Published online Dec 27, 2020. doi: 10.4254/wjh.v12.i12.1228
Peer-review started: July 9, 2020
First decision: September 11, 2020
Revised: September 28, 2020
Accepted: October 15, 2020
Article in press: October 15, 2020
Published online: December 27, 2020
Processing time: 163 Days and 13.3 Hours
Genetic factors play an important role in the pathogenesis and development of metabolic dysfunction-associated fatty liver disease (MAFLD).
To study the association of single nucleotide polymorphisms (SNPs), previously identified in Western populations, with the risk of MAFLD in a Singapore Chinese population and their interactions with environmental and medical risk factors.
A retrospective case-control study was conducted with 72 MAFLD cases and 72 controls with no hepatic steatosis on computed tomography, magnetic resonance imaging, or controlled attenuation parameter score. Subjects were recruited from two tertiary hospitals. Genetic alleles such as NCAN, GCKR, LYPLAL1, PNPLA3, PPP1R3B, FDFT1, COL13A1, EFCAB4B, PZP, and TM6SF2 were genotyped using the TaqMan® Predesigned SNP Genotyping Assay.
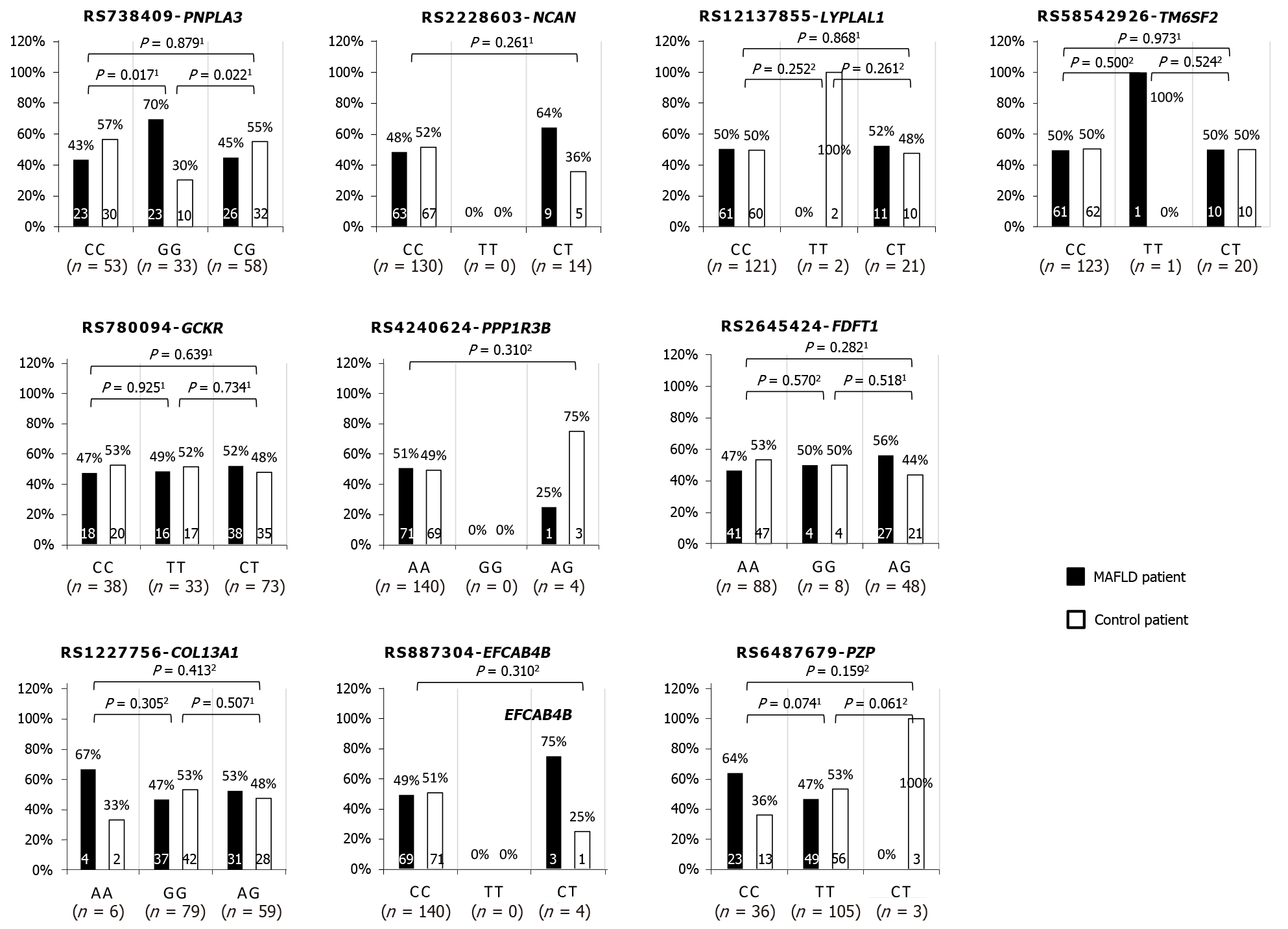
Weight and body mass index (BMI) were 1.2-times higher in patients (70.6 kg, 95% confidence interval [CI]: 57.1-84.1 vs 60.8 kg, 95%CI: 48.5-73.1, P < 0.001 and 26.9 kg, 95%CI: 23-40.8 vs 23.3 kg 95%CI: 19-27.6, P < 0.001 respectively). The prevalence of diabetes mellitus in patients was 40.3% and 20.8% in controls (P = 0.011). Patients had higher mean triglycerides than controls (P < 0.001). PNPLA3 GG was more likely to be associated with MAFLD (43.4% CC vs 69.7% GG, P = 0.017, and 44.8% CG vs 69.7% GG, P = 0.022). In multivariable analysis, hypertriglyceridemia (odds ratio [OR]: 2.04 95%CI: 1.3-3.1, P = 0.001), BMI (OR: 1.2 95%CI: 1.1-1.4, P < 0.001) and PNPLA3 GG (OR: 3.4 95%CI: 1.3-9.2, P = 0.014) were associated with MAFLD (area under the receiver operating characteristic curve of 0.823).
Among the Chinese population of Singapore, PNPLA3 homozygous GG allele is a strong predictor of MAFLD, whereas LYPLAL1, GCKR, FDFT1, COL13A1, PZP, and TM6SF2 are not significantly associated. Hypertriglyceridemia, high BMI, and PNPLA3 GG are independent predictors of MAFLD.
Core Tip: A number of genetic variations (known as single nucleotide polymorphism, SNPs) are reportedly associated with metabolic dysfunction-associated fatty liver disease (MAFLD), mostly by studies from Europe and America. This study examined 10 of the most important SNPs in a Chinese population in Singapore, and found that 1 such variation, the PNPLA3 GG variation, is strongly linked to MAFLD, whereas the rest are not significantly associated. PNPLA3, together with high triglyceride and elevated body mass index, are found to be independent, strong predictors of MAFLD.
- Citation: Lee GH, Phyo WW, Loo WM, Kwok R, Ahmed T, Shabbir A, So J, Koh CJ, Hartono JL, Muthiah M, Lim K, Tan PS, Lee YM, Lim SG, Dan YY. Validation of genetic variants associated with metabolic dysfunction-associated fatty liver disease in an ethnic Chinese population. World J Hepatol 2020; 12(12): 1228-1238
- URL: https://www.wjgnet.com/1948-5182/full/v12/i12/1228.htm
- DOI: https://dx.doi.org/10.4254/wjh.v12.i12.1228
Metabolic dysfunction-associated fatty liver disease (MAFLD) is increasingly recognized as a leading cause of liver morbidity and mortality, and has emerged as the most common chronic liver disease[1]. The increasing prevalence of MAFLD is associated with the epidemic surge in obesity and metabolic syndrome[2]. The estimated prevalence of MAFLD in Asia is about 27.4%[1]. MAFLD is a cli-nicopathological spectrum that consists of hepatic steatosis and non-alcoholic steatohepatitis (NASH), with up to 20% of NASH patients progressing to cirrhosis and end-stage liver complications[3-5]. This heralds an expectant future epidemic of hepatocellular carcinoma (HCC) caused by NASH, potentially overshadowing the role of viral hepatitis in the development of HCC[6-8]. Reflecting this trend, NASH is the most rapidly growing indication for liver transplantation in the United States, increasing 4-fold from 2002 to 2012, and is poised to become the leading indication[9].
The pathogenesis of MAFLD is multifactorial and complex, and both genetic and epigenetic factors appear to play vital roles in its development. These factors interact with environmental, dietary, and metabolic risk factors, which all contribute to the development of MAFLD and the risk of disease progression. Different genes encode proteins involved in the regulation of lipid metabolism in the liver[10]. Excess of hepatic triglycerides (TGs) associated with insulin resistance is a key mechanism in MAFLD pathophysiology. Adipose tissue insulin resistance is linked with lower circulating adipokines and this leads to increased lipolysis with resultant oxidative stress, lipotoxicity and apoptosis, thus inducing NASH[11,12].
A multitude of studies have demonstrated a heritable component and familial clustering as important factors in MAFLD. The majority of these studies have been published in the western population. First-degree relatives of individuals with MAFLD have conferred a higher risk of the disease compared to the general population[13]. Twin studies have shown that genetic factors contribute up to 60% of variability in alanine aminotransferase (ALT) levels and hepatic fat content in subjects[14]. Over the years, multiple genetic single nucleotide polymorphisms (SNPs) associated with MAFLD have been studied. PNPLA3 is the major gene associated with hepatic TG content, increasing susceptibility to more aggressive forms of MAFLD and HCC[15-18]. Specifically, the variant allele rs738409 C>G or I148M of PNPLA3 is deemed an important genetic factor for a predisposition towards progressive MAFLD and increases hepatic TG accumulation[15]. The genetic association with PNPLA3 is also evident in several Asian studies and is estimated to be present in 13%-19% of the general population[19-21]. In a Chinese study, PNPLA3 had a stronger correlation with hepatic steatosis in non-obese individuals without metabolic syndrome[20]. Other SNPs associated with progressive MAFLD include TM6SF2, LYPLAL1, NCAN, APOB, MBOAT7, LPIN1, GCKR, ENPP1[22]. The effect size and allele frequency of these genetic variants have shown much diversity across different ethnicities leading to racial and ethnic differences in MAFLD prevalence[23]. To date, these genetic variants have not been evaluated in our local population.
In this study, we evaluated the association of SNPs, previously identified in Western populations, and environmental and medical risk factors with the risk of MAFLD in a Singapore Chinese population.
The study was conducted using a retrospective case-control design recruiting subjects from the National University Hospital (NUH) and Khoo Teck Puat Hospital in Singapore. All participants gave informed consent and the study was approved by the Institutional Review Board.
Fatty liver was identified based on computed tomography (CT) or magnetic resonance imaging (MRI) or histopathology of liver (> 20% steatosis) or controlled attenuation parameter (CAP) score. CAP is a non-invasive tool for the detection of hepatic steatosis but is limited by body mass index (BMI). CAP value of S2-3 (≥ 34% steatosis) is considered as positive hepatic steatosis and S0 (< 5% steatosis) as no steatosis. The enrolled subjects in this study were Singapore residents of Chinese ancestry. These subjects also do not have other chronic liver etiology. The control group will either have a CT or MRI scan or CAP score showing the absence of hepatic steatosis (performed for non-liver disease indications) within 1 year of enrolment into the study.
Patients with secondary causes of steatosis including alcohol abuse, total parenteral nutrition, hepatitis B and hepatitis C virus infection, and the use of drugs known to precipitate hepatic steatosis were excluded. In addition, patients with any of the following diseases such as autoimmune hepatitis, drug-induced liver disease, primary biliary cirrhosis, and primary sclerosing cholangitis were excluded from participation in this study.
Subjects underwent an assessment comprising anthropometric measurements and a questionnaire on health-related behaviors such as smoking and alcohol drinking habits. Smokers are defined as subjects who had smoked at least 100 cigarettes during their lifetime and are henceforth classified as smokers vs non-smokers. With regards to drinking history, the study was confined to men and women who drank less than 140 g or 70 g of alcohol per week respectively. Body measurements including weight and height were measured in a standardized fashion by a trained examiner. BMI was calculated as follows: Weight (kg) divided by height squared (m2). Overweight and/or obesity were defined as having a BMI > 23 kg/m2 and > 28 kg/m2 respectively. Venous blood samples were drawn for biochemical and genotyping analyses. Total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), TG, fasting plasma glucose (FPG), and liver function tests were measured by standard clinical laboratory techniques in NUH Biochemistry Laboratory. Reduced HDL-C, hypertriglyceridemia, and raised FPG were diagnosed according to the International Diabetes Federation consensus worldwide definition of the metabolic syndrome.
Blood samples obtained from the subjects were centrifuged at 1500 rpm for 10 min. The buffy coat layer was separated and transferred into a 1.5-mL centrifuge tube. Genomic DNA was extracted from the concentrated lymphocytes of the buffy coat using the QIAamp DNA Mini Kit (Qiagen, Hilden, German). The related genetic alleles such as NCAN, GCKR, LYPLAL1, PNPLA3, PPP1R3B, FDFT1, COL13A1, EFCAB4B, PZP, and TM6SF2 were genotyped using the TaqMan® Predesigned SNP Genotyping Assay (Applied Biosystems, Foster City, CA, United States) on a step one real-time PCR instrument (Applied Biosystems). The total reaction volume for each well was 10 µL containing 5 µL universal mastermix (Applied Biosystems), 0.5 µL assay mix, 3.5 µL distilled water, and 1 mL genomic DNA. The plate was set up at 95 °C holding stage for 20 s, 45 cycles of 95 °C denaturation for 3 s and 60 °C annealing for 20 s and run on a fast reaction (40 min for each run). Negative controls were introduced for every run to ensure genotyping quality.
The pathologist, radiologist, and laboratory technologist, who performed the tests for hepatic steatosis and SNPs, were blinded to the patients’ participation in this study. The laboratory team performing and interpreting the SNP assay was blinded to the identity and grouping of the patients and samples.
The data were analyzed using SPSS (IBM Corp. Released 2011. IBM SPSS Statistics for Windows, Version 20.0. Armonk, NY, United States) and P < 0.05 was considered statistically significant. All values are presented as the mean ± standard deviation (SD) for continuous data and as frequency and percent for categorical data. For normally distributed variables (age, BMI, waist circumference, ALT, HDL cholesterol, LDL cholesterol, TC, TG, and systolic blood pressure), two independent sample t-test was performed to compare between NASH and simple steatosis. Categorical variables were compared between cases and controls using Chi-square or Fisher’s exact test, where appropriate. Multivariable logistic regression analysis was performed to investigate the association of risk factors and SNPs with MAFLD outcome. Area under the curve, positive predictive value and negative predictive value were assessed using receiver operating characteristic analysis.
In total, 150 patients were screened initially; however, 6 were lost to follow-up after recruitment and were removed from the analysis; 72 MAFLD subjects and 72 control subjects completed the study. Among the cases, the methods used to diagnose fatty liver disease were CT (54%); biopsy (24%); combination of ultrasound, laboratory results and clinical features (11%); MRI (10%); and CAP score (1%). Majority (99%) of controls were identified based on CT and MRI results. The mean age was 57 years and 61 years for the NAFLD and control group respectively. 59.7% of the patients in each group were male. 11.8% were smokers. None of the patients in both groups drank more than the 140 g (male) or 70 g (female) of alcohol per week.
The subjects who had MAFLD were more likely to have BMI higher than 24.9 kg/m2 (27, 95% confidence interval [CI]: 23-31 vs 23, 95%CI: 19-28, P < 0.001). While waist-hip ratio was similar between the two groups (0.94, 95%CI: 0.9-1.0 vs 0.95, 95%CI: 0.8-1.1), lipid profiles were significantly higher in the MAFLD group than in the control group. Mean TC in MAFLD group was 4.4 (95%CI: 2.9-5.8) while in control was 3.4 (95%CI: 1.2-5.7), P = 0.0032, and mean TG in MAFLD group was 1.7 (95%CI: 0.8-2.7) while in control was 1.0 (95%CI: 0.1-1.98), P < 0.0012, (Table 1). 36% of MAFLD patients were taking lipid-lowering agents while only 24% in the control group were. Similar proportion of each group had hypertension (35% in MAFLD group vs 31% in control group, P = 0.594) and were on anti-hypertensive medication. There were 40% of patients who had diabetes mellitus in MAFLD group compared to 21% in the control group (P = 0.011). However, there was no difference in fasting glucose between the two groups (6.1, 95%CI: 5.6-6.6 vs 5.9, 95%CI: 5.5-6.3) since most patients were treated with anti-diabetic medication. Both aspartate aminotransferase (AST) and ALT levels were observed to be higher in MAFLD patients than in the controls (Table 1).
| NAFLD patient, n = 72 | Control, n = 72 | Total, n = 144 | P value | |
| Clinical characteristics | ||||
| Age | 57 (46-68) | 61 (48-74) | 59 (46-71) | 0.0372 |
| Male, % | 43 (59.7%) | 43 (59.7%) | 86 (59.7%) | 1.01 |
| BMI, kg/m2 | 27 (23-31) | 23 (19-28) | 25 (21-30) | < 0.0012 |
| Waist-hip ratio | 0.94 (0.9-1.0) | 0.95 (0.8-1.1) | 0.9 (0.8-1.0) | 0.5872 |
| Smoker, % | 5 (6.9%) | 12 (16.7%) | 17 (11.8%) | 0.0711 |
| DM, % | 29 (40.3%) | 15 (20.8%) | 44 (30.6%) | 0.0111 |
| Hypertension, % | 25 (34.7) | 22 (30.6%) | 47 (32.6%) | 0.5941 |
| On anti-diabetic treatment | 26 (36.1) | 14 (19.4) | 40 (27.8) | 0.0261 |
| On lipid-lowering treatment | 26 (36.1) | 17 (23.6) | 43 (29.9) | 0.1011 |
| Laboratory measures | ||||
| Serum lipid levels and fasting glucose | ||||
| Total cholesterol, mmol/L | 4.4 (2.9-5.8) | 3.4 (1.2-5.7) | 3.9 (1.9-5.8) | 0.00322 |
| LDL-C, mmol/L | 2.5 (1.5-3.5) | 2.1 (0.7-3.5) | 2.3 (1.1-3.6) | 0.04922 |
| HDL-C, mmol/L | 1.3 (0.6-1.9) | 1.0 (0.3-1.7) | 1.6 (0.5-1.8) | 0.02922 |
| TG, mmol/L | 1.7 (0.8-2.7) | 1.0 (0.1-1.98) | 1.4 (0.4-2.4) | < 0.00122 |
| FPG, mmol/L | 6.1 (5.6-6.6) | 5.9 (5.5-6.3) | 6.0 (5.7-6.3) | 0.45922 |
| Liver function test | ||||
| Albumin, g/L | 40.9 (32.1-49.7) | 41.0 (34.6-47.3) | 40.9 (33.3-48.6) | 0.97422 |
| Bilirubin, mmol/L, Median | 13.1 (4.5-21.7), Median 11.0 | 14.7 (-15.6-44.97), Median 9.0 | 13.9 (-8.3-36.1), Median 10.0 | 0.66722 |
| AST, U/L | 37.1 (16.7-57.5) | 26.2 (7.4-44.9) | 31.7 (11.4-51.98) | 0.00122 |
| ALT, U/L | 46.2 (15.5-76.8) | 22.2 (8.2-36.1) | 34.3 (7.6-60.9) | < 0.00122 |
| ALP, U/L | 87.6 (36.2-138.9) | 77.4 (39.95-114.8) | 82.5 (37.4-127.6) | 0.17922 |
| INR | 0.9 (0.5-1.2) | 0.8 (0.2-1.3) | 0.8 (0.3-1.3) | 0.37322 |
| Platelet as 109/L | 223 (130-315) | 256 (162-350) | 240 (146-334) | 0.04022 |
A total of 9 SNPs were tested and PNPLA3 G allele is more likely to be associated with MAFLD compared to C allele (GG vs CC OR: 3.0 [95%CI: 1.2-7.5] and GG vs CG OR: 2.83 [95%CI: 1.1-6.9]). Some homozygous alleles of NCAN, PPP1R3B, and EFCAB4B that were proven significantly associated with MAFLD in the west are found to be absent among the study population. For instance, NCAN homozygous TT allele, PPP1R3B homozygous GG allele and EFXAB4B homozygous TT allele were not found in any of the recruited MAFLD and non-MAFLD participants. Furthermore, there were some alleles in which each was present only among the minority (< 3%) of the study population, particularly for allele PPP1R3B AG, EFCAB4B CT, PZP CT, LYPLAL1 TT and TM6SF2 TT. Despite the high presentation among the MAFLD patient group, four SNPs were not significantly associated with the presence of MAFLD. Two of the insignificant SNPs could be due to small sample sizes as mentioned above in EFCAB4B CT vs CC (odds ratio [OR]: 3.09, 95%CI: 0.31-30.40) and PPP1R3B AA vs AG (OR: 3.09, 95%CI: 0.31-30.4). Another two insignificant alleles were COL13A1 AA vs GG (OR: 2.3, 95%CI: 0.4-13.1) and PZP CC vs TT (OR: 2.0, 95%CI: 0.9-4.4) (Figure 1).
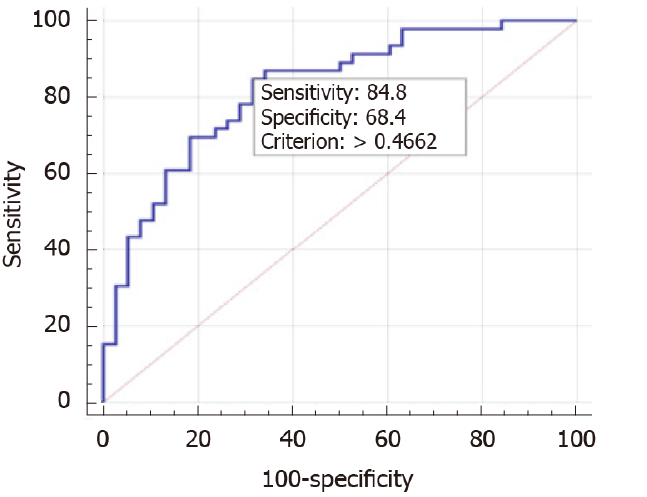
We performed multivariable analysis of risk factors and SNPs, independent predictors of MAFLD were hypertriglyceridemia (OR: 2.3 95%CI: 1.3-4.1), BMI (OR: 1.2 95%CI: 1.1-1.4) and PNPLA3 GG (OR: 4.1 95%CI: 1.3-12.9) (Table 2). This model has a cut-off value of 0.47 (based on Youden index) that can predict 73.8% of positive cases with an area under the receiver operating characteristic curve (AUROC) of 0.823 (95%CI: 0.73-0.91, P < 0.001) (Figure 2). Without the PNPLA3, the model performance in the prediction of MAFLD was lower with the AUROC of 0.789 (95%CI: 0.71-0.86, P < 0.001).
| Odds ratio | 95%CI | P value | ||
| Lower | Upper | |||
| TG, mmol/L | 2.338 | 1.3 | 4.1 | 0.003 |
| BMI, kg/m2 | 1.243 | 1.1 | 1.4 | 0.003 |
| PNPLA3 GG vs CC | 4.146 | 1.3 | 12.9 | 0.014 |
In this study, PNPLA3 GG allele was the only SNP found to be associated with progressive MAFLD in the Singaporean Chinese population. The surprising finding is none of the other SNP variants, including LYPLAL1, GCKR, FDFT1, COL13A1, PZP, and TM6SF2, which were identified in the Western population, demonstrate a similar association. On further analysis, the PNPLA3 GG variant, elevated BMI, and hypertriglyceridemia were independent predictors of MAFLD.
The PNPLA3 I148M variant is associated with increased severity of MAFLD, with a higher susceptibility to NASH, fibrosis, and HCC[24]. The I148M mutation confers double the risk for HCC for each variant allele[17]. PNPLA3 I148M predisposes to lower hepatic very-low-density lipoprotein (VLDL)[25] and also lower levels of adiponectin[26] which result in inflammation leading to the development of NASH[27]. Furthermore, this association is independent of predisposition to hepatic steatosis, insulin resistance, dyslipidemia or obesity[15]. A study of Italian and United Kingdom patients who carried the PNPLA3 GG genotype had a 3.3-fold risk of MAFLD compared to those with the CC genotype. This data is in line with our results of the predisposition of homozygous GG allele to MAFLD. The GG phenotype was linked with higher LDL levels, fasting insulin levels, homeostasis model assessment of insulin resistance (HOMA-IR) score. However, there was no association with type 2 diabetes. These subjects also had significantly higher ALT levels in comparison with those who possessed the CC or CG genotypes[28]. In Asians, there is a higher prevalence of the entity that is non-obese MAFLD defined as those with a BMI of less than 25 kg/m2. There is some evidence indicating a greater influence of the PNPLA3 variant in the non-obese MAFLD subjects. Wei et al[29] reported a higher proportion of non-obese MAFLD patients with the PNPLA3 variant (78.4%) compared with the obese MAFLD group (59.8%) in a population-based study from Hong Kong. But the mechanism linking the two is incompletely understood. On the contrary, these results were not replicated in our study.
Polymorphism in the PNPLA3 gene may also play an important role in the management of MAFLD as there is data suggesting that lifestyle modification may be more beneficial in this group compared to other genotypes. The effect of degree of weight loss on intrahepatic TG content and liver enzyme levels was more pronounced in MAFLD patients with the homozygous GG allele than in subjects with the homozygous CC allele[30,31]. Hence, lifestyle modification and weight loss can be strongly advocated in our patient cohort with a predominance of the GG genotype which may result in more optimal outcomes.
Variants in the GCKR gene (rs1260326 and rs780094) increase MAFLD susceptibility by inducing lipogenesis via activation of hepatic glucose uptake[22,32]. In a study on Italian MAFLD patients, GCKR rs780094 C>T was associated with higher serum TG levels and severity of liver fibrosis[33]. However, there was no observed association between the GCKR SNP and our study cohort. The FDFT1 gene is involved in the regulation of cholesterol biosynthesis and the rs2645424 SNP has been shown to demonstrate a positive correlation with the MAFLD activity score (NAS)[34]. Several genes with different SNPs including COL13A1 and EFCAB4B were associated with lobular inflammation in NASH whereas PZP SNPs had an association with serum AST levels[34]. A large GWAS study detected variants in NCAN (rs2228603) and LYPLAL1 correlating with histologic lobular inflammation and fibrosis, but not in PPP1R3B which was associated with liver steatosis only[22]. Another major genetic determinant of MAFLD is TM6SF2 which is associated with reduced hepatic secretion of VLDL and increased risk of myocardial infarction. However, a study from Hong Kong showed a prevalence of only 0.4% of Chinese who had the variant which conferred a higher risk of MAFLD, indicating a potentially limited role of this gene in Asian MAFLD[35]. Overall, none of these genetic SNPs except for PNPLA3 were related to MAFLD development in our study population.
This study had limitations. The general limitations of case-control design are well-documented. The sample size may not be large enough to detect the effects of some of the genetic polymorphisms tested, but the effect sizes will likely be too small for clinical application. Despite our best effort to match the control group with the MAFLD subjects, we were not able to completely balance all known risk factors and associated co-morbidities linked to MAFLD. Very few patients with high BMI in our population have completely normal findings for liver steatosis measurement, and also free from other features of metabolic syndrome. As a result, BMI and corresponding risk factors, such as diabetes mellitus and hyperlipidemia are more prevalent in the MAFLD group. The other major limitation is the lack of liver histology in the study population to have the gold standard diagnosis of hepatic steatosis and steatohepatitis. Surrogate markers, including CAP score (on FibroScan), CT, and MRI, are accepted. To avoid diagnostic uncertainties, we used more stringent criteria for defining hepatic steatosis. For CAP score, we accepted only subjects with S2 and above into the MAFLD group and only S0 into the control group. For histopathology, we included only those patients with higher liver steatosis of 20% or more into the MAFLD group and < 5% into the control group.
In conclusion, we demonstrated that the genetic variant in PNPLA3 is associated with an increased risk of MAFLD in the Singaporean Chinese population, but not with the other studied SNPs. While it is possible that with a much larger study population, some of these MAFLD-related SNPs may reach statistical significance, their effect size is likely to be much smaller than PNPLA3, and are unlikely to inspire lifestyle changes in affected individuals. Together with the other factors of TG level and BMI, PNPLA3 can potentially be used as a predictive tool to identify individuals in the community, who are at risk of more progressive forms of MAFLD for targeted close surveillance and early weight loss interventions. The influence of genetic variation can be translated into more precise clinical management, which should be tailored to each individual population in the country.
Metabolic dysfunction-associated fatty liver disease (MAFLD) is increasingly recognized as a leading cause of liver morbidity and mortality and has emerged as the most common chronic liver disease globally. The estimated prevalence of MAFLD in Asia is about 27.4%. Genetic factors play an important role in the pathogenesis and development of MAFLD.
A number of genetic variations (known as single nucleotide polymorphism, SNP) have been reported to be associated with MAFLD, mostly by studies from Europe and America. This study examines 10 of the most important SNPs in the Chinese population in Singapore.
To study the association of SNPs, previously identified in Western populations, with the risk for MAFLD in the Singapore Chinese population and their interactions with environmental and medical risk factors.
This is a retrospective case-control study of 72 MAFLD cases and 72 controls with no hepatic steatosis on imaging or controlled attenuation parameter score. Subjects were recruited from two tertiary hospitals in Singapore.
PNPLA3 GG was more likely to be associated with MAFLD (43.4% CC vs 69.7% GG, P = 0.017, and 44.8% CG vs 69.7% GG, P = 0.022). In multivariable analysis, hypertriglyceridemia (OR: 2.04 95%CI: 1.3-3.1, P = 0.001), body mass index (BMI, OR: 1.2 95% confidence interval [CI]: 1.1-1.4, P < 0.001) and PNPLA3 GG (odds ratio [OR]: 3.4 95%CI: 1.3-9.2, P = 0.014) were associated with MAFLD (area under the receiver operating characteristic curve of 0.823).
This study showed that PNPLA3 GG allele was the only SNP associated with progressive MAFLD in the Singaporean Chinese population. The PNPLA3 GG variant, elevated BMI, and hypertriglyceridemia were independent predictors of MAFLD.
PNPLA3, along with triglyceride level and BMI, can potentially be used as a predictive tool to identify and risk-stratify affected individuals in the community for early intervention and targeted surveillance.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Singapore
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Bubnov R, Ferrarese A, Liao R, Smolić M S-Editor: Huang P L-Editor: Filipodia P-Editor: Wu YXJ
| 1. | Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5322] [Cited by in RCA: 7517] [Article Influence: 835.2] [Reference Citation Analysis (0)] |
| 2. | Khashab MA, Liangpunsakul S, Chalasani N. Nonalcoholic fatty liver disease as a component of the metabolic syndrome. Curr Gastroenterol Rep. 2008;10:73-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 3. | Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, Harrison SA, Brunt EM, Sanyal AJ. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67:328-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3544] [Cited by in RCA: 4935] [Article Influence: 705.0] [Reference Citation Analysis (9)] |
| 4. | Farrell GC, Wong VW, Chitturi S. NAFLD in Asia--as common and important as in the West. Nat Rev Gastroenterol Hepatol. 2013;10:307-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 354] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 5. | Kim D, Kim WR, Kim HJ, Therneau TM. Association between noninvasive fibrosis markers and mortality among adults with nonalcoholic fatty liver disease in the United States. Hepatology. 2013;57:1357-1365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 626] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 6. | Goh LY, Leow AH, Goh KL. Observations on the epidemiology of gastrointestinal and liver cancers in the Asia-Pacific region. J Dig Dis. 2014;15:463-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (1)] |
| 7. | Goh KL. Changing trends in gastrointestinal disease in the Asia-Pacific region. J Dig Dis. 2007;8:179-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 52] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 8. | Younossi ZM, Otgonsuren M, Henry L, Venkatesan C, Mishra A, Erario M, Hunt S. Association of nonalcoholic fatty liver disease (NAFLD) with hepatocellular carcinoma (HCC) in the United States from 2004 to 2009. Hepatology. 2015;62:1723-1730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 616] [Article Influence: 61.6] [Reference Citation Analysis (0)] |
| 9. | Wong RJ, Cheung R, Ahmed A. Nonalcoholic steatohepatitis is the most rapidly growing indication for liver transplantation in patients with hepatocellular carcinoma in the U.S. Hepatology. 2014;59:2188-2195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 520] [Cited by in RCA: 585] [Article Influence: 53.2] [Reference Citation Analysis (0)] |
| 10. | Dongiovanni P, Romeo S, Valenti L. Genetic Factors in the Pathogenesis of Nonalcoholic Fatty Liver and Steatohepatitis. Biomed Res Int. 2015;2015:460190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 11. | Bugianesi E, Gastaldelli A, Vanni E, Gambino R, Cassader M, Baldi S, Ponti V, Pagano G, Ferrannini E, Rizzetto M. Insulin resistance in non-diabetic patients with non-alcoholic fatty liver disease: sites and mechanisms. Diabetologia. 2005;48:634-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 539] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 12. | Day CP. From fat to inflammation. Gastroenterology. 2006;130:207-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 300] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 13. | Schwimmer JB, Celedon MA, Lavine JE, Salem R, Campbell N, Schork NJ, Shiehmorteza M, Yokoo T, Chavez A, Middleton MS, Sirlin CB. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136:1585-1592. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 358] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 14. | Makkonen J, Pietiläinen KH, Rissanen A, Kaprio J, Yki-Järvinen H. Genetic factors contribute to variation in serum alanine aminotransferase activity independent of obesity and alcohol: a study in monozygotic and dizygotic twins. J Hepatol. 2009;50:1035-1042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 15. | Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461-1465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2701] [Cited by in RCA: 2592] [Article Influence: 152.5] [Reference Citation Analysis (0)] |
| 16. | Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883-1894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 756] [Cited by in RCA: 739] [Article Influence: 52.8] [Reference Citation Analysis (1)] |
| 17. | Liu YL, Patman GL, Leathart JB, Piguet AC, Burt AD, Dufour JF, Day CP, Daly AK, Reeves HL, Anstee QM. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol. 2014;61:75-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 413] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 18. | Singal AG, Manjunath H, Yopp AC, Beg MS, Marrero JA, Gopal P, Waljee AK. The effect of PNPLA3 on fibrosis progression and development of hepatocellular carcinoma: a meta-analysis. Am J Gastroenterol. 2014;109:325-334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 297] [Cited by in RCA: 275] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 19. | Rotman Y, Koh C, Zmuda JM, Kleiner DE, Liang TJ; NASH CRN. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52:894-903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 387] [Cited by in RCA: 381] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 20. | Li Y, Xing C, Cohen JC, Hobbs HH. Genetic variant in PNPLA3 is associated with nonalcoholic fatty liver disease in China. Hepatology. 2012;55:327-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Shen J, Wong GL, Chan HL, Chan HY, Yeung DK, Chan RS, Chim AM, Chan AW, Choi PC, Woo J, Chu WC, Wong VW. PNPLA3 gene polymorphism accounts for fatty liver in community subjects without metabolic syndrome. Aliment Pharmacol Ther. 2014;39:532-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 22. | Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, Gudnason V, Eiriksdottir G, Garcia ME, Launer LJ, Nalls MA, Clark JM, Mitchell BD, Shuldiner AR, Butler JL, Tomas M, Hoffmann U, Hwang SJ, Massaro JM, O'Donnell CJ, Sahani DV, Salomaa V, Schadt EE, Schwartz SM, Siscovick DS; NASH CRN; GIANT Consortium; MAGIC Investigators; Voight BF; Carr JJ; Feitosa MF; Harris TB; Fox CS; Smith AV; Kao WH; Hirschhorn JN; Borecki IB; GOLD Consortium. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 775] [Cited by in RCA: 761] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 23. | Hernaez R, McLean J, Lazo M, Brancati FL, Hirschhorn JN, Borecki IB, Harris TB; Genetics of Obesity-Related Liver Disease (GOLD) Consortium; Nguyen T; Kamel IR; Bonekamp S; Eberhardt MS; Clark JM; Kao WH; Speliotes EK. Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the third National Health and Nutrition Examination Survey. Clin Gastroenterol Hepatol 2013; 11: 1183-1190. e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 24. | Sookoian S, Castaño GO, Burgueño AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50:2111-2116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 312] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 25. | Pirazzi C, Adiels M, Burza MA, Mancina RM, Levin M, Ståhlman M, Taskinen MR, Orho-Melander M, Perman J, Pujia A, Andersson L, Maglio C, Montalcini T, Wiklund O, Borén J, Romeo S. Patatin-like phospholipase domain-containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J Hepatol. 2012;57:1276-1282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 230] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 26. | Valenti L, Rametta R, Ruscica M, Dongiovanni P, Steffani L, Motta BM, Canavesi E, Fracanzani AL, Mozzi E, Roviaro G, Magni P, Fargion S. The I148M PNPLA3 polymorphism influences serum adiponectin in patients with fatty liver and healthy controls. BMC Gastroenterol. 2012;12:111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 27. | Polyzos SA, Toulis KA, Goulis DG, Zavos C, Kountouras J. Serum total adiponectin in nonalcoholic fatty liver disease: a systematic review and meta-analysis. Metabolism. 2011;60:313-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 243] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 28. | Valenti L, Al-Serri A, Daly AK, Galmozzi E, Rametta R, Dongiovanni P, Nobili V, Mozzi E, Roviaro G, Vanni E, Bugianesi E, Maggioni M, Fracanzani AL, Fargion S, Day CP. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51:1209-1217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 477] [Cited by in RCA: 529] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 29. | Wei JL, Leung JC, Loong TC, Wong GL, Yeung DK, Chan RS, Chan HL, Chim AM, Woo J, Chu WC, Wong VW. Prevalence and Severity of Nonalcoholic Fatty Liver Disease in Non-Obese Patients: A Population Study Using Proton-Magnetic Resonance Spectroscopy. Am J Gastroenterol. 2015;110:1306-14; quiz 1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 241] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 30. | Shen J, Wong GL, Chan HL, Chan RS, Chan HY, Chu WC, Cheung BH, Yeung DK, Li LS, Sea MM, Woo J, Wong VW. PNPLA3 gene polymorphism and response to lifestyle modification in patients with nonalcoholic fatty liver disease. J Gastroenterol Hepatol. 2015;30:139-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 31. | Sevastianova K, Kotronen A, Gastaldelli A, Perttilä J, Hakkarainen A, Lundbom J, Suojanen L, Orho-Melander M, Lundbom N, Ferrannini E, Rissanen A, Olkkonen VM, Yki-Järvinen H. Genetic variation in PNPLA3 (adiponutrin) confers sensitivity to weight loss-induced decrease in liver fat in humans. Am J Clin Nutr. 2011;94:104-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 32. | Beer NL, Tribble ND, McCulloch LJ, Roos C, Johnson PR, Orho-Melander M, Gloyn AL. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18:4081-4088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 327] [Cited by in RCA: 315] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 33. | Petta S, Miele L, Bugianesi E, Cammà C, Rosso C, Boccia S, Cabibi D, Di Marco V, Grimaudo S, Grieco A, Pipitone RM, Marchesini G, Craxì A. Glucokinase regulatory protein gene polymorphism affects liver fibrosis in non-alcoholic fatty liver disease. PLoS One. 2014;9:e87523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 34. | Chalasani N, Guo X, Loomba R, Goodarzi MO, Haritunians T, Kwon S, Cui J, Taylor KD, Wilson L, Cummings OW, Chen YD, Rotter JI; Nonalcoholic Steatohepatitis Clinical Research Network. Genome-wide association study identifies variants associated with histologic features of nonalcoholic Fatty liver disease. Gastroenterology 2010; 139: 1567-1576, 1576.e1-1576. e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 256] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 35. | Wong VW, Wong GL, Tse CH, Chan HL. Prevalence of the TM6SF2 variant and non-alcoholic fatty liver disease in Chinese. J Hepatol. 2014;61:708-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |