Published online Dec 27, 2020. doi: 10.4254/wjh.v12.i12.1148
Peer-review started: July 20, 2020
First decision: September 24, 2020
Revised: October 1, 2020
Accepted: October 26, 2020
Article in press: October 26, 2020
Published online: December 27, 2020
Processing time: 150 Days and 14.4 Hours
Intrahepatic cholangiocarcinoma (iCCA) is a heterogeneous primary liver cancer, and currently there exist only a few options of targeted therapy. Histopathologically, iCCA is sub-classified according to morphology (mass forming type, periductal infiltrating type, and intraductal growing type) and histology (small duct type and large duct type). According to different histopathological types, clinical features such as risk factors and prognosis vary. Recent developments in genomic profiling have revealed several molecular markers for poor prognosis and activation of oncogenic pathways. Exploration of molecular characteristics of iCCA in each patient is a major challenge in a clinical setting, and there is no effective molecular-based targeted therapy. However, several recent studies suggested molecular-based subtypes with corresponding clinical and pathological features. Even though the subtypes have not yet been validated, it is possible that molecular features can be predicted based on clinicopathological characteristics and that this could be used for a more rational approach to integrative clinical and molecular subclassification and targeted therapy. In this review, we explored the genomic landscape of iCCA and attempted to find relevance between clinicopathologic and molecular features in molecular subtypes in several published studies. The results reveal future directions that may lead to a rational approach to the targeted therapy.
Core Tip: Intrahepatic cholangiocarcinoma (iCCA) is a histopathologically and molecularly heterogeneous tumor. Recent developments in genomic profiling have revealed several molecular markers for poor prognosis and activation of oncogenic pathways. Exploration of molecular characteristics of iCCA in each patient is a major challenge in a clinical setting, and there exists no effective molecular-based targeted therapy. Therefore, the analysis of relevance between molecular and clin-icopathological features is very important. The present analysis showed that the molecular subtypes of iCCA have distinct clinicopathologic features and prognostic differences. For developing effective targeted and personalized therapies based on clinical and molecular understanding, future additional large scale studies are needed.
- Citation: Ahn KS, Kang KJ. Molecular heterogeneity in intrahepatic cholangiocarcinoma. World J Hepatol 2020; 12(12): 1148-1157
- URL: https://www.wjgnet.com/1948-5182/full/v12/i12/1148.htm
- DOI: https://dx.doi.org/10.4254/wjh.v12.i12.1148
Cholangiocarcinoma (CCA) is a malignant tumor that arises from biliary epithelium in any portion of the bile duct. Intrahepatic cholangiocarcinoma (iCCA) arises from small peripheral bile duct to second-order segmental bile duct. Risk factors, clinical symptoms, type of surgical resection, and prognosis of iCCA are different from those of extrahepatic CCA (eCCA; Klatskin tumor and distal bile duct cancer)[1,2]. ICCA is the second most common primary malignant liver tumor and accounts for 10%-15% of hepatobiliary neoplasms[3]; however, recently, there has been an increase in the incidence and associated significance of pathogenic, clinical, and therapeutic challenges[4]. Based on morphological and gross appearance, iCCA can be further classified into three subtypes: Mass forming (MF), periductal infiltrating (PI), and intraductal growing (IG). The prognosis of the subtypes differ according to gross morphology[5]. Due to pathologic heterogeneity and lack of specific symptoms, iCCA is hard to diagnose at the early stages, and most of the patients are at an advanced stage at the time of diagnosis. Therefore, prognosis after curative surgical resection is dismal, and efficacy of chemotherapy or targeted therapy is limited[3].
Recent molecular analyses revealed several markers for poor prognosis and activation of oncogenic pathways (KRAS mutation, human epidermal growth factor receptor 2 (HER2), and epidermal growth factor receptor (EGFR) signaling)[6,7]. In addition, various recurrent mutations and fusions have been reported, including IDH1 and IDH2, BRAF, TP53, and FGFR2 genes[7,8]. These molecular findings demonstrate a more integrative analysis of clinical and molecular alterations in iCCA. However, there exist a few other challenges. First of all, the molecular characteristics of tumor heterogeneity are not yet clear. Second, integrative relevance between clinical and molecular characteristics is not enough; and finally, most prevalent oncogenic alterations in CCA are still undruggable. Understanding the molecular characteristics in these heterogeneous tumors may derive specific biologically meaningful subtypes that can be used to define more rational potential targeted therapy.
This review provides an overview of the genetic characteristics and heterogeneity of iCCA with a focus on molecular subtypes and their relevance with the clinicopathological phenotype. Furthermore, the role of molecular markers to stratify patients based on their prognosis and response to therapies is discussed.
Histological classification of iCCA is important for understanding the molecular heterogeneity of iCCA. Several investigations have revealed that a whole range of phenotypical traits of hepatocytes, cholangiocytes, and progenitor cells was seen in primary liver cancer [hepatocellular carcinoma (HCC) and iCCA]. It has been suggested that iCCA originated from biliary tree stem cells located within the peribiliary gland as well as hepatic progenitor cells within canals of Hering[2]. Hepatic bipotent progenitor cells along the small intrahepatic bile duct possibly differentiate not only into hepatocytes but also into cholangiocytes, which can lead to iCCA[9,10]. Consequently, two different histological types of iCCA may develop: One originating from hepatic stem cell-derived lineages with stem-like molecular characteristics similar to those in HCC or combined HCC-CCA and the other originating from biliary tree progenitor stem cell-derived cholangiocytes found along the large intrahepatic bile duct with characteristics similar to perihilar or extrahepatic CCA[2,11-13]. Histologically, iCCA is defined as an adenocarcinoma formed by columnar and cuboidal epithelial cells[1,14].
Based on the histological findings, conventional iCCA can be classified into two main subtypes. Small bile duct type iCCA may derive from small intrahepatic bile ducts; hepatic progenitor cells present as small-sized tubular or acinar adenocarcinoma with scant mucin production[11,14-16]. Small bile duct type is either represented as peripheral type or cholangiolar type[17,18]. Meanwhile, large duct type arises from biliary tree progenitor stem cell and is constituted by mucin-producing columnar tumor cells in large segmental bile ducts or papillary architecture[11,14-17,19]. Large duct type has been represented as a perihilar type or bile duct type in other studies[17,18]. The gross and histological features of large bile duct type iCCA are similar to those of perihilar CCA and distal CCA. In addition, the majority of PI and IG has large bile duct type[11,17,18]. However, the MF type, which is the most prominent morphologic type, is more heterogeneous as it comprises of both small duct type and large duct type[17,18].
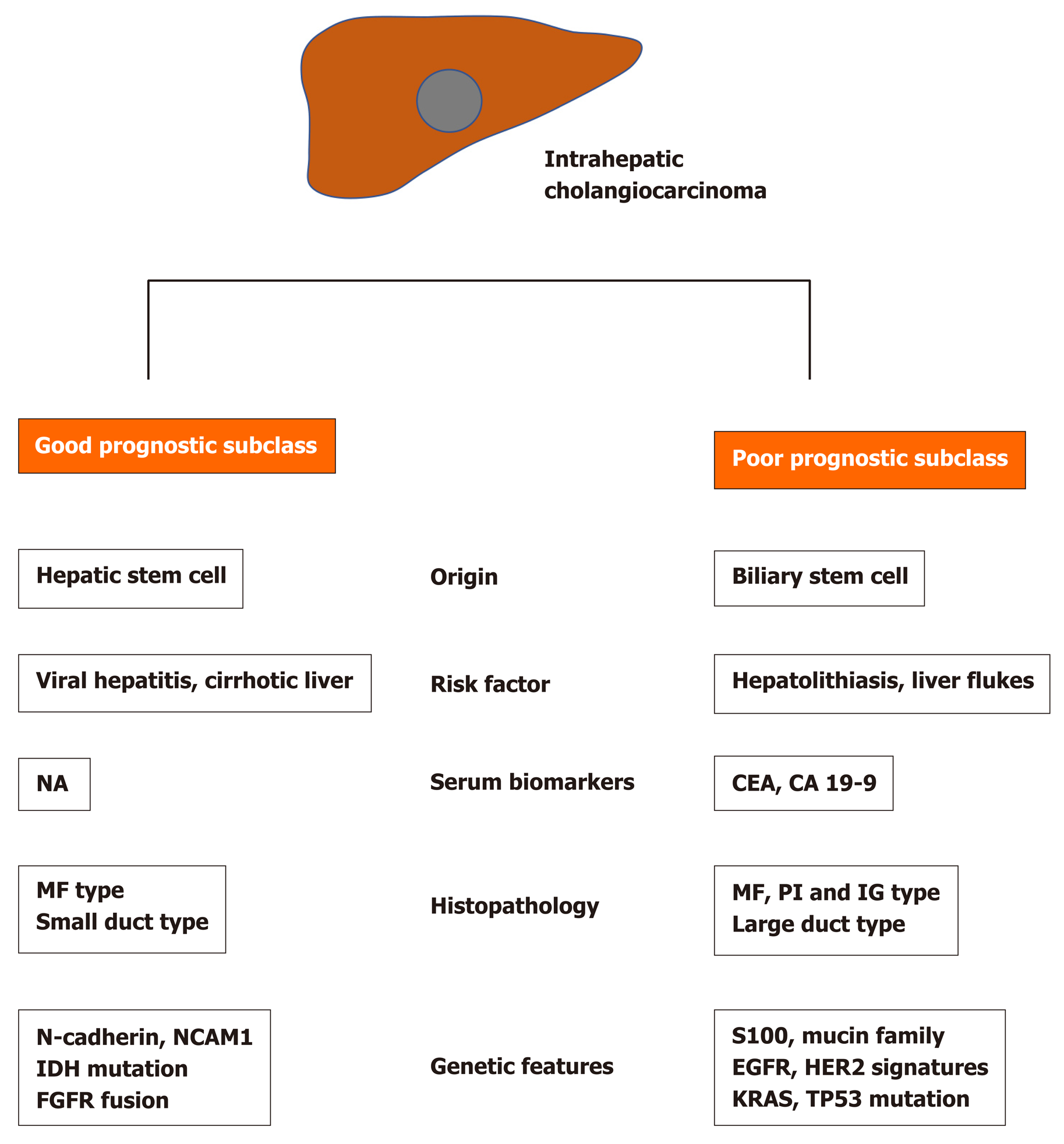
Although the two histological subtypes belong to iCCA, their clinical and molecular features are quite different. While viral hepatitis and cirrhosis are the risk factors of small duct type, cholangitis and parasite infection are the main cause of large duct type[17,18]. Both subtypes have different precursor lesions and show different survival outcomes[18]. Furthermore, they show different immunophenotypes like the abundant expression of mucin families, S100P, and anterior gradient homolog 2 in the large duct type, and N-cadherin and neural cell adhesion molecule 1 in the small duct type[11,17,18,20]. These histopathological heterogeneities based on cell origin are critical for understanding the heterogeneity of iCCA as well as heterogeneous molecular characteristics of iCCA.
Recent technological advancements have helped in understanding the mutational landscape of iCCA. Mutations in common driver oncogenes and suppressor genes are summarized in Table 1. Due to a small number of samples compared to other cancer and pathological heterogeneity, the prevalence of the mutation is variable across studies. However, several key driver somatic mutations commonly seen in other tumors, such as KRAS, BRAF, TP53, BAP1, and ARID1A, are also frequently identified in iCCA. Other driver genes like BRAF, PIK3CA, GNAS, EGFR, and ERBBR/HER2 have also been identified in iCCA, but at a much lower frequency in most of the cohorts[6,21-23]. The presence of EGFR, TP53, and KRAS mutation is known as poor prognostic factor[6,21,24]. Mutation of TERT promoter and ALB gene, which are frequently seen in HCC, are also detected in CCA, but only in iCCA or combined HCC and CCA samples with less frequency[23]. Meanwhile, isocitrate dehydrogenase (IDH)1 and IDH2 mutations have been reported in 10%-20% of iCCA cases[23]. Interestingly, a large extent of IDH mutation has been observed in iCCA and not in eCCA and rarely identified in HCC[23,25]. IDH mutation is associated with a better prognosis[26]. In one large scale study, IDH1/2 mutations were identified to be associated with improved overall survival[27]; however, as the incidence of IDH mutation is not frequent, survival impacts of IDH mutation is not yet clear[28]. In iCCA, frequency of fibroblast growth factor receptor 2 (FGFR2) fusion is reported as 10%-15%[29,30]. FGFR2 pairs with some genes such as TACC1, BICCI, PRKACA, AHCYL, and PRKACB. These fusions result in the constitutive activation of FGFR2 and its oncogenic functions[31]. The FGFR related pathway is involved in cellular migration and proliferation. Patients with FGFR2 fusion show good prognosis, which suggests that FGFR2 fusions can be a prognostic marker as well as potential target for therapy[21,32]. Altered genes involving chromatin remodeling, such as BAP1, ARID1A, and PBRM1, are also frequently found in iCCA[27]. Meanwhile, germline DNA mismatch repair deficiency (Lynch syndrome) has been reported to be associated with CCA[33]. There exists a report that deleterious germline mutations in breast cancer gene 1/2, RAD51D, MutL homolog 1, and MutS homolog 2 were detected in 11% of CCA patients[21].
| Pathway | Gene | Frequency of alteration |
| NADPH metabolism | IDH1/2 | 4-36 |
| Chromatic remodeling | BAP1 | 9%-25% |
| ARID1A | 11-36 | |
| PBRM1 | 11-17 | |
| Cell cycle regulation and DNA damage response | CDKN2A | 7 |
| CDK6 | 7 | |
| TP53 | 3-38 | |
| BRCA1/2 | 4 | |
| PI3K signaling | PIK3CA | 4-6 |
| PTEN | 1-11 | |
| Ras/Raf/MEK/ERK | EGFR | 2.2 |
| KRAS | 9-24 | |
| NRAS | 3.6 | |
| BRAF | 3-22 | |
| FGF | FGFR2 | 4-38 |
Genomic alteration in CCA is highly heterogeneous, like pathologic features. Several studies identified different gene alterations between iCCA and eCCA[21,34]. While alterations in IDH1/2, BRAF, FGFR2, BAP1, and NRAS are frequently found in iCCA, TP53, KRAS, SMAD4, and BRAF mutations are common in eCCA[35]. Interestingly, some of the altered genes commonly found in eCCA such as KRAS, SMAD4, and TP53 were also shared by large duct types of iCCA. Whereas, small duct iCCA has frequent IDH1/2 mutations and FGFR2 gene fusion[31,36]. Therefore, pathological characteristics and genetic alterations appear to be closely related to each other.
Epigenetic mechanisms of iCCA include histone modification, DNA methylation, and noncoding RNAs. In CCA, hypermethylation at the promotors of tumor suppressor genes has been reported[37]. ICCA is a highly epigenetic regulated tumor type.
DNA methylation is an early molecular lesion of carcinogenesis; tumor suppressor promoter hypermethylation of tumor suppressor gene leads to transcriptional modification and inactivation, and hypomethylation of oncogenes results in activation[38]. Most of the genes that were altered by CpG methylation belonged to wingless-related integration site (WNT), transforming growth factor beta, PI3K, MAPK, and NOTCH signaling pathways in iCCA[39]. Like other cancers, promoter hypermethylation of tumor suppressor genes, such as DAPK, SOX17, and RUNX3, has been commonly reported[40]. It is known that IDH mutations result in hypermethylation and induce silencing of ARID1A[8].
MicroRNA (miRNA) plays a crucial role in diverse cellular processes and regulates gene function. Several pieces of research revealed that overexpression of miR-21 inhibits TIMP3 and PDCD4 and sequentially leads to cancer progression[41]. Besides, miR-191, miR-200, miR-141, miR-204, miR-214, and miR-221 are involved in CCA development[42]. Among these miRNAs, miR-21, miuR-191, and miR-26a were identified as poor prognostic markers[43]. Meanwhile, the high expression of several lncRNA (H19, NEAT1, PVT1, CKDN2B-AS1, and HUILC) has been reported to be associated with poor survival of CCA[44,45].
However, most of the epigenetic mechanisms of iCCA have not been studied sufficiently, and their role as biomarkers and potential targeted therapies should be extensively investigated.
Several studies based on microarray or NGS revealed the expression profile and oncogenic pathway of iCCA. The major key oncogenic molecules, including tumor necrosis factor, transforming growth factor, extracellular regulated-signal kinase, epidermal growth factor, RAS, AKT, p53, NOTCH, and platelet-derived growth factor, are deregulated in iCCA. Immune response-related pathways and inflammation associated with signatures are also enriched[6,8,22,24]. Aberrant HER2 expression is seen in about 30% of iCCA, and it is related to poor prognosis with coactivation of ERBB3 and EGFR2 as well as mesenchymal epithelial transition factor and mammalian target of rapamycin[6]. Inflammation associated signatures are commonly activated in iCCA, but their oncologic and prognostic role is controversial[22,24]. Activation of the WNT pathway is often seen in iCCA, and it relates to inflammatory reaction because macrophages in the stroma surrounding the tumor are required for the maintenance of activated WNT pathway[46].
The oncogenic signatures are also found in many other cancers. However, deregulated pathways are different according to pathologic and molecular subtypes; therefore, subtype-specific activated pathways are important to assess biology.
Based on the genomic profile, a few studies have suggested molecular subtype of iCCA beyond anatomical and histological subclassification (Table 2). The recent advances in the molecular classification allow better characterization of heterogeneity of iCCA. Furthermore, they provide insight into the integrated approach of clinical and molecular characterization of iCCA.
| Good prognostic subclass | Poor prognostic subclass | |
| GSE26566 | Periductal infiltrating type, perineural invasion; KRAS mutation, EGFR and HER2 signatures | |
| GSE32225 | Well differentiated tumor; inflammation-related signatures | Poor differentiated tumor; RTK-related pathways (AKT, MET, RAS/RAF/MAPK); overexpression of EGFR; KRAS mutation |
| GSE32879 | EMT-related signatures; TGFβ1, NCAM1, CD133 | |
| GSE89749 | Fluke-negative; FGFR fusion; BAP1, IDH mutation | Fluke-positive; BRCA1/2, TP53 mutation; ERBB2 gain |
| GSE107943 | Small duct type (cholangiolar type); underlying hepatitis, cirrhosis; metabolism-related signatures; FGFR2 fusion | Large duct type (bile duct type); Elevated CEA, CA 19-9; underlying cholangitis; P53, inflammation-related signatures; KRAS mutation |
| TCGA1 | Mitochondria/metabolic-related signatures; IDH, BAP1 mutation | Inflammation-related pathways |
Although each subclassification has some heterogeneity, the molecular feature of iCCA is dichotomized two subtypes that have different survival and clinical outcomes[6,8,22,24,47]. Generally, the poor prognostic molecular subtype is associated with the KRAS mutation. Also, BRAF, ERBB2, and HER alterations are often seen in a poor prognostic subtype. On the other hand, IDH mutation and FGFR fusion are commonly seen in the good prognostic subtype. The molecular subclasses were reported to be rather related to clinical and pathologic features. While PI type, similar to eCCA, is commonly seen in the poor prognostic subtype, the MF type is almost evenly distributed in both good and poor prognostic subtypes[24]. Large duct type, history of cholangitis and parasite infection, and elevated levels of serum biomarkers (carcinoembryonic antigen and carbohydrate antigen 19-9) are associated with poor prognostic molecular subtype, while small duct type and history of viral hepatitis are associated with good prognostic molecular subtype[24] (Figure 1).
Several molecular subclassifications in the reported studies provide information about molecular heterogeneity in addition to histopathological heterogeneity. The integrated clinical and molecular subclassifications would be helpful to provide a more rational approach to overcome clinical and molecular heterogeneity. Molecular profile of iCCA is helpful for early diagnosis and prognosis prediction and may potentially provide personized treatment. However, exploration of molecular characteristics of iCCA in each patient is a major challenge in a clinical setting because of the high cost for evaluating molecular characterization. If molecular subtypes of iCCA have specific clinical and pathologic features, molecular subtypes can be predicted from clinical features. Although the subclassifications reported in several studies have a few differences based on demographic characteristics and study methods, there is still no consensus on the molecular subclass. The present review shows that clinical and molecular relevance based on molecular subclassification has been exploring and may establish integrative clinical and molecular subclassification soon. Since the number of patients is not sufficient in iCCA compared to other cancers, further large scale studies are necessary for validation and establishment of molecular classification.
Still, molecular-based target therapy is not considered to be effective in CCA due to molecular heterogeneity. However, the establishment of molecular subtypes can promote the development of effective subtype-specific therapeutic molecular targeted therapy. Lapatinib, a dual inhibitor of EGFR and HER2, has been reported to be effective in cell lines that had genetic characteristics similar to poor prognostic subtype[6], while gemcitabine was identified to be effective in cell lines with similar expression profile to good prognostic subtype, which had enriched gemcitabine sensitive genes[24]. Although these are the outcomes of cell line studies and not validated clinical data, it is hypothesized that additional applications of drug study on different subtype signaling pathways may be helpful to stratify patients for targeted approaches for the treatment of iCCA.
In the present study, we reviewed the molecular heterogeneity of iCCA in association with the clinicopathological features. Several recent studies have revealed molecular characteristics of iCCA and suggested several molecular subclassifications. Molecular study of iCCA may help identify patients at risk of developing iCCA, predicting prognosis, and targeting approach to treatment. However, molecular exploration in all patients is not feasible because of the high cost. Accordingly, analysis of relevance between molecular and clinicopathological features is considered as imperative because if clinicomolecular relevance is established, molecular characteristics can be predicted based on clinical features in each patient.
The present analysis showed that the molecular subtypes of iCCA have distinct clinicopathologic features and prognostic differences. However, integrative clinical and molecular subclassification is not yet validated. For developing effective targeted and personalized therapies based on clinical and molecular knowledge, future additional large scale studies are necessary.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and Hepatology
Country/Territory of origin: South Korea
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Huang CY, Leardkamolkarn V S-Editor: Zhang H L-Editor: Filipodia P-Editor: Zhang YL
| 1. | Krasinskas AM. Cholangiocarcinoma. Surg Pathol Clin. 2018;11:403-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 92] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 2. | Cardinale V, Semeraro R, Torrice A, Gatto M, Napoli C, Bragazzi MC, Gentile R, Alvaro D. Intra-hepatic and extra-hepatic cholangiocarcinoma: New insight into epidemiology and risk factors. World J Gastrointest Oncol. 2010;2:407-416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 138] [Cited by in RCA: 146] [Article Influence: 9.7] [Reference Citation Analysis (1)] |
| 3. | Blechacz BR, Gores GJ. Cholangiocarcinoma. Clin Liver Dis. 2008;12:131-150, ix. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 173] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 4. | Patel T. Worldwide trends in mortality from biliary tract malignancies. BMC Cancer. 2002;2:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 335] [Cited by in RCA: 359] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 5. | Bagante F, Spolverato G, Weiss M, Alexandrescu S, Marques HP, Aldrighetti L, Maithel SK, Pulitano C, Bauer TW, Shen F, Poultsides GA, Soubrane O, Martel G, Groot Koerkamp B, Guglielmi A, Itaru E, Pawlik TM. Impact of Morphological Status on Long-Term Outcome Among Patients Undergoing Liver Surgery for Intrahepatic Cholangiocarcinoma. Ann Surg Oncol. 2017;24:2491-2501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 6. | Andersen JB, Spee B, Blechacz BR, Avital I, Komuta M, Barbour A, Conner EA, Gillen MC, Roskams T, Roberts LR, Factor VM, Thorgeirsson SS. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology 2012; 142: 1021-1031. e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 422] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 7. | Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, Choo SP, Myint SS, Thanan R, Nagarajan S, Lim WK, Ng CCY, Boot A, Liu M, Ong CK, Rajasegaran V, Lie S, Lim AST, Lim TH, Tan J, Loh JL, McPherson JR, Khuntikeo N, Bhudhisawasdi V, Yongvanit P, Wongkham S, Totoki Y, Nakamura H, Arai Y, Yamasaki S, Chow PK, Chung AYF, Ooi LLPJ, Lim KH, Dima S, Duda DG, Popescu I, Broet P, Hsieh SY, Yu MC, Scarpa A, Lai J, Luo DX, Carvalho AL, Vettore AL, Rhee H, Park YN, Alexandrov LB, Gordân R, Rozen SG, Shibata T, Pairojkul C, Teh BT, Tan P. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov. 2017;7:1116-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 675] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 8. | Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, Hinoue T, Hoadley KA, Gibb EA, Roszik J, Covington KR, Wu CC, Shinbrot E, Stransky N, Hegde A, Yang JD, Reznik E, Sadeghi S, Pedamallu CS, Ojesina AI, Hess JM, Auman JT, Rhie SK, Bowlby R, Borad MJ; Cancer Genome Atlas Network; Zhu AX; Stuart JM; Sander C; Akbani R; Cherniack AD; Deshpande V; Mounajjed T; Foo WC; Torbenson MS; Kleiner DE; Laird PW; Wheeler DA; McRee AJ; Bathe OF; Andersen JB; Bardeesy N; Roberts LR; Kwong LN. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep. 2017;18:2780-2794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 379] [Article Influence: 47.4] [Reference Citation Analysis (1)] |
| 9. | Moeini A, Sia D, Bardeesy N, Mazzaferro V, Llovet JM. Molecular Pathogenesis and Targeted Therapies for Intrahepatic Cholangiocarcinoma. Clin Cancer Res. 2016;22:291-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 186] [Cited by in RCA: 175] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 10. | Kordes C, Häussinger D. Hepatic stem cell niches. J Clin Invest. 2013;123:1874-1880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 11. | Komuta M, Govaere O, Vandecaveye V, Akiba J, Van Steenbergen W, Verslype C, Laleman W, Pirenne J, Aerts R, Yano H, Nevens F, Topal B, Roskams T. Histological diversity in cholangiocellular carcinoma reflects the different cholangiocyte phenotypes. Hepatology. 2012;55:1876-1888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 250] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 12. | Cardinale V, Bragazzi MC, Carpino G, Torrice A, Fraveto A, Gentile R, Pasqualino V, Melandro F, Aliberti C, Bastianelli C, Brunelli R, Berloco PB, Gaudio E, Alvaro D. Cholangiocarcinoma: increasing burden of classifications. Hepatobiliary Surg Nutr. 2013;2:272-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 38] [Reference Citation Analysis (0)] |
| 13. | Wang Y, Lanzoni G, Carpino G, Cui CB, Dominguez-Bendala J, Wauthier E, Cardinale V, Oikawa T, Pileggi A, Gerber D, Furth ME, Alvaro D, Gaudio E, Inverardi L, Reid LM. Biliary tree stem cells, precursors to pancreatic committed progenitors: evidence for possible life-long pancreatic organogenesis. Stem Cells. 2013;31:1966-1979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 14. | Nakanuma Y, Kakuda Y. Pathologic classification of cholangiocarcinoma: New concepts. Best Pract Res Clin Gastroenterol. 2015;29:277-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 15. | Aishima S, Kuroda Y, Nishihara Y, Iguchi T, Taguchi K, Taketomi A, Maehara Y, Tsuneyoshi M. Proposal of progression model for intrahepatic cholangiocarcinoma: clinicopathologic differences between hilar type and peripheral type. Am J Surg Pathol. 2007;31:1059-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 107] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 16. | Hayashi A, Misumi K, Shibahara J, Arita J, Sakamoto Y, Hasegawa K, Kokudo N, Fukayama M. Distinct Clinicopathologic and Genetic Features of 2 Histologic Subtypes of Intrahepatic Cholangiocarcinoma. Am J Surg Pathol. 2016;40:1021-1030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 108] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 17. | Akita M, Fujikura K, Ajiki T, Fukumoto T, Otani K, Azuma T, Itoh T, Ku Y, Zen Y. Dichotomy in intrahepatic cholangiocarcinomas based on histologic similarities to hilar cholangiocarcinomas. Mod Pathol. 2017;30:986-997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 18. | Liau JY, Tsai JH, Yuan RH, Chang CN, Lee HJ, Jeng YM. Morphological subclassification of intrahepatic cholangiocarcinoma: etiological, clinicopathological, and molecular features. Mod Pathol. 2014;27:1163-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 199] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 19. | Cardinale V, Wang Y, Carpino G, Reid LM, Gaudio E, Alvaro D. Mucin-producing cholangiocarcinoma might derive from biliary tree stem/progenitor cells located in peribiliary glands. Hepatology. 2012;55:2041-2042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 20. | Nakanuma Y, Uchida T, Sato Y, Uesaka K. An S100P-positive biliary epithelial field is a preinvasive intraepithelial neoplasm in nodular-sclerosing cholangiocarcinoma. Hum Pathol. 2017;60:46-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 21. | Churi CR, Shroff R, Wang Y, Rashid A, Kang HC, Weatherly J, Zuo M, Zinner R, Hong D, Meric-Bernstam F, Janku F, Crane CH, Mishra L, Vauthey JN, Wolff RA, Mills G, Javle M. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One. 2014;9:e115383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 272] [Cited by in RCA: 360] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 22. | Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B, Peix J, Sole M, Tovar V, Alsinet C, Cornella H, Klotzle B, Fan JB, Cotsoglou C, Thung SN, Fuster J, Waxman S, Garcia-Valdecasas JC, Bruix J, Schwartz ME, Beroukhim R, Mazzaferro V, Llovet JM. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144:829-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 432] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 23. | Cancer Genome Atlas Research Network. Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017; 169: 1327-1341. e23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1578] [Cited by in RCA: 1725] [Article Influence: 215.6] [Reference Citation Analysis (1)] |
| 24. | Ahn KS, O'Brien D, Kang YN, Mounajjed T, Kim YH, Kim TS, Kocher JA, Allotey LK, Borad MJ, Roberts LR, Kang KJ. Prognostic subclass of intrahepatic cholangiocarcinoma by integrative molecular-clinical analysis and potential targeted approach. Hepatol Int. 2019;13:490-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 25. | Fujimoto A, Furuta M, Shiraishi Y, Gotoh K, Kawakami Y, Arihiro K, Nakamura T, Ueno M, Ariizumi S, Nguyen HH, Shigemizu D, Abe T, Boroevich KA, Nakano K, Sasaki A, Kitada R, Maejima K, Yamamoto Y, Tanaka H, Shibuya T, Shibata T, Ojima H, Shimada K, Hayami S, Shigekawa Y, Aikata H, Ohdan H, Marubashi S, Yamada T, Kubo M, Hirano S, Ishikawa O, Yamamoto M, Yamaue H, Chayama K, Miyano S, Tsunoda T, Nakagawa H. Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nat Commun. 2015;6:6120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 172] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 26. | Wang P, Dong Q, Zhang C, Kuan PF, Liu Y, Jeck WR, Andersen JB, Jiang W, Savich GL, Tan TX, Auman JT, Hoskins JM, Misher AD, Moser CD, Yourstone SM, Kim JW, Cibulskis K, Getz G, Hunt HV, Thorgeirsson SS, Roberts LR, Ye D, Guan KL, Xiong Y, Qin LX, Chiang DY. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32:3091-3100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 315] [Cited by in RCA: 318] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 27. | Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P, Offerhaus GJA, Roa JC, Roberts LR, Gores GJ, Popescu I, Alexandrescu ST, Dima S, Fassan M, Simbolo M, Mafficini A, Capelli P, Lawlor RT, Ruzzenente A, Guglielmi A, Tortora G, de Braud F, Scarpa A, Jarnagin W, Klimstra D, Karchin R, Velculescu VE, Hruban RH, Vogelstein B, Kinzler KW, Papadopoulos N, Wood LD. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470-1473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 545] [Cited by in RCA: 537] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 28. | Goyal L, Govindan A, Sheth RA, Nardi V, Blaszkowsky LS, Faris JE, Clark JW, Ryan DP, Kwak EL, Allen JN, Murphy JE, Saha SK, Hong TS, Wo JY, Ferrone CR, Tanabe KK, Chong DQ, Deshpande V, Borger DR, Iafrate AJ, Bardeesy N, Zheng H, Zhu AX. Prognosis and Clinicopathologic Features of Patients With Advanced Stage Isocitrate Dehydrogenase (IDH) Mutant and IDH Wild-Type Intrahepatic Cholangiocarcinoma. Oncologist. 2015;20:1019-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 29. | Graham RP, Barr Fritcher EG, Pestova E, Schulz J, Sitailo LA, Vasmatzis G, Murphy SJ, McWilliams RR, Hart SN, Halling KC, Roberts LR, Gores GJ, Couch FJ, Zhang L, Borad MJ, Kipp BR. Fibroblast growth factor receptor 2 translocations in intrahepatic cholangiocarcinoma. Hum Pathol. 2014;45:1630-1638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 238] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 30. | Mahipal A, Tella SH, Kommalapati A, Anaya D, Kim R. FGFR2 genomic aberrations: Achilles heel in the management of advanced cholangiocarcinoma. Cancer Treat Rev. 2019;78:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 31. | Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, Hiraoka N, Ojima H, Shimada K, Okusaka T, Kosuge T, Miyagawa S, Shibata T. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47:1003-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 651] [Cited by in RCA: 938] [Article Influence: 93.8] [Reference Citation Analysis (0)] |
| 32. | Borad MJ, Gores GJ, Roberts LR. Fibroblast growth factor receptor 2 fusions as a target for treating cholangiocarcinoma. Curr Opin Gastroenterol. 2015;31:264-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 33. | Mecklin JP, Järvinen HJ, Virolainen M. The association between cholangiocarcinoma and hereditary nonpolyposis colorectal carcinoma. Cancer. 1992;69:1112-1114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 37] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 34. | Kayhanian H, Smyth EC, Braconi C. Emerging molecular targets and therapy for cholangiocarcinoma. World J Gastrointest Oncol. 2017;9:268-280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Putra J, de Abreu FB, Peterson JD, Pipas JM, Mody K, Amos CI, Tsongalis GJ, Suriawinata AA. Molecular profiling of intrahepatic and extrahepatic cholangiocarcinoma using next generation sequencing. Exp Mol Pathol. 2015;99:240-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 36. | Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, Hezel AF, Ancukiewicz M, Liebman HM, Kwak EL, Clark JW, Ryan DP, Deshpande V, Dias-Santagata D, Ellisen LW, Zhu AX, Iafrate AJ. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 596] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 37. | Louis C, Papoutsoglou P, Coulouarn C. Molecular classification of cholangiocarcinoma. Curr Opin Gastroenterol. 2020;36: 57-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 38. | Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1431] [Cited by in RCA: 1264] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 39. | Goeppert B, Konermann C, Schmidt CR, Bogatyrova O, Geiselhart L, Ernst C, Gu L, Becker N, Zucknick M, Mehrabi A, Hafezi M, Klauschen F, Stenzinger A, Warth A, Breuhahn K, Renner M, Weichert W, Schirmacher P, Plass C, Weichenhan D. Global alterations of DNA methylation in cholangiocarcinoma target the Wnt signaling pathway. Hepatology. 2014;59:544-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 40. | Xiaofang L, Kun T, Shaoping Y, Zaiqiu W, Hailong S. Correlation between promoter methylation of p14(ARF), TMS1/ASC, and DAPK, and p53 mutation with prognosis in cholangiocarcinoma. World J Surg Oncol. 2012;10:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 41. | Selaru FM, Olaru AV, Kan T, David S, Cheng Y, Mori Y, Yang J, Paun B, Jin Z, Agarwal R, Hamilton JP, Abraham J, Georgiades C, Alvarez H, Vivekanandan P, Yu W, Maitra A, Torbenson M, Thuluvath PJ, Gores GJ, LaRusso NF, Hruban R, Meltzer SJ. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology. 2009;49:1595-1601. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 227] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 42. | O'Rourke CJ, Munoz-Garrido P, Aguayo EL, Andersen JB. Epigenome dysregulation in cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1423-1434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 43. | Salati M, Braconi C. Noncoding RNA in Cholangiocarcinoma. Semin Liver Dis. 2019;39:13-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 44. | Li J, Huang L, Li Z, Zhong X, Tai S, Jiang X, Cui Y. Functions and roles of long noncoding RNA in cholangiocarcinoma. J Cell Physiol. 2019;234:17113-17126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 45. | Angenard G, Merdrignac A, Louis C, Edeline J, Coulouarn C. Expression of long non-coding RNA ANRIL predicts a poor prognosis in intrahepatic cholangiocarcinoma. Dig Liver Dis. 2019;51:1337-1343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 46. | Loilome W, Bungkanjana P, Techasen A, Namwat N, Yongvanit P, Puapairoj A, Khuntikeo N, Riggins GJ. Activated macrophages promote Wnt/β-catenin signaling in cholangiocarcinoma cells. Tumour Biol. 2014;35:5357-5367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 82] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 47. | Oishi N, Kumar MR, Roessler S, Ji J, Forgues M, Budhu A, Zhao X, Andersen JB, Ye QH, Jia HL, Qin LX, Yamashita T, Woo HG, Kim YJ, Kaneko S, Tang ZY, Thorgeirsson SS, Wang XW. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology. 2012;56:1792-1803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 194] [Article Influence: 14.9] [Reference Citation Analysis (0)] |