for an effective vaccine to be validated, it should be tested in populations at risk for HCV infection[11,36]. This is an issue in developed countries where HCV infection incidence is low other than in PWID populations. Targeting this group of patients has ethical concerns and practical difficulties to be overcome[190]. Despite this, there are a few studies which have been successful in identifying, enrolling and monitoring PWID before developing an acute HCV infection[191,192], the latest completed phase I/II clinical trial with outcome results was able to enroll 548 active intravenous drug users (ClinicalTrials.gov Identifier: NCT01436357[193])[194]. On the other hand, large studies could be conducted where incidence is higher, such as some developing countries. However, logistical problems may arise due to the large number of patients needed and their appropriate follow up, specifically to detect acute cases of hepatitis, which usually course without any symptoms[36].
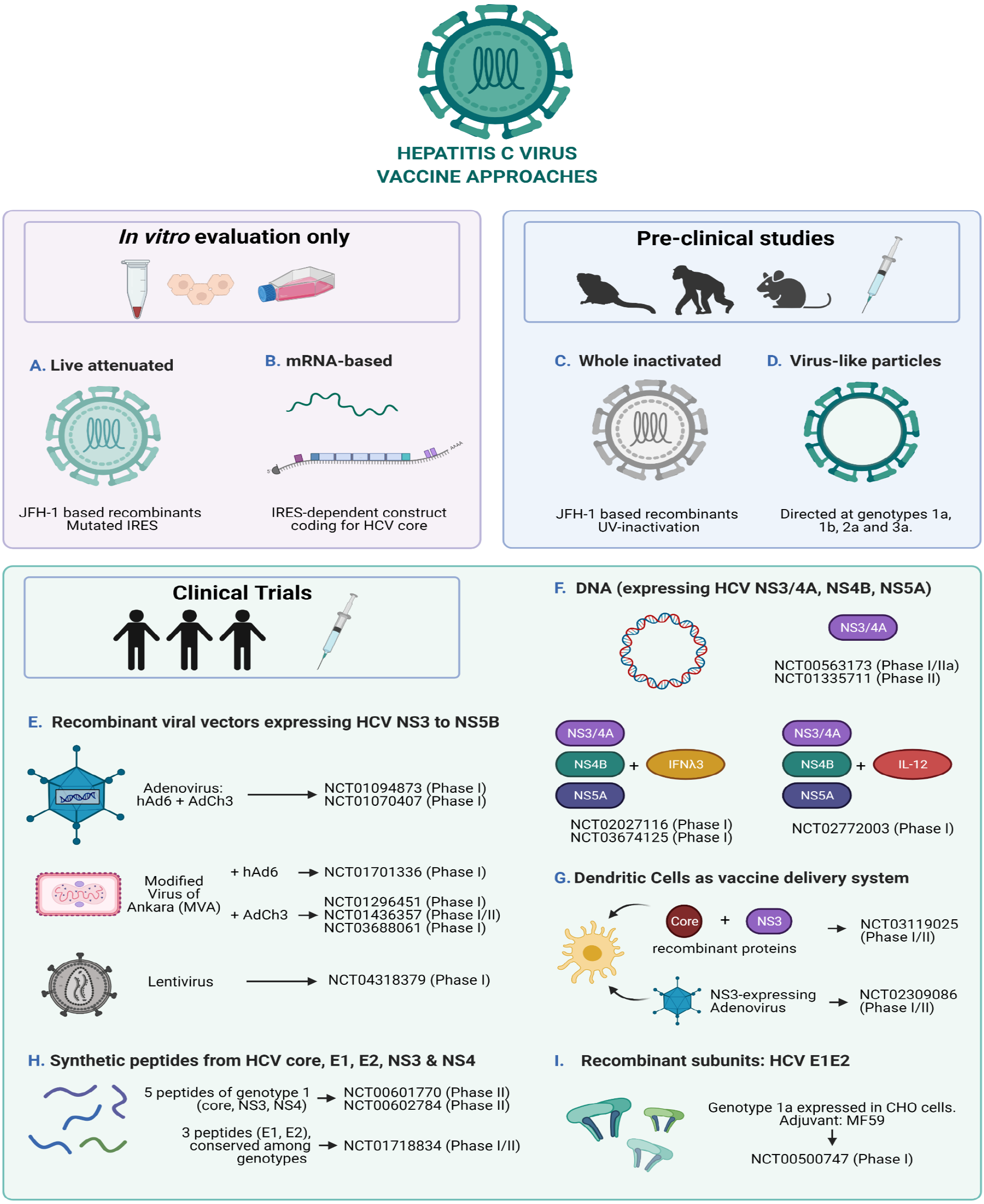
There are several traditional and newer approaches in vaccine development, and most of them have been explored for the design of HCV vaccine candidates (Figure 2), albeit the majority only directed at genotype 1.
Traditional vaccine approaches include whole-organisms vaccines containing either inactivated whole or live attenuated viruses. Live attenuated vaccines are potent in inducing CMI and humoral immunity and have been successful for many viral infections because they resemble what occurs naturally. Nevertheless, they have the potential risk of reverting to virulent wild-type strains. In contrast, inactivated viruses are noninfectious but have the downside of being less immunogenic than attenuated viruses. Therefore, when inactivated whole viruses are developed as vaccine candidates, they often include adjuvants and/or booster injections in order to enhance the immunogenicity[195].
Newer methods involve the use of one or more genes of the virus of interest to be incorporated into the genome of a nonpathogenic organism for amplification. In this way, mainly three different approaches have been developed: Subunits vaccines (by purifying the protein/s of interest generated in the heterologous organisms), DNA vaccines (usually by isolating a plasmid containing the gene/s of interest), and recombinant viruses (by using the entire host virus as a live vector)[195].
The latest method successfully explored has been the use of RNA-based vaccines, whose development is faster than other technologies, easily scalable, and of lower cost to manufacture. These characteristics have been essential to the development and recent authorization for emergency use of some of the vaccines currently available to control the COVID-19 pandemic[196].
In this section we will go over some of the vaccine candidates explored against HCV, and we will delve into nucleic acid-based and recombinant viral vector app
This traditional approach of inactivated virus was only feasible after the development of cell culture systems, with all the challenges that they impose even nowadays. This is partly the reason why there are only a few pre-clinical studies assessing the immunogenicity of inactivated HCVcc as vaccine candidates[197,198]. Both studies have shown the induction of humoral immune responses in chimeric mice[198] as well as in a non-human primate model[197]. The latter also elicited T cell responses. These findings are promising, but there are still some developmental challenges to overcome if this approach is to be considered for clinical trials, such as production in serum-free culture conditions and scalable and cost-efficient downstream processes. Fortunately, there are a few studies which have addressed these difficulties, and have shown that high titer serum-free HCVcc is possible for different intra and intergenotypic recombinants based on JFH-1 isolate[199] and that more efficient downstream processes based on ultracentrifugation and chromatography can be applied[200]. Nevertheless, the challenge of generating high titers of HCVcc of the most widespread genotypes and subtypes still remains.
Recombinant E1/E2 proteins were the first prophylactic vaccine candidates being tested since they are the major targets for nAb, in particular HVR1 region within E2. They were shown to be able to induce the generation of nAb in chimpanzees[201], yet only one candidate reached clinical trials in 2007 (ClinicalTrials.gov Identifier: NCT00500747[202]). Results of the phase I trial in healthy volunteers showed the vaccine was well-tolerated at different doses used, and that it was able to induce antibody production[203,204].
Whereas recombinant E1E2 vaccines were designed to elicit humoral immune response, synthetic peptide vaccines are more attractive since they can be designed to prime both arms of the immune response. Some peptide combinations targeting both cytotoxic lymphocytes and CD4+ T cell epitopes (core, NS3, NS4) have entered clinical trials. Results for the phase 2 trial NCT00602784[205] have shown that the peptide vaccine IC41 can trigger T cell responses in relapse patients after dual therapy, yet viral clearance was not achieved[206]. Unfortunately, humoral response was not analyzed. The results of the other studies remain to be published (ClinicalTrials.gov Identifier: NCT01718834[207] and NCT00601770[208]).
Of interest, computational identification of B and T cell epitopes has been explored as an alternative for the rational design of effective vaccine candidates. By means of different immune-bioinformatic and population dynamics simulation approaches, many predicted epitopes in E2, NS3/4A, NS5A and NS5B have been identified[209-212]. These approaches provided valuable information and in silico screening methods for highly conserved immunogen candidates with the putative ability to block escape mutations (for a detailed review please see[213]). These computational designs can help speed up vaccine development at the experimental stages by rationally selecting the most promising epitopes for subunit vaccine in vitro and ex vivo evaluation.
Virus-like particles (VLPs) are particles that resemble a virion but do not contain the viral genome, rather they are generated by the auto assembly of structural proteins in a manner that is genome-independent. In this way, the particle is similar to the native virus but it lacks the ability to replicate and for vaccine candidates is a very attractive technology since they are more immunogenic than soluble proteins and can prime both arms of the immune response[214].
The rationale behind this type of vaccines is supported by the successful deve
The use of live recombinant viral-based HCV vaccines as a genetic immunization approach has shown to be powerful for eliciting CMI[217]. For this purpose, different modified viruses are used as vectors to carry HCV genetic information[19].
Adenoviral vectors are the most widespread used in the vaccine developing industry. They are attractive models for different reasons: Adenoviral genomes are well characterized and are relatively easy to modify into replication-defective viruses, most human adenoviruses cause mild infections, they infect a broad number of cell types (dividing and non-dividing), they can be grown to high titers in tissue culture, and by deleting essential genes, genetic information of interest can be inserted[218]. The most frequently used in immunization studies is the human adenovirus serotype 5 (hAd5), which is included in at least 12 of the vaccines against SARS-CoV-2 that are currently on clinical trials and in one that already had authorization for emergency use (Sputnik V vaccine)[219,220]. Despite their benefits, individuals might exhibit preexisting anti hAd5 Abs, which could diminish the immune response to vaccines based on this viral vector. For this reason, less frequent serotypes such as hAd24, hAd6 or hAd26 have been employed in pre-clinical and clinical studies of vaccine candidates against different viruses[221-223]. Additionally, adenoviruses that infect chimpanzees (AdCh3) have been tested in conjunction with hAd6, both carrying HCV non-structural proteins NS3 to NS5B of genotype 1b, yet despite reaching clinical trials, they have only been evaluated in phase I studies (ClinicalTrials.gov Identifiers: NCT01094873[224] and NCT01070407[225]). The reason for not continuing these studies seemed to be the inability to restore CMI, and as a result, a non-significant effect on HCV viral load was observed[145].
In light of these drawbacks, another viral vector has been employed in prime/boost vaccination strategies against HCV: The Modified Virus of Ankara (MVA), an attenuated poxvirus strain which is immunogenic and safe since it lacks several immunomodulatory genes[226]. MVA vector together with hAd6, both expressing HCV non-structural proteins NS3 to NS5B have entered phase I clinical trials to evaluate the combination as a therapeutic vaccine to be used in conjunction with dual therapy (ClinicalTrials.gov Identifier: NCT01701336[227]). Even though the study is complete, no results have been disclosed, presumably due to the newer DAA treatments which have completely substituted classical therapy. The most promising trials currently in phase I and II use the combination of ChAd and MVA vectors harboring HCV NS3-NS5B genomic regions. A phase I study in healthy volunteers showed promising results in terms of eliciting T cell responses (ClinicalTrials.gov Identifier: NCT01296451[228])[229]. Unfortunately, a phase I/II study in PWID population showed that this vaccination strategy was not effective for preventing chronic infections since T cell exhaustion was not reversed (ClinicalTrials.gov Identifier: NCT01436357[193])[194,230]. These results highlight the need for a vaccine strategy that stimulates both humoral and T cell immunity[23,231]. However, attempts to enhance CMI without the need of boosting the generation of Abs, have been addressed in pre-clinical studies on non-human primates by fusing the HCV non-structural antigen to MHC class II-associated invariant chain[232]. The results showed enhanced and accelerated CD8+ T cell responses and paved the way to reach clinical trials. At the time of writing this manuscript, there is an actively recruiting phase I clinical trial (ClinicalTrials.gov Identifier: NCT03688061[233]) that seeks to enroll 25 healthy participants to assess the safety and immunogenicity of HCV prime/boost vaccination with both ChAd and MVA vectors expressing HCV non-structural antigens fused to a class II-invariant gene. Results from only 15 individuals seem promising, largely mimicking pre-clinical studies, but more participants are still needed and assessment of durability of the enhanced CMI needs to be further addressed[234].
The most recent vector-based therapeutic vaccine candidate entering phase I clinical trials is a lentiviral based HCV immunotherapy (HCVax) which aims to evaluate both the safety and the immune response in chronic HCV patients (ClinicalTrials.gov Identifier: NCT04318379[235]). Last generation lentiviral vectors are safer than first generation ones (previously used for gene therapy) and like adenoviral vectors, are capable of infecting both dividing and nondividing cells, and since they integrate into the host’s genome, expression of the transgene can be long-term, a characteristic which makes them attractive as vaccine strategy[236].
Nucleic acid based-vaccines present numerous advantages over traditional vaccine approaches: (1) No issues associated with misfolding of proteins in recombinant protein vaccines or with high manufacture costs; (2) No infectious risks that might be associated with live-attenuated or inactivated whole virus vaccines; (3) They are able to activate both arms of the immune response (humoral and cellular); (4) The expression of antigens resembles natural epitopes; (5) In a single injection, multiple genes can be delivered; and (6) If multiple doses are needed, unlike the use of recombinant virus-based vaccines, there is no risk of anti-vector immunity[39,237,238].
DNA-based vaccines have been in the picture for nearly 40 years now[239]. They usually consist of purified plasmids which harbor sequences of interest that are expressed under the control of a eukaryotic promoter for a robust expression in mammalian cells. They are inexpensive, easy to manufacture, and also important, stable at room temperature. All of which are features that make them an ideal technology in vaccine research, as distribution and access could be granted effortlessly even to developing countries[39].
RNA vaccines have been explored for around 25 years, beginning with studies of self-amplifying RNA vectors (modified RNA from viruses) as well as mRNA pulsed into dendritic cells (DCs)[240,241], and have been largely assessed for tumor vaccination[242]. They share some features with DNA vaccines, but they do not need to enter the nucleus to translate the genetic information into antigen proteins, which represents an advantage over DNA immunization since the barrier of the nuclear envelope is removed, and thus, their efficacy is higher[238]. However, RNA is more labile than DNA, which might yield less robust vaccines than DNA-based formu
The first approach for delivery of nucleic acid-based vaccines, was direct injection of naked DNA plasmid or mRNA (transdermally or intramuscularly), however, effici
Multiple pre-clinical studies in different animal models have been performed throug
The use of core as antigen, directly injected as naked DNA plasmid intramuscularly (IM) or intraperitoneally (IP) into different mice models, has evidenced a weak immunogenic capacity in terms of humoral response but strong CMI, even though at least 2 doses 2-4 wk apart were administered[244-248]. Using the same delivery method and injection scheme, HCV core and E2 sequences were fused to immu
Targeting structural proteins in DNA-based formulations employing injection of naked plasmid as the delivery method was thoroughly tested in animal models but the vast majority failed to enter clinical trials. With the increasing knowledge on immune correlates during acute infections, it became clear that non-structural proteins are the target of CMI during acute resolutions, and that other delivery methods such as electroporation or gene gun rendered broadly reactive CTL responses[254].
As a consequence, DNA-based vaccines encoding HCV non-structural proteins have become widely used approaches. Transdermal gene gun injection of DNA plasmid encoding NS3/4A proteins into mice has shown high titers of Abs and the ability to prime CD4+ T helper cells[255] and also a CD8+ T cells that were able to clear HCV protein-expressing hepatocytes and persist up to 12-18 mo after immunization[256,257]. When NS3 DNA vaccine was co-administered with interleukin-12 as adjuvant, strong immunogenicity was also displayed in murine models[258]. Several other adju
Even though pre-clinical results were promising, full-length NS3 protein exhibits immunosuppressive effects and it is possibly involved in the development of HCC due to its enzymatic activity which deregulates the normal functions of the host cells[263]. Even though DNA immunization renders antigen expression only transiently, and the adverse effects possibly caused by NS3 enzymatic activity would be marginal, alter
These findings seem to indicate that immunizing only with DNA-based formulations coding for NS3/4A or NS5A might not be sufficient to control viremia in HCV-infected patients, despite encouraging pre-clinical results in animal models.
In addition to NS3/4A or NS5A plasmid vaccination, IM injections followed by electroporation of constructs encoding NS3 to NS5B into Rhesus macaques and chim
Therefore, as an alternative, heterologous prime/boost vaccination strategies have also been explored in mice, in which immunization with DNA-based vaccines is followed by immunization with viral vectors such as MVA to enhance response levels[273]. Even though results provided proof-of-concept that 2 different HCV vaccine technologies can improve immunogenicity when used in combination, to the best of our knowledge, so far, no clinical trial has tested this approach.
As will be detailed in the section about vaccines against SARS-CoV-2, several mRNAs-based vaccine candidates have been intensely explored in clinical trials, in particular to fight the COVID-19 pandemic. However, so far none have been approved for human use, with the exception of some of the vaccines currently in phase 3 clinical trials which are undergoing assessment for WHO emergency use listing and prequalification[274-277] (ClinicalTrials.gov Identifier: NCT04368728[278] and NCT04713553[279]–Pfizer/BioNTech SE, ClinicalTrials.gov Identifier: NCT04470427[280] and NCT04649151[281]–Moderna TX, Inc).
On the contrary, with the exception of using mRNA to transfect DCs (which will be discussed in the next section), there have been no pre-clinical or clinical trials using mRNA-based vaccines against HCV. Interestingly, Sharifnia et al[282] (2019) have proposed for the first time that an RNA-based vaccine against HCV could be feasible since after in vitro generation of an mRNA coding for the core protein, they were able to detect core protein in monocyte-derived DCs which were previously transfected with this construct[282]. Unfortunately, no further animal studies were performed to assess the immunogenicity of this approach.
DCs are one of the most potent antigen-presenting cells needed to induce and maintain immune responses. Given their fundamental roles, DC-based vaccination strategies have been given special attention, in particular for cancer immunotherapy[283]. However, different approaches have also been explored in HCV vaccination both in pre-clinical studies as well as in clinical trials[284]. Strategies involve loading DCs with HCV core, NS3 or NS5 proteins[285,286], pulsing them with HCVpp[287], transfecting them with DNA[288] or mRNA[289], or transducing them with adenoviral vectors expressing HCV non-structural proteins[290-293].
Two recently concluded phase I/II clinical trials have enrolled chronically HCV-infected patients (HCV genotype 1b) to evaluate the safety and clinical efficacy of therapeutic vaccination using autologous DCs. Despite employing different strategies (autologous DCs loaded with recombinant HCV core and NS3 proteins vs transduced with a recombinant adenovirus encoding NS3), both studies revealed similar results in terms of immunogenicity and ability to reduce viral titers: T cell responses were generated albeit weakly, and these were insufficient to clear the virus or reduce viral loads[286,293] (ClinicalTrials.gov Identifier: NCT03119025[294] and NCT02309086[295]). These findings are somewhat discouraging since in order to design better vaccination strategies, attention will have to be placed on enhancing CMI so as to, at least partially, reduce viral titers.
As with whole inactivated virus vaccines against HCV, the limited in vitro culture systems have hampered studies on attenuated vaccines. In particular, attenuation has been achieved by serial passaging of a given virus in non-primate cells, which leads to the emergence of mutations that have low fitness in human cells. Yet HCV does not replicate efficiently in non-human cells, which poses problems for the identification and production of attenuated strains. Additionally, there is also the risk of causing an infection after the use of these types of vaccines, which in principle, limits their potential use[11,14]. However, it is worth noting that live-attenuated viral vaccines are licensed for human use for prevention of several viral diseases such as dengue, hepatitis A, measles, mumps, varicella, yellow fever and gastrointestinal disorders caused by rotaviruses[296]. Therefore, if properly designed, this technology offers safe and effective vaccines.
Considering the issue of identifying attenuating mutations in non-human cultures, an alternative is to detect mutations occurring naturally within the human host, present only as minority variants within the quasispecies, and exhibiting an attenuated phenotype.
HCV, as many members of the Flaviviridae family (all except for those within the Flavivirus genus), translate its polyprotein in a CAP-independent manner by recruiting the ribosome directly to the internal ribosome entry site (IRES), which is found in the 5’ non-coding region[297]. IRES structure and sequence are essential to its function, and any change can affect the translation process[298,299]. Therefore, investigating on mutations that might affect this process may enable an alternative approach for the design of live-attenuated vaccines against HCV. In this regard, our group has identified several mutations within the IRES of HCV isolates from chronically infected patients of genotype 1a and 3a, that are present in very low frequencies within the viral population, and that have evidenced a significant decrease in viral translation efficiency in vitro[300]. Studies in cell culture, using full-genome chimera replicons based on JFH-1 strain are underway in order to assess both translation efficiency as well as viral fitness.
It is important to mention, that one of the initial vaccines designed to fight polio was a formulation with poliovirus (PV) strains where, through successive passages in non-human cells, mutations were selected along the whole genome[301]. Of those, a mutation within PV IRES which drastically diminishes the translation efficiency, is the main responsible for the attenuated phenotype[302]. Unfortunately, live-attenuated PV vaccines have shown to be genetically unstable, and some of the mutations that confer the attenuated phenotype can reverse during replication in humans, causing rare cases of vaccine-associated paralytic poliomyelitis[303]. Thus, if the aim were to design a safe live-attenuated HCV vaccine with mutations in the IRES region, perhaps additional approaches would need to be considered so as to minimize the chances for reversion or enhancing the resulting immune response. One such approach could be constructing a bicistronic vector co-expressing an antiviral protein (for example IFN-β), which has already been proven effective to limit viral spread and to induce antiviral immunity in animal models when assessing a Flavivirus vaccine candidate[304].
On the other hand, a rational synthetic design of attenuated strains might be a new and achievable approach to employ based on the newest infectious replicons that harbor almost the entire genome sequence from non-JFH-1 strains, covering in this way most of the circulating HCV genotypes. This strategy has been successfully developed and tested in mice for other RNA viruses such as Influenza A virus and Coxsackievirus[305]. It consisted of engineering codons that were more prone to generate a Stop mutation after a single nucleotide change in as many positions as possible, without changing the amino acid identity. This strategy proved that the synthetic and rational generation of self-limiting vaccines is possible in different RNA viruses and thus, could represent an alternative way of generating HCV attenuated vaccines as well, provided that the issues with in vitro scaling-up production can be overcome in the near future.
COVID-19, caused by the SARS-CoV-2[306], has become a major health concern all over the world and has spawned challenges to develop safe and effective antiviral drugs and vaccines for preventive use. Vaccine development is a complex and time-consuming process, that typically requires years of research and testing before reaching the clinic. But in 2020, in an unprecedented effort due to the synergy between academia, researchers, and pharmacists, added to financial support and guided by cumulative knowledge from many years of scientific work, scientists were able to produce safe and effective coronavirus vaccines in record time[307]. Coronavirus vaccine types include inactivated vaccines, nucleic acid vaccines, adenovirus vector -based vaccines, and recombinant subunits vaccines. Up until February 18th researchers were testing 70 vaccine candidates in clinical trials, and 20 have reached the final stages of testing. Over 10 have been approved for emergency use in several countries around the word. Among these, it seems important to highlight the Emergency Use Authorization for 2 highly effective mRNA COVID-19 vaccines from Pfizer-BioNTech and Moderna. This is the first time that mRNA-based vaccines have ever been approved for human use, and marks a critical milestone for achievement in both science and public health[275,308,309]. As previously mentioned, mRNA vaccines trigger immune responses by transfecting synthetic mRNA encoding viral antigens (in this case spike protein or protein motifs) into human cells. Once the nucleic acid enters the cytosol of the cell, the mRNA vaccine temporarily induces the cell to produce specific viral antigens coded by the mRNA[308,310]. The major breakthroughs of these two vaccines were: (1) The mRNA modifications and purification process to reduce the innate immune response and to improve mRNA stability; and (2) The effective intracellular delivery to facilitate cellular uptake of mRNA and to protect it from RNase degradation.
These RNA vaccines generate powerful antibody responses to the SARS-CoV-2 coronavirus, but they have not proven to be as good as the AstraZeneca/Oxford vaccine (adenoviral vector vaccine) at stimulating CD8+ T cells. Recently animal studies suggest that a combination of an RNA coronavirus vaccine and a adenoviral vector vaccine (AstraZeneca/Oxford vaccine) could strengthen immune response by rousing CD8+ T cells in mice better than either vaccine alone[311,312]. This preliminary data should be confirmed in upcoming clinical trials.
Thus, what can we learn about SARS-CoV-2 impressive vaccine development? Firstly, that when there is interest and resources, the development and production times of a vaccine can be significantly reduced. Secondly, that mRNA vaccines have a high potency, ability for rapid development, and cost-efficient production. Thirdly, that preliminary data suggests that mixing COVID vaccines technologies boosts the immune response at a cellular level.
Is it possible, therefore, to apply all the knowledge gained from COVID-19 vaccines to accelerate HCV vaccine development? Unfortunately, only partially. As mentioned in the section about challenges, many hurdles remain since HCV biology and immunology differ greatly from that of SARS-CoV-2. However, the so far unexplored possibility of an HCV mRNA-based vaccine could certainly benefit from the experiences and developments in the field of RNA-based vaccines against SARS-CoV-2.
HCV is an insidious virus, which, since its discovery, has caused enormous difficulty to be kept under control. The successful introduction of DAAs has become a milestone in keeping the epidemic in line, however it has proven to be insufficient to achieve global eradication of this virus and all the health complications derived from the infection. Therefore, numerous approaches have been explored in order to design an effective vaccine, either prophylactic or therapeutic. Unfortunately, to date, none of these attempts have rendered a viable vaccine for human use. Several drawbacks have hampered its development, among which, to our understanding, one of the most difficult to override is T cell exhaustion, the main cause of therapeutic vaccines failure. However, many other challenges related to a still incomplete understanding of HCV immunology remain to be overcome. Noteworthy among these, is the insufficiency of CMI to control infections and the need for a joint humoral response, as well as the necessity for characterization of better epitopes for nAbs. An approach that might prove effective in the future, is the use of heterologous prime/boost vaccination, where two different technologies can be employed to enhance the immune responses. Additionally, we believe that ongoing efforts to develop improved and more suitable in vivo systems should be a priority, since many of the successful pre-clinical studies have possibly failed in clinical trials due to the differences in immunopathology between the used animal models and humans. All of the hard work that has enabled the rapid and effective development of vaccines against SARS-CoV-2 should be taken as an example of what can be achieved if the interest and the efforts are focused on tackling a health burden. In particular, the advances on mRNA-based vaccine technology, which so far has not been explored in HCV vaccine candidates, would be a good starting point if the aim is to explore alternatives not investigated so far. Additionally, different methodologies which have been shown to be efficacious against other RNA viruses, are available for the design of live-attenuated strains as vaccines against HCV. Following this line of thought, and likely fueled both by the success of COVID-19 vaccines[313] and by the Nobel Prize in Physiology or Medicine 2020 (awarded to three scientists for the discovery of HCV)[28], last year, the NIH opened a grant opportunity for projects concerning HCV vaccine design[30]. As a result, it is expected that more research will be focused on this subject in the upcoming years, and hopefully, auspicious findings will follow. This renewed interest in funding HCV vaccines might be what is needed to achieve HCV global eradication, as has been proposed by the WHO a few years ago. Allocating funds for this purpose boosts the research area that has been left behind in terms of breakthroughs that can be effectively translated to public health benefits.
We would like to thank MSc. Fabiana Gámbaro for collaborating with in vitro translational studies of HCV IRES mutants during her work at the Laboratorio de Virología Molecular (Facultad de Ciencias, Universidad de la República). We also thank Megan Lasako, RN, for reviewing and revising the manuscript for grammar and syntax.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Uruguay
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ahmad S, Tajiri K S-Editor: Fan JR L-Editor: A P-Editor: Wu RR
| 1. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4996] [Cited by in RCA: 4655] [Article Influence: 129.3] [Reference Citation Analysis (0)] |
| 2. | WHO. Global Hepatitis Report 2017. Geneva: World Health Organization. [cited 10 February 2021]. Available from: https://www.who.int/publications/i/item/global-hepatitis-report-2017. |
| 3. | WHO. Fact Sheet: Hepatitis C [Internet]. World Heal. Organ.2018. [cited 10 January 2021]. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c. |
| 4. | Cholankeril G, Ahmed A. Alcoholic Liver Disease Replaces Hepatitis C Virus Infection as the Leading Indication for Liver Transplantation in the United States. Clin Gastroenterol Hepatol. 2018;16:1356-1358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 195] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 5. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Liver transplantation. J Hepatol. 2016;64:433-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 703] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 6. | Graziadei I, Zoller H, Fickert P, Schneeberger S, Finkenstedt A, Peck-Radosavljevic M, Müller H, Kohl C, Sperner-Unterweger B, Eschertzhuber S, Hofer H, Öfner D, Tilg H, Vogel W, Trauner M, Berlakovich G. Indications for liver transplantation in adults: Recommendations of the Austrian Society for Gastroenterology and Hepatology (ÖGGH) in cooperation with the Austrian Society for Transplantation, Transfusion and Genetics (ATX). Wien Klin Wochenschr. 2016;128:679-690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 7. | Debes JD, Chan AJ, Balderramo D, Kikuchi L, Gonzalez Ballerga E, Prieto JE, Tapias M, Idrovo V, Davalos MB, Cairo F, Barreyro FJ, Paredes S, Hernandez N, Avendaño K, Diaz Ferrer J, Yang JD, Carrera E, Garcia JA, Mattos AZ, Hirsch BS, Gonçalves PT, Carrilho FJ, Roberts LR. Hepatocellular carcinoma in South America: Evaluation of risk factors, demographics and therapy. Liver Int. 2018;38:136-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 8. | Piñero F, Costa P, Boteon YL, Duque SH, Marciano S, Anders M, Varón A, Zerega A, Poniachik J, Soza A, Padilla Machaca M, Menéndez J, Zapata R, Vilatoba M, Muñoz L, Maraschio M, Podestá LG, McCormack L, Gadano A, Boin ISFF, García P, Silva M; Latin American Liver Research, Education, Awareness Network (LALREAN). A changing etiologic scenario in liver transplantation for hepatocellular carcinoma in a multicenter cohort study from Latin America. Clin Res Hepatol Gastroenterol. 2018;42:443-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (1)] |
| 9. | Gravitz L. Introduction: a smouldering public-health crisis. Nature. 2011;474:S2-S4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 137] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 10. | Bartenschlager R, Baumert TF, Bukh J, Houghton M, Lemon SM, Lindenbach BD, Lohmann V, Moradpour D, Pietschmann T, Rice CM, Thimme R, Wakita T. Critical challenges and emerging opportunities in hepatitis C virus research in an era of potent antiviral therapy: Considerations for scientists and funding agencies. Virus Res. 2018;248:53-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 115] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 11. | Bailey JR, Barnes E, Cox AL. Approaches, Progress, and Challenges to Hepatitis C Vaccine Development. Gastroenterology. 2019;156:418-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 172] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 12. | Sorbo MC, Cento V, Di Maio VC, Howe AYM, Garcia F, Perno CF, Ceccherini-Silberstein F. Hepatitis C virus drug resistance associated substitutions and their clinical relevance: Update 2018. Drug Resist Updat. 2018;37:17-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 150] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 13. | Midgard H, Weir A, Palmateer N, Lo Re V 3rd, Pineda JA, Macías J, Dalgard O. HCV epidemiology in high-risk groups and the risk of reinfection. J Hepatol. 2016;65:S33-S45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 136] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 14. | Cox AL. Challenges and Promise of a Hepatitis C Virus Vaccine. Cold Spring Harb Perspect Med. 2020;10:a036947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 15. | Chaudhari R, Fouda S, Sainu A, Pappachan JM. Metabolic complications of hepatitis C virus infection. World J Gastroenterol. 2021;27:1267-1282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 38] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (3)] |
| 16. | Ioannou GN, Feld JJ. What Are the Benefits of a Sustained Virologic Response to Direct-Acting Antiviral Therapy for Hepatitis C Virus Infection? Gastroenterology. 2019;156:446-460.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 149] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 17. | Tapper EB, Catana AM, Sethi N, Mansuri D, Sethi S, Vong A, Afdhal NH. Direct costs of care for hepatocellular carcinoma in patients with hepatitis C cirrhosis. Cancer. 2016;122:852-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 18. | Bardach A, Hernández-Vásquez A, Palacios A, Calderón M, Soto N, Balan D, Augustovski F. Epidemiología, consumo de recursos y costos del manejo médico de la Hepatitis C en Argentina, Colombia, Uruguay y Venezuela. Value Health Reg Issues. 2019;20:180-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 19. | Roohvand F, Kossari N. Advances in hepatitis C virus vaccines, part two: advances in hepatitis C virus vaccine formulations and modalities. Expert Opin Ther Pat. 2012;22:391-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 20. | Hill AM, Nath S, Simmons B. The road to elimination of hepatitis C: analysis of cures versus new infections in 91 countries. J Virus Erad. 2017;3:117-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 105] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 21. | Feinstone SM, Hu DJ, Major ME. Prospects for prophylactic and therapeutic vaccines against hepatitis C virus. Clin Infect Dis. 2012;55 Suppl 1:S25-S32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 22. | Massoud O. Hepatitis C: looking into the future. Expert Rev Gastroenterol Hepatol. 2020;14:367-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 23. | Duncan JD, Urbanowicz RA, Tarr AW, Ball JK. Hepatitis C Virus Vaccine: Challenges and Prospects. Vaccines (Basel). 2020;8:90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 24. | Roingeard P, Beaumont E. Hepatitis C Vaccine: 10 Good Reasons for Continuing. Hepatology. 2020;71:1845-1850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 25. | Fine P, Eames K, Heymann DL. "Herd immunity": a rough guide. Clin Infect Dis. 2011;52:911-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 692] [Cited by in RCA: 670] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 26. | Mennini FS, Marcellusi A, Andreoni M, Gasbarrini A, Salomone S, Craxi A. Health policy model: long-term predictive results associated with the management of hepatitis C virus-induced diseases in Italy. Clin Outcomes Res. 2014;6:303-310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 27. | Cipriano LE, Goldhaber-Fiebert JD. Population Health and Cost-Effectiveness Implications of a “Treat All” Recommendation for HCV: A Review of the Model-Based Evidence. MDM Policy Pract. 2018;3:2381468318776634. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 28. | Nobelprize.org. Press release: The Nobel Prize in Physiology or Medicine 2020 [Internet]. Nobel Media AB 20212020. [cited 10 February 2021]. Available from: https://www.nobelprize.org/prizes/medicine/2020/press-release/. |
| 29. | Nobelprize.org. Streams during Nobel Week 2020 [Internet]. Nobel Lect. Physiol. or Med.2020 [cited 10 February 2021]. Available from: https://www.nobelprize.org/nobel-week-live-streams-2020/. |
| 30. | National Institutes of Health (NIH). Funding Opportunity: Rational Design of Vaccines Against Hepatitis C Virus [Internet]. 2020. [cited 10 February 2021]. Available from: https://grants.nih.gov/grants/guide/rfa-files/RFA-AI-20-019.html. |
| 31. | Farlex Partner Medical Dictionary. Prophylactic vaccination. (n.d.) [Internet]. 2012 [cited 10 February 2021]. Available from: https://medical-dictionary.thefreedictionary.com/prophylactic+vaccination. |
| 32. | McGraw-Hill Dictionary of Scientific & Technical Terms 6E. Therapeutic vaccination. (n.d.) [Internet]. 2003. [cited 10 February 2021]. Available from: https://encyclopedia2.thefreedictionary.com/therapeutic+vaccination. |
| 33. | Major M, Gutfraind A, Shekhtman L, Cui Q, Kachko A, Cotler SJ, Hajarizadeh B, Sacks-Davis R, Page K, Boodram B, Dahari H. Modeling of patient virus titers suggests that availability of a vaccine could reduce hepatitis C virus transmission among injecting drug users. Sci Transl Med. 2018;10:eaao4496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 34. | Scott N, McBryde E, Vickerman P, Martin NK, Stone J, Drummer H, Hellard M. The role of a hepatitis C virus vaccine: modelling the benefits alongside direct-acting antiviral treatments. BMC Med. 2015;13:198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 35. | Stone J, Martin NK, Hickman M, Hellard M, Scott N, McBryde E, Drummer H, Vickerman P. The Potential Impact of a Hepatitis C Vaccine for People Who Inject Drugs: Is a Vaccine Needed in the Age of Direct-Acting Antivirals? PLoS One. 2016;11:e0156213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 36. | Halliday J, Klenerman P, Barnes E. Vaccination for hepatitis C virus: closing in on an evasive target. Expert Rev Vaccines. 2011;10:659-672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 37. | Huang CF, Yu ML. Unmet needs of chronic hepatitis C in the era of direct-acting antiviral therapy. Clin Mol Hepatol. 2020;26:251-260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 38. | Torresi J, Johnson D, Wedemeyer H. Progress in the development of preventive and therapeutic vaccines for hepatitis C virus. J Hepatol. 2011;54:1273-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 39. | Mekonnen ZA, Grubor-Bauk B, Masavuli MG, Shrestha AC, Ranasinghe C, Bull RA, Lloyd AR, Gowans EJ, Wijesundara DK. Toward DNA-Based T-Cell Mediated Vaccines to Target HIV-1 and Hepatitis C Virus: Approaches to Elicit Localized Immunity for Protection. Front Cell Infect Microbiol. 2019;9:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 40. | Wolski D, Foote PK, Chen DY, Lewis-Ximenez LL, Fauvelle C, Aneja J, Walker A, Tonnerre P, Torres-Cornejo A, Kvistad D, Imam S, Waring MT, Tully DC, Allen TM, Chung RT, Timm J, Haining WN, Kim AY, Baumert TF, Lauer GM. Early Transcriptional Divergence Marks Virus-Specific Primary Human CD8+ T Cells in Chronic versus Acute Infection. Immunity. 2017;47:648-663.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 41. | Kared H, Fabre T, Bédard N, Bruneau J, Shoukry NH. Galectin-9 and IL-21 mediate cross-regulation between Th17 and Treg cells during acute hepatitis C. PLoS Pathog. 2013;9:e1003422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 116] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 42. | Schulze zur Wiesch J, Lauer GM, Day CL, Kim AY, Ouchi K, Duncan JE, Wurcel AG, Timm J, Jones AM, Mothe B, Allen TM, McGovern B, Lewis-Ximenez L, Sidney J, Sette A, Chung RT, Walker BD. Broad repertoire of the CD4+ Th cell response in spontaneously controlled hepatitis C virus infection includes dominant and highly promiscuous epitopes. J Immunol. 2005;175:3603-3613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 151] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 43. | Pestka JM, Zeisel MB, Bläser E, Schürmann P, Bartosch B, Cosset FL, Patel AH, Meisel H, Baumert J, Viazov S, Rispeter K, Blum HE, Roggendorf M, Baumert TF. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A. 2007;104:6025-6030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 444] [Cited by in RCA: 434] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 44. | Walker MR, Leung P, Eltahla AA, Underwood A, Abayasingam A, Brasher NA, Li H, Wu BR, Maher L, Luciani F, Lloyd AR, Bull RA. Clearance of hepatitis C virus is associated with early and potent but narrowly-directed, Envelope-specific antibodies. Sci Rep. 2019;9:13300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 45. | Micallef JM, Kaldor JM, Dore GJ. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J Viral Hepat. 2006;13:34-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 589] [Cited by in RCA: 634] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 46. | Smyk-Pearson S, Tester IA, Klarquist J, Palmer BE, Pawlotsky JM, Golden-Mason L, Rosen HR. Spontaneous recovery in acute human hepatitis C virus infection: functional T-cell thresholds and relative importance of CD4 help. J Virol. 2008;82:1827-1837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 65] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 47. | Baumert TF, Fauvelle C, Chen DY, Lauer GM. A prophylactic hepatitis C virus vaccine: a distant peak still worth climbing. J Hepatol. 2014;61:S34-S44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 48. | Bailey JR, Flyak AI, Cohen VJ, Li H, Wasilewski LN, Snider AE, Wang S, Learn GH, Kose N, Loerinc L, Lampley R, Cox AL, Pfaff JM, Doranz BJ, Shaw GM, Ray SC, Crowe JE Jr. Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance. JCI Insight. 2017;2:e92872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 49. | Kaplan DE, Sugimoto K, Newton K, Valiga ME, Ikeda F, Aytaman A, Nunes FA, Lucey MR, Vance BA, Vonderheide RH, Reddy KR, McKeating JA, Chang KM. Discordant role of CD4 T-cell response relative to neutralizing antibody and CD8 T-cell responses in acute hepatitis C. Gastroenterology. 2007;132:654-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 118] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 50. | Cooper S, Erickson AL, Adams EJ, Kansopon J, Weiner AJ, Chien DY, Houghton M, Parham P, Walker CM. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10:439-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 610] [Cited by in RCA: 589] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 51. | Lechner F, Wong DK, Dunbar PR, Chapman R, Chung RT, Dohrenwend P, Robbins G, Phillips R, Klenerman P, Walker BD. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191:1499-1512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1003] [Cited by in RCA: 1000] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 52. | Luxenburger H, Neumann-Haefelin C, Thimme R, Boettler T. HCV-Specific T Cell Responses During and After Chronic HCV Infection. Viruses. 2018;10:645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 53. | Spada E, Mele A, Berton A, Ruggeri L, Ferrigno L, Garbuglia AR, Perrone MP, Girelli G, Del Porto P, Piccolella E, Mondelli MU, Amoroso P, Cortese R, Nicosia A, Vitelli A, Folgori A. Multispecific T cell response and negative HCV RNA tests during acute HCV infection are early prognostic factors of spontaneous clearance. Gut. 2004;53:1673-1681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 82] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 54. | Smyk-Pearson S, Tester IA, Lezotte D, Sasaki AW, Lewinsohn DM, Rosen HR. Differential antigenic hierarchy associated with spontaneous recovery from hepatitis C virus infection: implications for vaccine design. J Infect Dis. 2006;194:454-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 54] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 55. | Badr G, Bédard N, Abdel-Hakeem MS, Trautmann L, Willems B, Villeneuve JP, Haddad EK, Sékaly RP, Bruneau J, Shoukry NH. Early interferon therapy for hepatitis C virus infection rescues polyfunctional, long-lived CD8+ memory T cells. J Virol. 2008;82:10017-10031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 115] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 56. | Chigbu DI, Loonawat R, Sehgal M, Patel D, Jain P. Hepatitis C Virus Infection: Host⁻Virus Interaction and Mechanisms of Viral Persistence. Cells. 2019;8:376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 92] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 57. | Schulze Zur Wiesch J, Ciuffreda D, Lewis-Ximenez L, Kasprowicz V, Nolan BE, Streeck H, Aneja J, Reyor LL, Allen TM, Lohse AW, McGovern B, Chung RT, Kwok WW, Kim AY, Lauer GM. Broadly directed virus-specific CD4+ T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J Exp Med. 2012;209:61-75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 172] [Cited by in RCA: 187] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 58. | Day CL, Lauer GM, Robbins GK, McGovern B, Wurcel AG, Gandhi RT, Chung RT, Walker BD. Broad specificity of virus-specific CD4+ T-helper-cell responses in resolved hepatitis C virus infection. J Virol. 2002;76:12584-12595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 198] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 59. | Chang KM, Thimme R, Melpolder JJ, Oldach D, Pemberton J, Moorhead-Loudis J, McHutchison JG, Alter HJ, Chisari FV. Differential CD4(+) and CD8(+) T-cell responsiveness in hepatitis C virus infection. Hepatology. 2001;33:267-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 267] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 60. | Semmo N, Day CL, Ward SM, Lucas M, Harcourt G, Loughry A, Klenerman P. Preferential loss of IL-2-secreting CD4+ T helper cells in chronic HCV infection. Hepatology. 2005;41:1019-1028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 134] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 61. | Semmo N, Klenerman P. CD4+ T cell responses in hepatitis C virus infection. World J Gastroenterol. 2007;13:4831-4838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 44] [Cited by in RCA: 44] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 62. | Bowen DG, Walker CM. Mutational escape from CD8+ T cell immunity: HCV evolution, from chimpanzees to man. J Exp Med. 2005;201:1709-1714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 169] [Cited by in RCA: 165] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 63. | Abdel-Hakeem MS, Shoukry NH. Protective immunity against hepatitis C: many shades of gray. Front Immunol. 2014;5:274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 64. | Gruener NH, Lechner F, Jung MC, Diepolder H, Gerlach T, Lauer G, Walker B, Sullivan J, Phillips R, Pape GR, Klenerman P. Sustained dysfunction of antiviral CD8+ T lymphocytes after infection with hepatitis C virus. J Virol. 2001;75:5550-5558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 395] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 65. | Wedemeyer H, He XS, Nascimbeni M, Davis AR, Greenberg HB, Hoofnagle JH, Liang TJ, Alter H, Rehermann B. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J Immunol. 2002;169:3447-3458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 504] [Cited by in RCA: 490] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 66. | Sayers TJ, Brooks AD, Seki N, Smyth MJ, Yagita H, Blazar BR, Malyguine AM. T cell lysis of murine renal cancer: multiple signaling pathways for cell death via Fas. J Leukoc Biol. 2000;68:81-86. [PubMed] |
| 67. | Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter HJ, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci U S A. 1996;93:15394-15399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 454] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 68. | Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991;175:220-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 236] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 69. | Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, Shapiro M, Purcell RH. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci U S A. 1994;91:7792-7796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 347] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 70. | Cuevas JM, González-Candelas F, Moya A, Sanjuán R. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J Virol. 2009;83:5760-5764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 117] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 71. | Gal-Tanamy M, Keck ZY, Yi M, McKeating JA, Patel AH, Foung SK, Lemon SM. In vitro selection of a neutralization-resistant hepatitis C virus escape mutant. Proc Natl Acad Sci U S A. 2008;105:19450-19455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 67] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 72. | Broering TJ, Garrity KA, Boatright NK, Sloan SE, Sandor F, Thomas WD Jr, Szabo G, Finberg RW, Ambrosino DM, Babcock GJ. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J Virol. 2009;83:12473-12482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 151] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 73. | Bartosch B, Bukh J, Meunier JC, Granier C, Engle RE, Blackwelder WC, Emerson SU, Cosset FL, Purcell RH. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc Natl Acad Sci U S A. 2003;100:14199-14204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 245] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 74. | Keck ZY, Li TK, Xia J, Gal-Tanamy M, Olson O, Li SH, Patel AH, Ball JK, Lemon SM, Foung SK. Definition of a conserved immunodominant domain on hepatitis C virus E2 glycoprotein by neutralizing human monoclonal antibodies. J Virol. 2008;82:6061-6066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 106] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 75. | Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI, Ball JK, McKeating JA, Kneteman NM, Burton DR. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14:25-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 492] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 76. | Zibert A, Meisel H, Kraas W, Schulz A, Jung G, Roggendorf M. Early antibody response against hypervariable region 1 is associated with acute self-limiting infections of hepatitis C virus. Hepatology. 1997;25:1245-1249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 78] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 77. | Dowd KA, Netski DM, Wang XH, Cox AL, Ray SC. Selection pressure from neutralizing antibodies drives sequence evolution during acute infection with hepatitis C virus. Gastroenterology. 2009;136:2377-2386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 184] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 78. | Osburn WO, Snider AE, Wells BL, Latanich R, Bailey JR, Thomas DL, Cox AL, Ray SC. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology. 2014;59:2140-2151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 216] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 79. | Strasak AM, Kim AY, Lauer GM, de Sousa PS, Ginuino CF, Fernandes CA, Velloso CE, de Almeida AJ, de Oliveira JM, Yoshida CF, Schulze zur Wiesch J, Paranhos-Baccalá G, Lang S, Brant LJ, Ulmer H, Strohmaier S, Kaltenbach L, Lampe E, Lewis-Ximenez LL. Antibody dynamics and spontaneous viral clearance in patients with acute hepatitis C infection in Rio de Janeiro, Brazil. BMC Infect Dis. 2011;11:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 80. | Lavillette D, Morice Y, Germanidis G, Donot P, Soulier A, Pagkalos E, Sakellariou G, Intrator L, Bartosch B, Pawlotsky JM, Cosset FL. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J Virol. 2005;79:6023-6034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 214] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 81. | Deng K, Liu R, Rao H, Jiang D, Wang J, Xie X, Wei L. Antibodies Targeting Novel Neutralizing Epitopes of Hepatitis C Virus Glycoprotein Preclude Genotype 2 Virus Infection. PLoS One. 2015;10:e0138756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 82. | Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, Rice CM, Ploss A, Burton DR, Law M. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci. 2012;109:6205-6210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 283] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 83. | Thomas DL, Astemborski J, Rai RM, Anania FA, Schaeffer M, Galai N, Nolt K, Nelson KE, Strathdee SA, Johnson L, Laeyendecker O, Boitnott J, Wilson LE, Vlahov D. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284:450-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 758] [Cited by in RCA: 777] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 84. | Mehta B, Kumar V, Chawla S, Jindal H, Bhatt B. Hepatitis C: is a vaccine the solution? Hum Vaccin Immunother. 2014;10:417-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 85. | Osburn WO, Fisher BE, Dowd KA, Urban G, Liu L, Ray SC, Thomas DL, Cox AL. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology. 2010;138:315-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 287] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 86. | Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J, Murthy KK, Rice CM, Walker CM. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302:659-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 632] [Cited by in RCA: 633] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 87. | Lanford RE, Guerra B, Chavez D, Bigger C, Brasky KM, Wang XH, Ray SC, Thomas DL. Cross-genotype immunity to hepatitis C virus. J Virol. 2004;78:1575-1581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 142] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 88. | Shata MT, Tricoche N, Perkus M, Tom D, Brotman B, McCormack P, Pfahler W, Lee DH, Tobler LH, Busch M, Prince AM. Exposure to low infective doses of HCV induces cellular immune responses without consistently detectable viremia or seroconversion in chimpanzees. Virology. 2003;314:601-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 63] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 89. | Shoukry NH, Grakoui A, Houghton M, Chien DY, Ghrayeb J, Reimann KA, Walker CM. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J Exp Med. 2003;197:1645-1655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 492] [Cited by in RCA: 494] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 90. | Sacks-Davis R, Grebely J, Dore GJ, Osburn W, Cox AL, Rice TM, Spelman T, Bruneau J, Prins M, Kim AY, McGovern BH, Shoukry NH, Schinkel J, Allen TM, Morris M, Hajarizadeh B, Maher L, Lloyd AR, Page K, Hellard M; InC3 study group. Hepatitis C Virus Reinfection and Spontaneous Clearance of Reinfection--the InC3 Study. J Infect Dis. 2015;212:1407-1419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 91. | Takaki A, Wiese M, Maertens G, Depla E, Seifert U, Liebetrau A, Miller JL, Manns MP, Rehermann B. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat Med. 2000;6:578-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 563] [Cited by in RCA: 559] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 92. | Institute of Medicine (US) Committee on the Evaluation of Vaccine Purchase Financing in the United States. Financing Vaccines in the 21st Century: Assuring Access and Availability. Washington (DC): National Academies Press (US). 2003;. [PubMed] |
| 93. | Rémy V, Zöllner Y, Heckmann U. Vaccination: the cornerstone of an efficient healthcare system. J Mark Access Heal Policy. 2015;3:27041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 94. | Park M, Jit M, Wu JT. Cost-benefit analysis of vaccination: a comparative analysis of eight approaches for valuing changes to mortality and morbidity risks. BMC Med. 2018;16:139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 95. | Bukh J. The history of hepatitis C virus (HCV): Basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J Hepatol. 2016;65:S2-S21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 175] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 96. | Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631-1648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 450] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 97. | Echeverría N, Moratorio G, Cristina J, Moreno P. Hepatitis C virus genetic variability and evolution. World J Hepatol. 2015;7:831-845. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (2)] |
| 98. | Ribeiro RM, Li H, Wang S, Stoddard MB, Learn GH, Korber BT, Bhattacharya T, Guedj J, Parrish EH, Hahn BH, Shaw GM, Perelson AS. Quantifying the diversification of hepatitis C virus (HCV) during primary infection: estimates of the in vivo mutation rate. PLoS Pathog. 2012;8:e1002881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 122] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 99. | Geller R, Estada Ú, Peris JB, Andreu I, Bou JV, Garijo R, Cuevas JM, Sabariegos R, Mas A, Sanjuán R. Highly heterogeneous mutation rates in the hepatitis C virus genome. Nat Microbiol. 2016;1:16045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 100. | Martell M, Esteban JI, Quer J, Genescà J, Weiner A, Esteban R, Guardia J, Gómez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225-3229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 674] [Cited by in RCA: 665] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 101. | Domingo E, Baranowski E, Ruiz-Jarabo CM, Martín-Hernández AM, Sáiz JC, Escarmís C. Quasispecies structure and persistence of RNA viruses. Emerg Infect Dis. 1998;4:521-527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 140] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 102. | Keck ZY, Li SH, Xia J, von Hahn T, Balfe P, McKeating JA, Witteveldt J, Patel AH, Alter H, Rice CM, Foung SK. Mutations in hepatitis C virus E2 located outside the CD81 binding sites lead to escape from broadly neutralizing antibodies but compromise virus infectivity. J Virol. 2009;83:6149-6160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 103. | El-Diwany R, Cohen VJ, Mankowski MC, Wasilewski LN, Brady JK, Snider AE, Osburn WO, Murrell B, Ray SC, Bailey JR. Extra-epitopic hepatitis C virus polymorphisms confer resistance to broadly neutralizing antibodies by modulating binding to scavenger receptor B1. PLoS Pathog. 2017;13:e1006235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 104. | Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. HCV Classification: A web resource to manage the classification and genotype and subtype assignments of hepatitis C virus [Internet]. ICTV2017. [cited 10 January 2021]. Available from: https://talk.ictvonline.org/ictv_wikis/flaviviridae/w/sg_flavi/56/hcv-classification. |
| 105. | Borgia SM, Hedskog C, Parhy B, Hyland RH, Stamm LM, Brainard DM, Subramanian MG, McHutchison JG, Mo H, Svarovskaia E, Shafran SD. Identification of a Novel Hepatitis C Virus Genotype From Punjab, India: Expanding Classification of Hepatitis C Virus Into 8 Genotypes. J Infect Dis. 2018;218:1722-1729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 147] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 106. | Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 965] [Cited by in RCA: 978] [Article Influence: 88.9] [Reference Citation Analysis (1)] |
| 107. | Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol. 2016;22:7824-7840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 514] [Cited by in RCA: 565] [Article Influence: 62.8] [Reference Citation Analysis (7)] |
| 108. | Pawlotsky JM. Hepatitis C Virus Resistance to Direct-Acting Antiviral Drugs in Interferon-Free Regimens. Gastroenterology. 2016;151:70-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 411] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 109. | Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, Balestrieri A, Purcell RH, Alter HJ. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 680] [Cited by in RCA: 647] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 110. | Farci P, Bukh J, Purcell RH. The quasispecies of hepatitis C virus and the host immune response. Springer Semin Immunopathol. 1997;19:5-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 65] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 111. | Forns X, Purcell RH, Bukh J. Quasispecies in viral persistence and pathogenesis of hepatitis C virus. Trends Microbiol. 1999;7:402-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 100] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 112. | Timm J, Lauer GM, Kavanagh DG, Sheridan I, Kim AY, Lucas M, Pillay T, Ouchi K, Reyor LL, Schulze zur Wiesch J, Gandhi RT, Chung RT, Bhardwaj N, Klenerman P, Walker BD, Allen TM. CD8 epitope escape and reversion in acute HCV infection. J Exp Med. 2004;200:1593-1604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 247] [Cited by in RCA: 241] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 113. | Lavie M, Hanoulle X, Dubuisson J. Glycan Shielding and Modulation of Hepatitis C Virus Neutralizing Antibodies. Front Immunol. 2018;9:article910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 114. | Helle F, Goffard A, Morel V, Duverlie G, McKeating J, Keck ZY, Foung S, Penin F, Dubuisson J, Voisset C. The neutralizing activity of anti-hepatitis C virus antibodies is modulated by specific glycans on the E2 envelope protein. J Virol. 2007;81:8101-8111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 170] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 115. | Ren Y, Min YQ, Liu M, Chi L, Zhao P, Zhang XL. N-glycosylation-mutated HCV envelope glycoprotein complex enhances antigen-presenting activity and cellular and neutralizing antibody responses. Biochim Biophys Acta. 2016;1860:1764-1775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 116. | Helle F, Vieyres G, Elkrief L, Popescu C-I, Wychowski C, Descamps V, Castelain S, Roingeard P, Duverlie G, Dubuisson J. Role of N-Linked Glycans in the Functions of Hepatitis C Virus Envelope Proteins Incorporated into Infectious Virions. J Virol. 2010;84:11905-11915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 175] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 117. | Pantua H, Diao J, Ultsch M, Hazen M, Mathieu M, McCutcheon K, Takeda K, Date S, Cheung TK, Phung Q, Hass P, Arnott D, Hongo JA, Matthews DJ, Brown A, Patel AH, Kelley RF, Eigenbrot C, Kapadia SB. Glycan shifting on hepatitis C virus (HCV) E2 glycoprotein is a mechanism for escape from broadly neutralizing antibodies. J Mol Biol. 2013;425:1899-1914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 94] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 118. | Timpe JM, Stamataki Z, Jennings A, Hu K, Farquhar MJ, Harris HJ, Schwarz A, Desombere I, Roels GL, Balfe P, McKeating JA. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology. 2008;47:17-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 271] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 119. | Brimacombe CL, Grove J, Meredith LW, Hu K, Syder AJ, Flores MV, Timpe JM, Krieger SE, Baumert TF, Tellinghuisen TL, Wong-Staal F, Balfe P, McKeating JA. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J Virol. 2011;85:596-605. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 198] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 120. | Zhang P, Zhong L, Struble EB, Watanabe H, Kachko A, Mihalik K, Virata ML, Alter HJ, Feinstone S, Major M. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc Natl Acad Sci. 2009;106:7537-7541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 117] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 121. | Bankwitz D, Steinmann E, Bitzegeio J, Ciesek S, Friesland M, Herrmann E, Zeisel MB, Baumert TF, Keck ZY, Foung SK, Pécheur EI, Pietschmann T. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J Virol. 2010;84:5751-5763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 176] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 122. | Prentoe J, Verhoye L, Velázquez Moctezuma R, Buysschaert C, Farhoudi A, Wang R, Alter H, Meuleman P, Bukh J. HVR1-mediated antibody evasion of highly infectious in vivo adapted HCV in humanised mice. Gut. 2016;65:1988-1997. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 123. | Fauvelle C, Felmlee DJ, Crouchet E, Lee J, Heydmann L, Lefèvre M, Magri A, Hiet MS, Fofana I, Habersetzer F, Foung SK, Milne R, Patel AH, Vercauteren K, Meuleman P, Zeisel MB, Bartenschlager R, Schuster C, Baumert TF. Apolipoprotein E Mediates Evasion From Hepatitis C Virus Neutralizing Antibodies. Gastroenterology. 2016;150:206-217.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 124. | Bankwitz D, Doepke M, Hueging K, Weller R, Bruening J, Behrendt P, Lee JY, Vondran FWR, Manns MP, Bartenschlager R, Pietschmann T. Maturation of secreted HCV particles by incorporation of secreted ApoE protects from antibodies by enhancing infectivity. J Hepatol. 2017;67:480-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 125. | Deng L, Jiang W, Wang X, Merz A, Hiet MS, Chen Y, Pan X, Jiu Y, Yang Y, Yu B, He Y, Tu Z, Niu J, Bartenschlager R, Long G. Syntenin regulates hepatitis C virus sensitivity to neutralizing antibody by promoting E2 secretion through exosomes. J Hepatol. 2019;71:52-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 126. | Ströh LJ, Nagarathinam K, Krey T. Conformational Flexibility in the CD81-Binding Site of the Hepatitis C Virus Glycoprotein E2. Front Immunol. 2018;9:article1396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 127. | Grove J, Nielsen S, Zhong J, Bassendine MF, Drummer HE, Balfe P, McKeating JA. Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies. J Virol. 2008;82:12020-12029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 134] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 128. | Voisset C, de Beeck AO, Horellou P, Dreux M, Gustot T, Duverlie G, Cosset FL, Vu-Dac N, Dubuisson J. High-density lipoproteins reduce the neutralizing effect of hepatitis C virus (HCV)-infected patient antibodies by promoting HCV entry. J Gen Virol. 2006;87:2577-2581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 81] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 129. | Dreux M, Pietschmann T, Granier C, Voisset C, Ricard-Blum S, Mangeot PE, Keck Z, Foung S, Vu-Dac N, Dubuisson J, Bartenschlager R, Lavillette D, Cosset FL. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J Biol Chem. 2006;281:18285-18295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 172] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 130. | Fofana I, Fafi–Kremer S, Carolla P, Fauvelle C, Zahid MN, Turek M, Heydmann L, Cury K, Hayer J, Combet C, Cosset F, Pietschmann T, Hiet M, Bartenschlager R, Habersetzer F, Doffoël M, Keck Z, Foung SKH, Zeisel MB, Stoll–Keller F, Baumert TF. Mutations That Alter Use of Hepatitis C Virus Cell Entry Factors Mediate Escape From Neutralizing Antibodies. Gastroenterology. 2012;143:223-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 131. | Tarr AW, Owsianka AM, Timms JM, McClure CP, Brown RJ, Hickling TP, Pietschmann T, Bartenschlager R, Patel AH, Ball JK. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology. 2006;43:592-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 134] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 132. | Tarr AW, Urbanowicz RA, Hamed MR, Albecka A, McClure CP, Brown RJ, Irving WL, Dubuisson J, Ball JK. Hepatitis C patient-derived glycoproteins exhibit marked differences in susceptibility to serum neutralizing antibodies: genetic subtype defines antigenic but not neutralization serotype. J Virol. 2011;85:4246-4257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 133. | Costers S, Vanhee M, Van Breedam W, Van Doorsselaere J, Geldhof M, Nauwynck HJ. GP4-specific neutralizing antibodies might be a driving force in PRRSV evolution. Virus Res. 2010;154:104-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 58] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 134. | Keck ZY, Xia J, Wang Y, Wang W, Krey T, Prentoe J, Carlsen T, Li AY, Patel AH, Lemon SM, Bukh J, Rey FA, Foung SK. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 2012;8:e1002653. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 189] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 135. | Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, Wilson IA, Law M. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090-1094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 328] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 136. | Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, Grakoui A, Marcotrigiano J. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature. 2014;509:381-384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 230] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 137. | Keck ZY, Angus AG, Wang W, Lau P, Wang Y, Gatherer D, Patel AH, Foung SK. Non-random escape pathways from a broadly neutralizing human monoclonal antibody map to a highly conserved region on the hepatitis C virus E2 glycoprotein encompassing amino acids 412-423. PLoS Pathog. 2014;10:e1004297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 138. | Bailey JR, Wasilewski LN, Snider AE, El-Diwany R, Osburn WO, Keck Z, Foung SK, Ray SC. Naturally selected hepatitis C virus polymorphisms confer broad neutralizing antibody resistance. J Clin Invest. 2015;125:437-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 139. | Fuller MJ, Shoukry NH, Gushima T, Bowen DG, Callendret B, Campbell KJ, Hasselschwert DL, Hughes AL, Walker CM. Selection-driven immune escape is not a significant factor in the failure of CD4 T cell responses in persistent hepatitis C virus infection. Hepatology. 2010;51:378-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 140. | Cox AL, Mosbruger T, Lauer GM, Pardoll D, Thomas DL, Ray SC. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology. 2005;42:104-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 183] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 141. | Neumann-Haefelin C, Frick DN, Wang JJ, Pybus OG, Salloum S, Narula GS, Eckart A, Biezynski A, Eiermann T, Klenerman P, Viazov S, Roggendorf M, Thimme R, Reiser M, Timm J. Analysis of the evolutionary forces in an immunodominant CD8 epitope in hepatitis C virus at a population level. J Virol. 2008;82:3438-3451. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 53] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 142. | Neumann-Haefelin C, Oniangue-Ndza C, Kuntzen T, Schmidt J, Nitschke K, Sidney J, Caillet-Saguy C, Binder M, Kersting N, Kemper MW, Power KA, Ingber S, Reyor LL, Hills-Evans K, Kim AY, Lauer GM, Lohmann V, Sette A, Henn MR, Bressanelli S, Thimme R, Allen TM. Human leukocyte antigen B27 selects for rare escape mutations that significantly impair hepatitis C virus replication and require compensatory mutations. Hepatology. 2011;54:1157-1166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 143. | Söderholm J, Ahlén G, Kaul A, Frelin L, Alheim M, Barnfield C, Liljeström P, Weiland O, Milich DR, Bartenschlager R, Sällberg M. Relation between viral fitness and immune escape within the hepatitis C virus protease. Gut. 2006;55:266-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 80] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 144. | Uebelhoer L, Han JH, Callendret B, Mateu G, Shoukry NH, Hanson HL, Rice CM, Walker CM, Grakoui A. Stable cytotoxic T cell escape mutation in hepatitis C virus is linked to maintenance of viral fitness. PLoS Pathog. 2008;4:e1000143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 145. | Kelly C, Swadling L, Capone S, Brown A, Richardson R, Halliday J, von Delft A, Oo Y, Mutimer D, Kurioka A, Hartnell F, Collier J, Ammendola V, Del Sorbo M, Grazioli F, Esposito ML, Di Marco S, Siani L, Traboni C, Hill AV, Colloca S, Nicosia A, Cortese R, Folgori A, Klenerman P, Barnes E. Chronic hepatitis C viral infection subverts vaccine-induced T-cell immunity in humans. Hepatology. 2016;63:1455-1470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 146. | Thomas E, Liang TJ. Experimental models of hepatitis B and C - new insights and progress. Nat Rev Gastroenterol Hepatol. 2016;13:362-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 147. | Wakita T. Cell Culture Systems of HCV Using JFH-1 and Other Strains. Cold Spring Harb Perspect Med. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (1)] |
| 148. | Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2294] [Cited by in RCA: 2251] [Article Influence: 86.6] [Reference Citation Analysis (0)] |
| 149. | Kato T, Furusaka A, Miyamoto M, Date T, Yasui K, Hiramoto J, Nagayama K, Tanaka T, Wakita T. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J Med Virol. 2001;64:334-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 188] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 150. | Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125:1808-1817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 476] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 151. | Kato T, Date T, Miyamoto M, Sugiyama M, Tanaka Y, Orito E, Ohno T, Sugihara K, Hasegawa I, Fujiwara K, Ito K, Ozasa A, Mizokami M, Wakita T. Detection of anti-hepatitis C virus effects of interferon and ribavirin by a sensitive replicon system. J Clin Microbiol. 2005;43:5679-5684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 87] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 152. | Lohmann V. Hepatitis C virus cell culture models: an encomium on basic research paving the road to therapy development. Med Microbiol Immunol. 2019;208:3-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 153. | Ramirez S, Bukh J. Current status and future development of infectious cell-culture models for the major genotypes of hepatitis C virus: Essential tools in testing of antivirals and emerging vaccine strategies. Antiviral Res. 2018;158:264-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 154. | Luna JM, Saeed M, Rice CM. Taming a beast: lessons from the domestication of hepatitis C virus. Curr Opin Virol. 2019;35:27-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 155. | Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1843] [Cited by in RCA: 1848] [Article Influence: 92.4] [Reference Citation Analysis (0)] |
| 156. | Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci. 2005;102:9294-9299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1475] [Cited by in RCA: 1466] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 157. | Desombere I, Mesalam AA, Urbanowicz RA, Van Houtte F, Verhoye L, Keck ZY, Farhoudi A, Vercauteren K, Weening KE, Baumert TF, Patel AH, Foung SKH, Ball J, Leroux-Roels G, Meuleman P. A novel neutralizing human monoclonal antibody broadly abrogates hepatitis C virus infection in vitro and in vivo. Antiviral Res. 2017;148:53-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 158. | Pedersen J, Carlsen TH, Prentoe J, Ramirez S, Jensen TB, Forns X, Alter H, Foung SK, Law M, Gottwein J, Weis N, Bukh J. Neutralization resistance of hepatitis C virus can be overcome by recombinant human monoclonal antibodies. Hepatology. 2013;58:1587-1597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 159. | Pedersen J, Jensen TB, Carlsen TH, Schønning K, Christensen PB, Laursen AL, Krarup H, Bukh J, Weis N. Neutralizing antibodies in patients with chronic hepatitis C, genotype 1, against a panel of genotype 1 culture viruses: lack of correlation to treatment outcome. PLoS One. 2013;8:e62674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 160. | Tarr AW, Urbanowicz RA, Jayaraj D, Brown RJ, McKeating JA, Irving WL, Ball JK. Naturally occurring antibodies that recognize linear epitopes in the amino terminus of the hepatitis C virus E2 protein confer noninterfering, additive neutralization. J Virol. 2012;86:2739-2749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 161. | Fauvelle C, Colpitts CC, Keck ZY, Pierce BG, Foung SK, Baumert TF. Hepatitis C virus vaccine candidates inducing protective neutralizing antibodies. Expert Rev Vaccines. 2016;15:1535-1544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 162. | Merz A, Long G, Hiet MS, Brügger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem. 2011;286:3018-3032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 284] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 163. | Prentoe J, Bukh J. Hepatitis C virus expressing flag-tagged envelope protein 2 has unaltered infectivity and density, is specifically neutralized by flag antibodies and can be purified by affinity chromatography. Virology. 2011;409:148-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 164. | Gottwein JM, Jensen TB, Mathiesen CK, Meuleman P, Serre SB, Lademann JB, Ghanem L, Scheel TK, Leroux-Roels G, Bukh J. Development and application of hepatitis C reporter viruses with genotype 1 to 7 core-nonstructural protein 2 (NS2) expressing fluorescent proteins or luciferase in modified JFH1 NS5A. J Virol. 2011;85:8913-8928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 165. | Vietheer PT, Boo I, Gu J, McCaffrey K, Edwards S, Owczarek C, Hardy MP, Fabri L, Center RJ, Poumbourios P, Drummer HE. The core domain of hepatitis C virus glycoprotein E2 generates potent cross-neutralizing antibodies in guinea pigs. Hepatology. 2017;65:1117-1131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 166. | Gottwein JM, Scheel TK, Hoegh AM, Lademann JB, Eugen-Olsen J, Lisby G, Bukh J. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology. 2007;133:1614-1626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 163] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 167. | Date T, Morikawa K, Tanaka Y, Tanaka-Kaneko K, Sata T, Mizokami M, Wakita T. Replication and infectivity of a novel genotype 1b hepatitis C virus clone. Microbiol Immunol. 2012;56:308-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 168. | Mori K, Matsumoto A, Maki N, Ichikawa Y, Tanaka E, Yagi S. Production of infectious HCV genotype 1b virus in cell culture using a novel Set of adaptive mutations. BMC Microbiol. 2016;16:224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 169. | Pham LV, Ramirez S, Gottwein JM, Fahnøe U, Li YP, Pedersen J, Bukh J. HCV Genotype 6a Escape From and Resistance to Velpatasvir, Pibrentasvir, and Sofosbuvir in Robust Infectious Cell Culture Models. Gastroenterology. 2018;154:2194-2208.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 170. | Gottwein JM, Jensen SB, Li YP, Ghanem L, Scheel TK, Serre SB, Mikkelsen L, Bukh J. Combination treatment with hepatitis C virus protease and NS5A inhibitors is effective against recombinant genotype 1a, 2a, and 3a viruses. Antimicrob Agents Chemother. 2013;57:1291-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 171. | Bailey JR, Urbanowicz RA, Ball JK, Law M, Foung SKH. Standardized Method for the Study of Antibody Neutralization of HCV Pseudoparticles (HCVpp). Methods Mol Biol. 2019;1911:441-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 172. | Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633-642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 885] [Cited by in RCA: 876] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 173. | Flint M, Logvinoff C, Rice CM, McKeating JA. Characterization of infectious retroviral pseudotype particles bearing hepatitis C virus glycoproteins. J Virol. 2004;78:6875-6882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 74] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 174. | Urbanowicz RA, McClure CP, King B, Mason CP, Ball JK, Tarr AW. Novel functional hepatitis C virus glycoprotein isolates identified using an optimized viral pseudotype entry assay. J Gen Virol. 2016;97:2265-2279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 175. | Underwood AP, Walker MR, Brasher NA, Eltahla AA, Maher L, Luciani F, Lloyd AR, Bull RA. Understanding the Determinants of BnAb Induction in Acute HCV Infection. Viruses. 2018;10:659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 176. | Soares HR, Ferreira-Fernandes M, Almeida AI, Marchel M, Alves PM, Coroadinha AS. Enhancing Hepatitis C virus pseudoparticles infectivity through p7NS2 cellular expression. J Virol Methods. 2019;274:113714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 177. | Knight A. The beginning of the end for chimpanzee experiments? Philos Ethics Humanit Med. 2008;3:16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 31] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 178. | Ploss A, Kapoor A. Animal Models of Hepatitis C Virus Infection. Cold Spring Harb Perspect Med. 2020;10:a036970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 179. | Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR, Tyrrell DL, Kneteman NM. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927-933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 693] [Cited by in RCA: 685] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 180. | Ding Q, von Schaewen M, Hrebikova G, Heller B, Sandmann L, Plaas M, Ploss A. Mice Expressing Minimally Humanized CD81 and Occludin Genes Support Hepatitis C Virus Uptake In Vivo. J Virol. 2017;91:e01799-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 181. | Yong KSM, Her Z, Chen Q. Humanized Mouse Models for the Study of Hepatitis C and Host Interactions. Cells. 2019;8:604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 182. | Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, Law M, Rice CM, Ploss A. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208-211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 306] [Cited by in RCA: 287] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 183. | Chen J, Zhao Y, Zhang C, Chen H, Feng J, Chi X, Pan Y, Du J, Guo M, Cao H, Wang Z, Pei R, Wang Q, Pan L, Niu J, Chen X, Tang H. Persistent hepatitis C virus infections and hepatopathological manifestations in immune-competent humanized mice. Cell Res. 2014;24:1050-1066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 184. | Keng CT, Sze CW, Zheng D, Zheng Z, Yong KS, Tan SQ, Ong JJ, Tan SY, Loh E, Upadya MH, Kuick CH, Hotta H, Lim SG, Tan TC, Chang KT, Hong W, Chen J, Tan YJ, Chen Q. Characterisation of liver pathogenesis, human immune responses and drug testing in a humanised mouse model of HCV infection. Gut. 2016;65:1744-1753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 185. | Amako Y, Tsukiyama-Kohara K, Katsume A, Hirata Y, Sekiguchi S, Tobita Y, Hayashi Y, Hishima T, Funata N, Yonekawa H, Kohara M. Pathogenesis of hepatitis C virus infection in Tupaia belangeri. J Virol. 2010;84:303-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 96] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 186. | Firth C, Bhat M, Firth MA, Williams SH, Frye MJ, Simmonds P, Conte JM, Ng J, Garcia J, Bhuva NP, Lee B, Che X, Quan PL, Lipkin WI. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. mBio. 2014;5:e01933-e01914. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 258] [Cited by in RCA: 295] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 187. | Billerbeck E, Wolfisberg R, Fahnøe U, Xiao JW, Quirk C, Luna JM, Cullen JM, Hartlage AS, Chiriboga L, Ghoshal K, Lipkin WI, Bukh J, Scheel TKH, Kapoor A, Rice CM. Mouse models of acute and chronic hepacivirus infection. Science. 2017;357:204-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 188. | Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N, Wagner J, Tokarz R, Cullen JM, Iadarola MJ, Rice CM, Lipkin WI, Kapoor A. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J Virol. 2012;86:6171-6178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 193] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 189. | Yu Y, Scheel TKH, Luna JM, Chung H, Nishiuchi E, Scull MA, Echeverría N, Ricardo-Lax I, Kapoor A, Lipkin WI, Divers TJ, Antczak DF, Tennant BC, Rice CM. miRNA independent hepacivirus variants suggest a strong evolutionary pressure to maintain miR-122 dependence. PLoS Pathog. 2017;13:e1006694. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 190. | Maher L, White B, Hellard M, Madden A, Prins M, Kerr T, Page K. Candidate hepatitis C vaccine trials and people who inject drugs: challenges and opportunities. Vaccine. 2010;28:7273-7278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 191. | Cox AL, Page K, Bruneau J, Shoukry NH, Lauer GM, Kim AY, Rosen HR, Radziewicz H, Grakoui A, Fierer DS, Branch AD, Kaplan DE, Chang KM. Rare birds in North America: acute hepatitis C cohorts. Gastroenterology. 2009;136:26-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 192. | Garfein RS, Rondinelli A, Barnes RF, Cuevas J, Metzner M, Velasquez M, Rodriguez D, Reilly M, Xing J, Teshale EH. HCV infection prevalence lower than expected among 18-40-year-old injection drug users in San Diego, CA. J Urban Health. 2013;90:516-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 193. | National Institute of Allergy and Infectious Diseases. Staged Phase I/II Hepatitis C Prophylactic Vaccine (accessed 2021 Feb 18). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 10 January 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT01436357. |
| 194. | Page K, Melia MT, Veenhuis RT, Winter M, Rousseau KE, Massaccesi G, Osburn WO, Forman M, Thomas E, Thornton K, Wagner K, Vassilev V, Lin L, Lum PJ, Giudice LC, Stein E, Asher A, Chang S, Gorman R, Ghany MG, Liang TJ, Wierzbicki MR, Scarselli E, Nicosia A, Folgori A, Capone S, Cox AL. Randomized Trial of a Vaccine Regimen to Prevent Chronic HCV Infection. N Engl J Med. 2021;384:541-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 115] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 195. | Lechmann M, Liang TJ. Vaccine development for hepatitis C. Semin Liver Dis. 2000;20:211-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 196. | Sandbrink JB, Shattock RJ. RNA Vaccines: A Suitable Platform for Tackling Emerging Pandemics? Front Immunol. 2020;11:608460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 197. | Yokokawa H, Higashino A, Suzuki S, Moriyama M, Nakamura N, Suzuki T, Suzuki R, Ishii K, Kobiyama K, Ishii KJ, Wakita T, Akari H, Kato T. Induction of humoural and cellular immunity by immunisation with HCV particle vaccine in a non-human primate model. Gut. 2018;67:372-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 198. | Akazawa D, Moriyama M, Yokokawa H, Omi N, Watanabe N, Date T, Morikawa K, Aizaki H, Ishii K, Kato T, Mochizuki H, Nakamura N, Wakita T. Neutralizing Antibodies Induced by Cell Culture–Derived Hepatitis C Virus Protect Against Infection in Mice. Gastroenterology. 2013;145:447-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 199. | Mathiesen CK, Jensen TB, Prentoe J, Krarup H, Nicosia A, Law M, Bukh J, Gottwein JM. Production and characterization of high-titer serum-free cell culture grown hepatitis C virus particles of genotype 1-6. Virology. 2014;458-459:190-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 200. | Lothert K, Offersgaard AF, Pihl AF, Mathiesen CK, Jensen TB, Alzua GP, Fahnøe U, Bukh J, Gottwein JM, Wolff MW. Development of a downstream process for the production of an inactivated whole hepatitis C virus vaccine. Sci Rep. 2020;10:16261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 201. | Choo QL, Kuo G, Ralston R, Weiner A, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci U S A. 1994;91:1294-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 413] [Cited by in RCA: 397] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 202. | National Institute of Allergy and Infectious Diseases. Chiron Corp HCV E1/E2 Vaccine (accessed 2021 Feb 18). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 10 January 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT00500747. |
| 203. | Frey SE, Houghton M, Coates S, Abrignani S, Chien D, Rosa D, Pileri P, Ray R, Di Bisceglie AM, Rinella P, Hill H, Wolff MC, Schultze V, Han JH, Scharschmidt B, Belshe RB. Safety and immunogenicity of HCV E1E2 vaccine adjuvanted with MF59 administered to healthy adults. Vaccine. 2010;28:6367-6373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 207] [Cited by in RCA: 192] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 204. | Chen F, Nagy K, Chavez D, Willis S, McBride R, Giang E, Honda A, Bukh J, Ordoukhanian P, Zhu J, Frey S, Lanford R, Law M. Antibody Responses to Immunization With HCV Envelope Glycoproteins as a Baseline for B-Cell-Based Vaccine Development. Gastroenterology. 2020;158:1058-1071.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 205. | Tauber E. Phase II Study of Immunization With a Hepatitis C Virus (HCV) Antigen Peptide Vaccine (accessed 2021 Feb 18). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 10 January 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT00602784. |
| 206. | Klade CS, Wedemeyer H, Berg T, Hinrichsen H, Cholewinska G, Zeuzem S, Blum H, Buschle M, Jelovcan S, Buerger V, Tauber E, Frisch J, Manns MP. Therapeutic vaccination of chronic hepatitis C nonresponder patients with the peptide vaccine IC41. Gastroenterology. 2008;134:1385-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 119] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 207. | El Awady MK. Safety and Efficacy Study of CENV3 Vaccine to Protect Against HCV Infection (CENV3) (accessed 2021 Feb 18). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 10 January 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT01718834. |
| 208. | Ernsthofer S. Virological Response Study of the HCV Vaccine IC41 (accessed 2021 Feb 18). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 10 January 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT00601770. |
| 209. | Ikram A, Anjum S, Tahir M. In Silico Identification and Conservation Analysis of B-cell and T-Cell Epitopes of Hepatitis C Virus 3a Genotype Enveloped Glycoprotein 2 From Pakistan: A Step Towards Heterologous Vaccine Design. Hepat Mon. 2014;14:1-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 210. | Meshram RJ, Gacche RN. Effective epitope identification employing phylogenetic, mutational variability, sequence entropy, and correlated mutation analysis targeting NS5B protein of hepatitis C virus: from bioinformatics to therapeutics. J Mol Recognit. 2015;28:492-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 211. | Hart GR, Ferguson AL. Empirical fitness models for hepatitis C virus immunogen design. Phys Biol. 2015;12:066006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 212. | Ikram A, Zaheer T, Awan FM, Obaid A, Naz A, Hanif R, Paracha RZ, Ali A, Naveed AK, Janjua HA. Exploring NS3/4A, NS5A and NS5B proteins to design conserved subunit multi-epitope vaccine against HCV utilizing immunoinformatics approaches. Sci Rep. 2018;8:16107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 213. | Guest JD, Pierce BG. Structure-Based and Rational Design of a Hepatitis C Virus Vaccine. Viruses. 2021;13:837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 214. | Masavuli MG, Wijesundara DK, Torresi J, Gowans EJ, Grubor-Bauk B. Preclinical Development and Production of Virus-Like Particles As Vaccine Candidates for Hepatitis C. Front Microbiol. 2017;8:article2413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 215. | Christiansen D, Earnest-Silveira L, Grubor-Bauk B, Wijesundara DK, Boo I, Ramsland PA, Vincan E, Drummer HE, Gowans EJ, Torresi J. Pre-clinical evaluation of a quadrivalent HCV VLP vaccine in pigs following microneedle delivery. Sci Rep. 2019;9:9251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 216. | Torresi J. The Rationale for a Preventative HCV Virus-Like Particle (VLP) Vaccine. Front Microbiol. 2017;8:article2163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 217. | Ura T, Okuda K, Shimada M. Developments in Viral Vector-Based Vaccines. Vaccines. 2014;2:624-41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 299] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 218. | Tatsis N, Ertl HCJ. Adenoviruses as vaccine vectors. Mol Ther. 2004;10:616-629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 428] [Cited by in RCA: 494] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 219. | Bhatta M, Nandi S, Dutta S, Saha MK. Coronavirus (SARS-CoV-2): a systematic review for potential vaccines. Hum Vaccin Immunother. 2021;1-18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 220. | US National Library of Medicine. ClinicalTrials.gov [Internet]. [cited 18 February 2021]. [cited 18 February 2021]. Available from: https://www.clinicaltrials.gov/. |
| 221. | Folgori A, Capone S, Ruggeri L, Meola A, Sporeno E, Ercole BB, Pezzanera M, Tafi R, Arcuri M, Fattori E, Lahm A, Luzzago A, Vitelli A, Colloca S, Cortese R, Nicosia A. A T-cell HCV vaccine eliciting effective immunity against heterologous virus challenge in chimpanzees. Nat Med. 2006;12:190-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 238] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 222. | Penaloza-MacMaster P, Provine NM, Ra J, Borducchi EN, McNally A, Simmons NL, Iampietro MJ, Barouch DH. Alternative serotype adenovirus vaccine vectors elicit memory T cells with enhanced anamnestic capacity compared to Ad5 vectors. J Virol. 2013;87:1373-1384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 223. | Bos R, Rutten L, van der Lubbe JEM, Bakkers MJG, Hardenberg G, Wegmann F, Zuijdgeest D, de Wilde AH, Koornneef A, Verwilligen A, van Manen D, Kwaks T, Vogels R, Dalebout TJ, Myeni SK, Kikkert M, Snijder EJ, Li Z, Barouch DH, Vellinga J, Langedijk JPM, Zahn RC, Custers J, Schuitemaker H. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccines. 2020;5:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 284] [Cited by in RCA: 273] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 224. | Mutimer D. Study of a Novel Therapeutic Vaccine for Hepatitis C Virus (accessed 2021 Feb 18). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 18 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT01094873. |
| 225. | Adams D. A Study of a New Candidate Vaccine Against Hepatitis C Virus (HCV) (accessed 2021 Feb 18). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 18 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT01070407. |
| 226. | Blanchard TJ, Alcami A, Andrea P, Smith GL. Modified vaccinia virus Ankara undergoes limited replication in human cells and lacks several immunomodulatory proteins: implications for use as a human vaccine. J Gen Virol. 1998;79 ( Pt 5):1159-1167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 270] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 227. | Brunetto M. Study of a Novel Therapeutic Vaccine Against Hepatitis C Using Ad6NSmut and MVA-NSmut in Chronically Infected Patients (HCV004) (accessed 2021 Feb 18). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 18 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT01701336. |
| 228. | Klenerman P, Gorard D. Study of a New MVA Vaccine for Hepatitis C Virus (accessed 2021 Feb 18). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 18 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT01296451. |
| 229. | Swadling L, Capone S, Antrobus RD, Brown A, Richardson R, Newell EW, Halliday J, Kelly C, Bowen D, Fergusson J, Kurioka A, Ammendola V, Del Sorbo M, Grazioli F, Esposito ML, Siani L, Traboni C, Hill A, Colloca S, Davis M, Nicosia A, Cortese R, Folgori A, Klenerman P, Barnes E. A human vaccine strategy based on chimpanzee adenoviral and MVA vectors that primes, boosts, and sustains functional HCV-specific T cell memory. Sci Transl Med. 2014;6:261ra153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 265] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 230. | Cox AL, Page K, Melia M, Veenhuis R, Massaccesi G, Osburn W, Wagner K, Giudice L, Stein E, Asher AK, Vassilev V, Lin L, Nicosia A, Capone S, Scarselli E, Folgori A, Gorman R, Chang S, Wolff P, Liang TJ, Ghany M, Wierzbicki M, Lum P. A Randomized, Double-Blind, Placebo-Controlled Efficacy Trial of a Vaccine to Prevent Chronic Hepatitis C Virus Infection in an at-Risk Population. Abstract LB10. Open Forum Infect Dis. 2019;6:S997-S997. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 231. | Sepulveda-Crespo D, Resino S, Martinez I. Innate immune response against hepatitis c virus: Targets for vaccine adjuvants. Vaccines. 2020;8:1-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 232. | Capone S, Naddeo M, D'Alise AM, Abbate A, Grazioli F, Del Gaudio A, Del Sorbo M, Esposito ML, Ammendola V, Perretta G, Taglioni A, Colloca S, Nicosia A, Cortese R, Folgori A. Fusion of HCV nonstructural antigen to MHC class II-associated invariant chain enhances T-cell responses induced by vectored vaccines in nonhuman primates. Mol Ther. 2014;22:1039-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 233. | Dorrell L. Class II Invariant Chain HCV Vaccine Study (accessed 2021 Feb 18). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 18 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT03688061. |
| 234. | Esposito I, Cicconi P, D'Alise AM, Brown A, Esposito M, Swadling L, Holst PJ, Bassi MR, Stornaiuolo M, Mori F, Vassilev V, Li W, Donnison T, Gentile C, Turner B, von Delft A, Del Sorbo M, Barra F, Contino AM, Abbate A, Novellino E, Thomsen AR, Christensen JP, Lahm A, Grazioli F, Ammendola V, Siani L, Colloca S, Klenerman P, Nicosia A, Dorrell L, Folgori A, Capone S, Barnes E; PEACHI Consortium. MHC class II invariant chain-adjuvanted viral vectored vaccines enhances T cell responses in humans. Sci Transl Med. 2020;12:eaaz7715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 235. | Barger G, Sheikh A. Therapeutic Hepatitis C Virus Vaccine (accessed 2021 Feb 18). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 18 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT04318379. |
| 236. | Martínez-Molina E, Chocarro-Wrona C, Martínez-Moreno D, Marchal JA, Boulaiz H. Large-Scale Production of Lentiviral Vectors: Current Perspectives and Challenges. Pharmaceutics. 2020;12:1051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 237. | Ebrahim GJ. DNA vaccines. J Trop Pediatr. 1998;44:64-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 238. | Ho W, Gao M, Li F, Li Z, Zhang X, Xu X. Next‐Generation Vaccines: Nanoparticle‐Mediated DNA and mRNA Delivery. Adv Healthc Mater. 2021;10:e2001812. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 162] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 239. | Malik VS. Recombinant DNA Technology. Adv App Micro. 1981;1-84. [DOI] [Full Text] |
| 240. | Strong TV, Hampton TA, Louro I, Bilbao G, Conry RM, Curiel DT. Incorporation of beta-globin untranslated regions into a Sindbis virus vector for augmentation of heterologous mRNA expression. Gene Ther. 1997;4:624-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 241. | Boczkowski D, Nair SK, Snyder D, Gilboa E. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J Exp Med. 1996;184:465-472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 671] [Cited by in RCA: 673] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 242. | Kreiter S, Diken M, Selmi A, Türeci Ö, Sahin U. Tumor vaccination using messenger RNA: prospects of a future therapy. Curr Opin Immunol. 2011;23:399-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 97] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 243. | Moderna biotech Spain SL. Annex I-Summary of product characteriatics-COVID-19 mRNA vaccine (nucleoside modified) [Internet]. 2021. [cited 18 February 2021]. Available from: https://www.ema.europa.eu/en/documents/product-information/covid-19-vaccine-moderna-epar-product-information_en.pdf. |
| 244. | Tokushige K, Wakita T, Pachuk C, Moradpour D, Weiner DB, Zurawski VR Jr, Wands JR. Expression and immune response to hepatitis C virus core DNA-based vaccine constructs. Hepatology. 1996;24:14-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 245. | Vidalin O, Tanaka E, Spengler U, Trépo C, Inchauspé G. Targeting of hepatitis C virus core protein for MHC I or MHC II presentation does not enhance induction of immune responses to DNA vaccination. DNA Cell Biol. 1999;18:611-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 246. | Hu GJ, Wang RY, Han DS, Alter HJ, Shih JW. Characterization of the humoral and cellular immune responses against hepatitis C virus core induced by DNA-based immunization. Vaccine. 1999;17:3160-3170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 247. | Acosta-Rivero N, Dueñas-Carrera S, Alvarez-Lajonchere L, Morales-Grillo J. HCV core protein-expressing DNA vaccine induces a strong class I-binding peptide DTH response in mice. Biochem Biophys Res Commun. 2004;314:781-786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 248. | Encke J, Geissler M, Stremmel W, Wands JR. DNA-based immunization breaks tolerance in a hepatitis C virus transgenic mouse model. Hum Vaccin. 2006;2:78-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 249. | Major ME, Vitvitski L, Mink MA, Schleef M, Whalen RG, Trépo C, Inchauspé G. DNA-based immunization with chimeric vectors for the induction of immune responses against the hepatitis C virus nucleocapsid. J Virol. 1995;69:5798-5805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 117] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 250. | Inchauspé G, Major ME, Nakano I, Vitvitski L, Trépo C. DNA vaccination for the induction of immune responses against hepatitis C virus proteins. Vaccine. 1997;15:853-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 251. | Lee SW, Cho JH, Lee KJ, Sung YC. Hepatitis C virus envelope DNA-based immunization elicits humoral and cellular immune responses. Mol Cells. 1998;8:444-451. [PubMed] |
| 252. | Ma X, Forns X, Gutierrez R, Mushahwar IK, Wu T, Payette PJ, Bukh J, Purcell RH, Davis HL. DNA-based vaccination against hepatitis C virus (HCV): effect of expressing different forms of HCV E2 protein and use of CpG-optimized vectors in mice. Vaccine. 2002;20:3263-3271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 253. | Gehring S, Gregory SH, Kuzushita N, Wands JR. Type 1 interferon augments DNA-based vaccination against hepatitis C virus core protein. J Med Virol. 2005;75:249-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 254. | Kim Y, Vaughan K, Greenbaum J, Peters B, Law M, Sette A. A meta-analysis of the existing knowledge of immunoreactivity against hepatitis C virus (HCV). PLoS One. 2012;7:e38028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 255. | Frelin L, Alheim M, Chen A, Söderholm J, Rozell B, Barnfield C, Liljeström P, Sällberg M. Low dose and gene gun immunization with a hepatitis C virus nonstructural (NS) 3 DNA-based vaccine containing NS4A inhibit NS3/4A-expressing tumors in vivo. Gene Ther. 2003;10:686-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 256. | Ahlén G, Holmström F, Gibbs A, Alheim M, Frelin L. Long-term functional duration of immune responses to HCV NS3/4A induced by DNA vaccination. Gene Ther. 2014;21:739-750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 257. | Ahlén G, Nyström J, Pult I, Frelin L, Hultgren C, Sällberg M. In Vivo Clearance of Hepatitis C Virus Nonstructural 3/4A–Expressing Hepatocytes by DNA Vaccine–Primed Cytotoxic T Lymphocytes. J Infect Dis. 2005;192: 2112-2116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 258. | Naderi M, Saeedi A, Moradi A, Kleshadi M, Zolfaghari MR, Gorji A, Ghaemi A. Interleukin-12 as a genetic adjuvant enhances hepatitis C virus NS3 DNA vaccine immunogenicity. Virol Sin. 2013;28:167-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 259. | Holmström F, Pasetto A, Nähr V, Brass A, Kriegs M, Hildt E, Broderick KE, Chen M, Ahlén G, Frelin L. A synthetic codon-optimized hepatitis C virus nonstructural 5A DNA vaccine primes polyfunctional CD8+ T cell responses in wild-type and NS5A-transgenic mice. J Immunol. 2013;190:1113-1124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 260. | Weiland OR. Phase I/IIa Dose Ranging CHRONVAC-C® Study in Chronic HCV Patients (accessed 2021 Feb 19). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 18 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT00563173. |
| 261. | Weiland OR. CHRONVAC-C Study Followed by Standard of Care in Chronic Hepatitis C Virus (HCV) Subjects (accessed 2021 Feb 19). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 18 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT01335711. |
| 262. | Weiland O, Ahlén G, Diepolder H, Jung MC, Levander S, Fons M, Mathiesen I, Sardesai NY, Vahlne A, Frelin L, Sällberg M. Therapeutic DNA vaccination using in vivo electroporation followed by standard of care therapy in patients with genotype 1 chronic hepatitis C. Mol Ther. 2013;21:1796-1805. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 263. | Morikawa K, Lange CM, Gouttenoire J, Meylan E, Brass V, Penin F, Moradpour D. Nonstructural protein 3-4A: the Swiss army knife of hepatitis C virus. J Viral Hepat. 2011;18:305-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 264. | Ratnoglik SL, Jiang DP, Aoki C, Sudarmono P, Shoji I, Deng L, Hotta H. Induction of cell-mediated immune responses in mice by DNA vaccines that express hepatitis C virus NS3 mutants lacking serine protease and NTPase/RNA helicase activities. PLoS One. 2014;9:e98877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 265. | Pouriayevali MH, Bamdad T, Aghasadeghi MR, Sadat SM, Sabahi F. Construction and immunogenicity analysis of hepatitis C virus (HCV) truncated non-structural protein 3 (NS3) plasmid vaccine. Jundishapur J Microbiol. 2016;9:e33909. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 266. | Latimer B, Toporovski R, Yan J, Pankhong P, Morrow MP, Khan AS, Sardesai NY, Welles SL, Jacobson JM, Weiner DB, Kutzler MA. Strong HCV NS3/4a, NS4b, NS5a, NS5b-specific cellular immune responses induced in Rhesus macaques by a novel HCV genotype 1a/1b consensus DNA vaccine. Hum Vaccin Immunother. 2014;10:2357-2365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 267. | Capone S, Zampaglione I, Vitelli A, Pezzanera M, Kierstead L, Burns J, Ruggeri L, Arcuri M, Cappelletti M, Meola A, Ercole BB, Tafi R, Santini C, Luzzago A, Fu T, Colloca S, Ciliberto G, Cortese R, Nicosia A, Fattori E, Folgori A. Modulation of the Immune Response Induced by Gene Electrotransfer of a Hepatitis C Virus DNA Vaccine in Nonhuman Primates. J Immunol. 2006;177:7462-7471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 64] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 268. | Lee H, Jeong M, Oh J, Cho Y, Shen X, Stone J, Yan J, Rothkopf Z, Khan AS, Cho BM, Park YK, Weiner DB, Son WC, Maslow JN. Preclinical evaluation of multi antigenic HCV DNA vaccine for the prevention of Hepatitis C virus infection. Sci Rep. 2017;7:43531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 269. | Hoon S, Heo J. Phase I Trial to Evaluate the Safety, Tolerability and Immunogenicity of VGX-6150 for Second-line Therapy of Chronic Hepatitis C Infection (VGX-6150-01) (accessed 2021 Feb 19). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 18 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT02027116. |
| 270. | Han JW, Sung PS, Hong SH, Lee H, Koh JY, White S, Maslow JN, Weiner DB, Park SH, Jeong M, Heo J, Ahn SH, Shin EC. IFNL3-adjuvanted HCV DNA vaccine reduces regulatory T cell frequency and increases virus-specific T cell responses. J Hepatol. 2020;73:72-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 271. | GeneOne Life Science Inc. Evaluation of Safety, Tolerability, and Immunogenicity Study of GLS-6150 in Healthy Volunteers and in Persons Previously Treated for Hepatitis C Virus Infection (accessed 2021 Feb 19). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 18 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT03674125. |
| 272. | Jacobson JM. DNA Vaccine Therapy in Treating Patients With Chronic Hepatitis C Virus Infection (accessed 2021 Feb 19). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 18 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT02772003. |
| 273. | Fournillier A, Frelin L, Jacquier E, Ahlén G, Brass A, Gerossier E, Holmström F, Broderick KE, Sardesai NY, Bonnefoy JY, Inchauspé G, Sällberg M. A heterologous prime/boost vaccination strategy enhances the immunogenicity of therapeutic vaccines for hepatitis C virus. J Infect Dis. 2013;208:1008-1019. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 274. | World Health Organization (WHO). Status of COVID-19 Vaccines within WHO EUL/PQ evaluation process [Internet]. 2021. [cited 24 February 2021]. Available from: https://extranet.who.int/pqweb/sites/default/files/documents/Status_COVID_VAX_24Feb2021.pdf. |
| 275. | Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW Jr, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC; C4591001 Clinical Trial Group. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020;383:2603-2615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10556] [Cited by in RCA: 10641] [Article Influence: 2128.2] [Reference Citation Analysis (1)] |
| 276. | Holzgreve H. COVID-19: Nobelpreiswürdiger Erfolg in der Impfstoff-Forschung. MMW Fortschr Med. 2021;163:24-25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 277. | Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Raabe V, Bailey R, Swanson KA, Li P, Koury K, Kalina W, Cooper D, Fontes-Garfias C, Shi PY, Türeci Ö, Tompkins KR, Walsh EE, Frenck R, Falsey AR, Dormitzer PR, Gruber WC, Şahin U, Jansen KU. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1003] [Cited by in RCA: 1050] [Article Influence: 210.0] [Reference Citation Analysis (0)] |
| 278. | BioNTech SE. Study to Describe the Safety, Tolerability, Immunogenicity, and Efficacy of RNA Vaccine Candidates Against COVID-19 in Healthy Individuals (accessed 2021 Feb 20). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 24 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT04368728. |
| 279. | BioNTech SE. A Phase 3 Study to Evaluate the Safety, Tolerability, and Immunogenicity of Multiple Production Lots and Dose Levels of BNT162b2 Against COVID-19 in Healthy Participants (accessed 2021 Feb 20). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 24 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT04713553. |
| 280. | Moderna Tx Inc. A Study to Evaluate Efficacy, Safety, and Immunogenicity of mRNA-1273 Vaccine in Adults Aged 18 Years and Older to Prevent COVID-19 (accessed 2021 Feb 20). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 24 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT04470427. |
| 281. | Moderna Tx Inc. A Study to Evaluate the Safety, Reactogenicity, and Effectiveness of mRNA-1273 Vaccine in Adolescents 12 to <18 Years Old to Prevent COVID-19 (TeenCove) (accessed 2021 Feb 20). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 24 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT04649151. |
| 282. | Sharifnia Z, Bandehpour M, Kazemi B, Zarghami N. Design and Development of Modified mRNA Encoding Core Antigen of Hepatitis C Virus: a Possible Application in Vaccine Production. Iran Biomed J. 2019;23:57-67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 283. | Fu C, Zhou L, Mi Q-S, Jiang A. DC-Based Vaccines for Cancer Immunotherapy. Vaccines. 2020;8:706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 91] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 284. | Zhou Y, Zhang Y, Yao Z, Moorman JP, Jia Z. Dendritic cell-based immunity and vaccination against hepatitis C virus infection. Immunology. 2012;136:385-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 285. | Wintermeyer P, Gehring S, Eken A, Wands JR. Generation of cellular immune responses to HCV NS5 protein through in vivo activation of dendritic cells. J Viral Hepat. 2010;17:705-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 286. | Chernykh E, Leplina O, Oleynik E, Tikhonova M, Tyrinova T, Starostina N, Ostanin A. Immunotherapy with interferon-α-induced dendritic cells for chronic HCV infection (the results of pilot clinical trial). Immunol Res. 2018;66:31-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 287. | Weigand K, Voigt F, Encke J, Hoyler B, Stremmel W, Eisenbach C. Vaccination with dendritic cells pulsed with hepatitis C pseudo particles induces specific immune responses in mice. World J Gastroenterol. 2012;18:785-793. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 288. | Mishra S, Lavelle BJ, Desrosiers J, Ardito MT, Terry F, Martin WD, De Groot AS, Gregory SH. Dendritic cell-mediated, DNA-based vaccination against hepatitis C induces the multi-epitope-specific response of humanized, HLA transgenic mice. PLoS One. 2014;9:e104606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 289. | Yu H, Babiuk LA, van Drunen Littel-van den Hurk S. Strategies for loading dendritic cells with hepatitis C NS5a antigen and inducing protective immunity. J Viral Hepat. 2008;15:459-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 290. | Tian Y, Zhang HH, Wei L, Du SC, Chen HS, Fei R, Liu F. The functional evaluation of dendritic cell vaccines based on different hepatitis C virus nonstructural genes. Viral Immunol. 2007;20:553-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 291. | Echeverria I, Pereboev A, Silva L, Zabaleta A, Riezu-Boj JI, Bes M, Cubero M, Borras-Cuesta F, Lasarte JJ, Esteban JI, Prieto J, Sarobe P. Enhanced T cell responses against hepatitis C virus by ex vivo targeting of adenoviral particles to dendritic cells. Hepatology. 2011;54:28-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 292. | Zhou Y, Zhao F, Chen L, Ma L, Wang Y, He Y, Ma Z, Liu H, Guo Y, Zhang Y, Yao Z, Hao C, Jia Z. Development of a dendritic cell vaccine encoding multiple cytotoxic T lymphocyte epitopes targeting hepatitis C virus. Int J Mol Med. 2013;32:901-909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 293. | Zabaleta A, D’Avola D, Echeverria I, Llopiz D, Silva L, Villanueva L, Riezu-Boj JI, Larrea E, Pereboev A, Lasarte JJ, Rodriguez-Lago I, Iñarrairaegui M, Sangro B, Prieto J, Sarobe P. Clinical testing of a dendritic cell targeted therapeutic vaccine in patients with chronic hepatitis C virus infection. Mol Ther - Methods Clin Dev. 2015;2:15006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 294. | Ostanin AA. Autologous Dendritic Cell Vaccine for Treatment of Patients With Chronic HCV-Infection. In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 24 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT03119025. |
| 295. | Clinica Universidad de Navarra. Phase I-II Vaccination of Autologous Dendritic Cells Transduced With Adenoviral Vector Encoding NS3 in Hepatitis C Encoding NS3 in Hepatitis C (accessed 2021 Feb 21). In: ClinicalTrials.gov [internet]. Bethesda (MD): U.S. National Library of Medicine. [cited 24 February 2021]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT02309086. |
| 296. | World Health Organization (WHO). Vaccines & Diseases [Internet]. [cited 18 February 2021]. Available from: https://www.who.int/teams/immunization-vaccines-and-biologicals/diseases. |
| 297. | Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 574] [Cited by in RCA: 581] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 298. | Khawaja A, Vopalensky V, Pospisek M. Understanding the potential of hepatitis C virus internal ribosome entry site domains to modulate translation initiation via their structure and function. Wiley Interdiscip Rev RNA. 2015;6:211-224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 299. | Angulo J, Ulryck N, Deforges J, Chamond N, Lopez-Lastra M, Masquida B, Sargueil B. LOOP IIId of the HCV IRES is essential for the structural rearrangement of the 40S-HCV IRES complex. Nucleic Acids Res. 2016;44:1309-1325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 300. | Echeverría N. Variabilidad genética, resistencia al tratamiento y eficiencia traduccional del Virus de la Hepatitis C: Caracterización de factores virales y del hospedero. PhD Thesis. Universidad de la República, Uruguay. 2019. [cited 18 February 2021]. Available from: https://biur.edu.u/F/SLYLNHRAXQD9KU34M9XC3N2E55S2CQV55UJJSTBBUNPH68XPGJ-18147?func=service&doc_library=URE01&doc_number=000508554&line_number=0001&func_code=WEB-BRIEF&service_type=MEDIA. |
| 301. | Sabin AB, Boulger LR. History of Sabin attenuated poliovirus oral live vaccine strains. J Biol Stand. 1973;1:115-118. [RCA] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 197] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 302. | Macadam AJ, Pollard SR, Ferguson G, Dunn G, Skuce R, Almond JW, Minor PD. The 5' noncoding region of the type 2 poliovirus vaccine strain contains determinants of attenuation and temperature sensitivity. Virology. 1991;181:451-458. [PubMed] |
| 303. | Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu Rev Microbiol. 2005;59:587-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 480] [Cited by in RCA: 486] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 304. | Frese M, Lee E, Larena M, Lim PS, Rao S, Matthaei KI, Khromykh A, Ramshaw I, Lobigs M. Internal ribosome entry site-based attenuation of a flavivirus candidate vaccine and evaluation of the effect of beta interferon coexpression on vaccine properties. J Virol. 2014;88:2056-2070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 305. | Moratorio G, Henningsson R, Barbezange C, Carrau L, Bordería AV, Blanc H, Beaucourt S, Poirier EZ, Vallet T, Boussier J, Mounce BC, Fontes M, Vignuzzi M. Attenuation of RNA viruses by redirecting their evolution in sequence space. Nat Microbiol. 2017;2:17088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 306. | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15248] [Cited by in RCA: 14083] [Article Influence: 2816.6] [Reference Citation Analysis (1)] |
| 307. | Defendi HGT, da Silva Madeira L, Borschiver S. Analysis of the COVID-19 Vaccine Development Process: an Exploratory Study of Accelerating Factors and Innovative Environments. J Pharm Innov. 2021;1-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 308. | Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1620] [Cited by in RCA: 2760] [Article Influence: 394.3] [Reference Citation Analysis (0)] |
| 309. | Dong Y, Dai T, Wei Y, Zhang L, Zheng M, Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct Target Ther. 2020;5:237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 394] [Cited by in RCA: 379] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 310. | Cao Y, Gao GF. mRNA vaccines: A matter of delivery. EClinicalMedicine. 2021;32:100746. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 311. | Ledford H. Could mixing COVID vaccines boost immune response? Nature. 2021;590:375-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 312. | Spencer AJ, McKay PF, Belij-Rammerstorfer S, Ulaszewska M, Bissett CD, Hu K, Samnuan K, Wright D, Sharpe HR, Gilbride C, Truby A, Allen ER, Gilbert SC, Shattock RJ, Lambe T. Heterologous vaccination regimens with self-amplifying RNA and Adenoviral COVID vaccines induce robust immune responses in mice. Nat Commun. 2021;12: 2893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 90] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 313. | Prüβ BM. Current State of the First COVID-19 Vaccines. Vaccines (Basel). 2021;9:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |