Published online Oct 27, 2020. doi: 10.4254/wjh.v12.i10.792
Peer-review started: April 30, 2020
First decision: May 24, 2020
Revised: May 28, 2020
Accepted: September 2, 2020
Article in press: September 2, 2020
Published online: October 27, 2020
Processing time: 177 Days and 0 Hours
Nonalcoholic fatty liver disease (NAFLD) is becoming the most common chronic liver disease worldwide, with significant morbidity associated with nonalcoholic steatohepatitis (NASH). Genome-wide association studies demonstrated that the variants rs738409 C/G in the PNPLA3 and rs58542926 C/T in the TM6SF2 genes are determinants of inter-individual and ethnicity-related differences in hepatic fat content and NAFLD progression.
To investigate PNPLA3 and TM6SF2 genotype frequency and their association with NAFLD development and progression in Brazilian patients.
This cross-sectional case-control study enrolled 285 individuals from the Gastroenterology and Hepatology clinics at a university hospital in Brazil. The case patients (n = 148) were confirmed to have NAFLD by the identification of hepatic steatosis on ultrasonography and exclusion of other causes of liver disease. According to the clinical protocol, patients underwent liver biopsy when at high risk for NASH and/or advanced fibrosis (n = 65). Steatohepatitis was confirmed in 54 patients. Individuals who did not have biopsy indication or NASH on histology were considered to have simple steatosis (n = 94). The control group (n = 137) was selected among patients that attended the Intestinal Disease clinic and was composed of subjects without abnormalities on abdominal ultrasonography and normal liver biochemical tests. All individuals underwent PNPLA3 and TM6SF2 genotype analysis.
PNPLA3 CC, CG and GG genotype frequencies were 37%, 44% and 19%, respectively, in NAFLD patients and were 58%, 31% and 10% in controls (P < 0.001). In a model adjusted for gender, age, body mass index and type 2 diabetes mellitus, the G allele increased the chance of NAFLD (OR = 1.69, 95%CI: 1.21-2.36, P = 0.002) and NASH (OR = 3.50, 95%CI: 1.84-6.64, P < 0.001). The chance of NASH was even higher with GG homozygosis (OR = 5.53, 95%CI: 2.04-14.92, P = 0.001). No association was found between G allele and the features of metabolic syndrome. In histological assessment, PNPLA3 genotype was not associated with steatosis grade, although GG homozygosis increased the chance of significant NASH activity (OR = 17.11, 95%CI: 1.87-156.25, P = 0.01) and fibrosis (OR = 7.42, 95%CI: 1.55-34.47, P = 0.01) in the same adjusted model. TM6SF2 CC, CT and TT genotype frequencies were 83%, 15% and 0.7%, respectively, in NAFLD patients and were 84%, 16% and 0.7% in controls (P = 0.78). The T allele presence was not associated with NAFLD or NASH, and was not associated with histological features.
PNPLA3 may be involved in susceptibility and progression of NAFLD and NASH in the Brazilian population. More advanced histological liver disease was associated with the G allele. The TM6SF2 genetic variants were not associated with NAFLD susceptibility and progressive histological forms in the population studied, but further studies are required to confirm these findings.
Core Tip: The rs738409 C/G mutation in the PNPLA3 gene may be involved in the pathogenesis of nonalcoholic fatty liver disease (NAFLD) in the Brazilian population. We found an association between this polymorphism and higher susceptibility to NAFLD occurrence, disease progression to nonalcoholic steatohepatitis, more severe histological activity scores and the presence of liver fibrosis. The TM6SF2 gene variants were also evaluated in the Brazilian population, although they were not associated with NAFLD susceptibility and different histological forms.
- Citation: Lisboa QC, Nardelli MJ, Pereira PA, Miranda DM, Ribeiro SN, Costa RSN, Versiani CA, Vidigal PVT, Ferrari TCA, Couto CA. PNPLA3 and TM6SF2 polymorphisms in Brazilian patients with nonalcoholic fatty liver disease. World J Hepatol 2020; 12(10): 792-806
- URL: https://www.wjgnet.com/1948-5182/full/v12/i10/792.htm
- DOI: https://dx.doi.org/10.4254/wjh.v12.i10.792
Nonalcoholic fatty liver disease (NAFLD) is characterized by hepatocellular accumulation of triglycerides (TG) in the absence of excess alcohol consumption or any other cause of secondary liver steatosis[1,2]. Currently, it is the most common chronic liver disease in Western countries, with an estimated prevalence of 25% globally and 32% in South America[3,4]. NAFLD is strongly associated with the metabolic syndrome [obesity, insulin resistance/type 2 diabetes mellitus (T2DM) and dyslipidemia][4,5] and encompasses a spectrum of liver diseases ranging from simple steatosis (SS), which is usually benign, to nonalcoholic steatohepatitis (NASH), which may lead to liver cirrhosis and hepatocellular carcinoma[6,7]. In fact, liver fibrosis is an independent risk factor of NAFLD severity and liver-related mortality[8,9].
Although highly prevalent, only a minority of NAFLD patients develop significant fibrosis and associated morbidity[10,11]. Thus, NAFLD is considered a complex disorder in which the disease phenotype results from an interaction between environmental exposure and susceptible polygenic background that comprises multiple independent modifiers[12,13].
In recent years, genome-wide association (GWA) and candidate gene studies have greatly contributed to understanding the genetics of NAFLD and their influence on prognosis[14-16]. In 2008, Romeo et al[17] identified that the mutation I148M encoded by the G allele at rs738409 in the gene patatin-like phospholipase domain-containing 3 (PNPLA3) is a major determinant of inter-individual and ethnicity-related differences in hepatic fat content. These findings have been confirmed in different populations through GWA[18-20] and candidate gene[21-26] studies, which demonstrated that PNPLA3 polymorphism was associated with greater susceptibility to NAFLD development and more severe histological and clinical forms.
In 2014, Kozlitina et al[27] identified that the E167K (rs58542926 C/T) variant in the TM6SF2 gene was also associated with increased liver fat content. This polymorphism was later associated with greater susceptibility to NAFLD and advanced histological NASH[28,29]. In this context, there is only one genetic study in Brazilian NAFLD patients[30], and TM6SF2 polymorphism has not yet been analyzed in this population.
Therefore, in this study, we investigated the association between PNPLA3 and TM6SF2 genotypes and clinical parameters of NAFLD, and analyzed the genotype variations as markers of liver histological features in Brazilian adult NAFLD patients. We also investigated the distribution of these genotype variations among Brazilians.
A cross-sectional study was performed at the Outpatient NAFLD Clinic, Medical School of Federal University of Minas Gerais, Belo Horizonte, Brazil. A total of 285 individuals (208 females and 77 males) were enrolled; of which, 148 patients had features of NAFLD (case patients) and 137 were non-NAFLD control subjects.
The case patients were confirmed to have hepatic steatosis by liver ultrasonography (US) according to established criteria[31]; additionally, other causes of liver disease were excluded, including elevated alcohol intake (men, > 30 g/d; women, > 20 g/d), autoimmune disorders (autoimmune hepatitis, primary biliary cholangitis and primary sclerosing cholangitis), hereditary hemochromatosis, alpha-1-antitrypsin deficiency, Wilson disease and hepatitis B or C virus infection. Patients who had decompensated cirrhosis or were taking drugs that induce steatosis were excluded. The NAFLD patients underwent liver biopsy according to the clinical protocol suggested by Chalasani et al[2]. Increased risk of NASH and/or advanced fibrosis included the presence of metabolic syndrome or significant fibrosis predicted by noninvasive methods. Subjects who had a liver biopsy showing steatosis, hepatocyte ballooning and lobular inflammation[1] were classified into the NASH group (n = 54) and individuals without an indication for liver biopsy and those who did not fulfill the NASH criteria on biopsy were classified into the SS group (n = 94).
Control subjects were selected from patients attending our institution at the Outpatient Intestinal Disease Clinic whose gender matched the NAFLD patients (97 females and 40 males). Individuals in the control group underwent a standard health examination, liver US and liver biochemistry. They were selected if there was no evidence of fatty liver on US and no liver biochemical abnormalities. Furthermore, the control subjects did not present any of the features of the metabolic syndrome as defined by the International Diabetes Federation criteria[32] and did not abuse alcohol.
Subjects from whom deoxyribonucleic acid (DNA) could be obtained for genotyping PNPLA3 at rs738409 and TM6SF2 at rs58542926 were included in the analyses. The case participants and the controls were selected from patients attending our institution from January 2017 to December 2018.
All the investigations performed in this study were conducted in accordance with the guidelines of the 1975 Declaration of Helsinki and its subsequent amendments. Written informed consent was obtained from each subject, and the study was approved by the Ethics Committee of the Federal University of Minas Gerais (CAAE 23610614.0.0000.5149).
Liver biopsy was performed in patients with NAFLD who were at increased risk of having NASH and/or advanced fibrosis (i.e., presence of metabolic syndrome or advanced fibrosis indicated by the NAFLD Fibrosis Score[33])[2]. Liver biopsy specimens were routinely fixed in 40 g/L formaldehyde, embedded in paraffin, and stained with hematoxylin and eosin, Masson trichrome and silver impregnation for reticular fibers analysis. The same liver pathologist, who was blinded to the patients’ information, analyzed all the exams. All the biopsies were at least 2 cm in length and contained a minimum of 8 portal tracts. The histological criterion for the diagnosis of NAFLD was macrovesicular fatty deposit in more than 5% of the hepatocytes. The NASH diagnostic criterion was the simultaneous presence of steatosis, ballooning and lobular inflammation in the liver biopsy, according to the Fatty Liver Inhibition of Progression algorithm[1]. Steatosis was graded based on the percentage of hepatocytes containing large and medium-sized intracytoplasmic lipid droplets, on a scale of 0 to 3 (S0: < 5%; S1: 5%-33%, S2: 34%-66%, S3: > 67%). Lobular inflammation was defined as a focus of 2 or more inflammatory cells within the lobule organized either as microgranulomas or located within the sinusoids. Foci were counted at × 20 magnification (grade 0: none; 1: ≤ 2 foci per lobule; 2: > 2 foci per lobule). Hepatocyte ballooning was graded from 0 to 2 (0: Normal hepatocytes with cuboidal shape, sharp angles and pink eosinophilic cytoplasm; 1: Presence of clusters of hepatocytes with a rounded shape and pale cytoplasm; 2: As for grade 1, associated with at least one enlarged ballooned hepatocyte). Severity of fibrosis was scored according to the Pathology Committee of the NASH Clinical Research Network method[34] and was expressed on a 4-point scale, as follows: 0, none; 1, perivenular and/or perisinusoidal fibrosis in zone 3; 2, combined pericellular and portal fibrosis; 3, septal/bridging fibrosis; 4, cirrhosis. The grade of NASH activity (A) was calculated by the addition of the grades of ballooning and lobular inflammation (from A0 to A4). Based on the Steatosis, Activity, and Fibrosis score evaluation[35], NASH was also classified histologically into mild (A ≤ 2 and F ≤ 2) and significant (A > 2 and/or F > 2).
Patient weight and height were measured using a calibrated scale after removing shoes and heavy clothing, if any. Body mass index (BMI) was calculated as the ratio of weight (in kilograms)/height (in square meters). Waist circumference was measured at the mid-level between the lower rib and the iliac crest[36]. Venous blood samples were obtained from the subjects after an overnight fast (12 h) to measure plasma glucose, serum aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP), gamma glutamyltranspeptidase, total cholesterol, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol (LDL), TG, total bilirubin, platelets, and ferritin concentrations. Serum vitamin D and fasting insulin levels were obtained only from the NAFLD patients. Homeostasis Model Assessment was used to evaluate insulin resistance and was calculated as fasting serum insulin (μU/mL) × fasting plasma glucose (mmol/L)/22.5[37]. All biochemical parameters were measured in a conventional automated analyzer. A blood sample was also obtained for DNA extraction and genotyping.
Genomic DNA was extracted using the high salt method[38]. A predesigned TaqMan® probe (Applied Biosystems, Foster City, CA, United States) was purchased for genotyping rs738409 (C_7241_10) and rs58542926 (C_89463510_20). Genotyping was performed by real-time PCR in allelic discrimination mode, using the Strategene® Mx3005 equipment (MxPro QPCR System, 2007 Software, La Jolla, CA, United States). PCR protocols were carried out according to the TaqMan® Genotyping Master Mix manufacturer’s instructions (Applied Biosystems, Foster City, CA, United States). The success rates of these assays were > 99%.
Statistical analyses were performed using SPSS 23.0 software (IBM, United States). Data are expressed as mean ± SD for normally distributed continuous variables, as median and interquartile range when the distribution was skewed, or as number and percentage for qualitative variables. Continuous variables distribution was assessed by the Shapiro-Wilk test. The Hardy-Weinberg equilibrium was checked for all individual single nucleotide polymorphisms (SNPs)[39]. The Student’s t-test or a non-parametric test, i.e., Mann-Whitney U-test were used to compare quantitative data, as appropriate. χ2 test or Fisher’s exact tests were used for comparison of categorical data. All tests were two-tailed and P values < 0.05 were considered significant. Variables associated with a P value < 0.20 in univariate analysis were included in multivariate analysis.
Multinomial binary logistic regression analysis was performed to detect the effect of a SNP mutation on liver histology. For association analysis, the PNPLA3 rs738409 and the TM6SF2 rs58542926 variants were coded in an additive and dominant genetic model. The model fit was assessed by the Hosmer-Lemeshow test.
Clinical features, anthropometric variables and laboratory findings of the 148 NAFLD patients and 137 controls are shown in Table 1.
| Variables | NAFLD, n = 148 | Controls, n = 137 | P value |
| Female | 111 (75.0%) | 97 (70.8%) | 0.426 |
| Age (yr) | 57 (46.3-63) | 55 (43 61.5) | 0.610 |
| BMI (kg/m2) | 32.8 ± 4.8 | 25.4 ± 3.3 | < 0.0001 |
| Hypertension | 98 (66.2%) | 24 (17.5%) | < 0.0001 |
| Diabetes mellitus | 69 (46.6%) | 12 (8.8%) | < 0.0001 |
| Dyslipidemia | 108 (73.0%) | 19 (13.5%) | < 0.0001 |
| Fasting glucose (mg/dL) | 101 (90-124.8) | 88 (83-98) | < 0.0001 |
| AST (U/L) | 34 (25.5-56.5) | 24 (20.3-29) | < 0.0001 |
| ALT (U/L) | 38 (28-70.5) | 29.5 (23-38) | < 0.0001 |
| GGT (U/L) | 53 (29-116) | 28 (18.5-55.5) | < 0.0001 |
| ALP (U/L) | 97 (74-116.8) | 75 (58.3-83.8) | < 0.0001 |
| Total bilirubin (mg/dL) | 0.6 (0.5-0.9) | 0.7 (0.5-0.8) | 0.438 |
| Triglycerides (mg/dL) | 162.5 (110-205.3) | 126.5 (93-176) | 0.006 |
| Total cholesterol (mg/dL) | 195.8 ± 41.5 | 179.9 ± 47.1 | 0.006 |
| VLDL (mg/dL) | 32 (22-42.3) | 24.5 (17.3-33.8) | 0.001 |
| HDL (mg/dL) | 48 (39.5-54) | 47 (40.5-60.8) | 0.404 |
| LDL (mg/dL) | 107.5 (89.5-137.9) | 96 (67.8-117.5) | 0.009 |
| Platelets (109/L) | 219.4 ± 81.8 | 254.6 ± 82.4 | 0.439 |
| Ferritin (ng/mL) | 89 (42.7-168.5) | 32 (32-32) | 0.349 |
| D vitamin (ng/mL) | 22 (19-27.7) | 26.95 (20.5-32.5) | 0.139 |
Comparing the NAFLD group with the controls, there were no significant differences regarding gender and age distributions. NAFLD patients showed a significantly higher frequency of the metabolic syndrome components–T2DM, arterial hypertension and dyslipidemia–and significantly higher values of BMI. Fasting glucose, TG, AST, ALT, ALP and gamma glutamyltranspeptidase serum concentrations were also significantly higher in the NAFLD subjects (Table 1).
Liver biopsy was performed in 65 of the NAFLD patients (43.9%). Based on histological findings, 54 (36.5%) individuals (7 with cirrhosis) were included in the NASH group. The SS group included 94 (63.5%) NAFLD subjects without a clinical indication for histological examination (i.e. patients without metabolic syndrome and/or advanced fibrosis by noninvasive methods) and those without NASH on biopsy. Clinical and demographic characteristics of the NASH and SS patients are depicted in Table 2.
| Variables | NASH, n = 54 | Simple steatosis, n = 94 | P value |
| Female | 44 (81.5%) | 67 (71.3%) | 0.169 |
| Age (yr) | 59 (45.8-63) | 57 (47.8-64) | 0.935 |
| BMI (kg/m2) | 32.7 ± 4.8 | 32.8 ± 4.8 | 0.968 |
| WC | 106.6 (100-114) | 105.3 (96-113.8) | 0.306 |
| Arterial hypertension | 39 (72.2%) | 59 (62.8%) | 0.886 |
| Diabetes mellitus | 35 (64.8%) | 34 (36.2%) | 0.007 |
| Dyslipidemia | 45 (83.3%) | 63 (67.0%) | 0.248 |
| Fasting insulin | 16.5 (11.9-31) | 14 (8-18) | 0.055 |
| Fasting glucose (mg/dL) | 112.5 (91-151.5) | 98 (90-112) | 0.019 |
| AST (U/L) | 53.5 (31.7-69) | 30 (23-38) | < 0.001 |
| ALT (U/L) | 61 (36-79.3) | 33 (24-49) | < 0.001 |
| GGT (U/L) | 87 (51.7-164.3) | 39 (27-73.3) | < 0.001 |
| ALP (U/L) | 99 (78-127.5) | 88 (72-116) | 0.075 |
| Total bilirubin (mg/dL) | 0.59 (0.5-0.9) | 0.6 (0.5-0.9) | 0.986 |
| Triglycerides (mg/dL) | 162 (113-227.8) | 163 (109.25-201) | 0.544 |
| Total cholesterol (mg/dL) | 196.4 ± 39.9 | 195.4 ± 42.7 | 0.793 |
| VLDL (mg/dL) | 32 (22.8-45.3) | 32 (22-40.8) | 0.440 |
| HDL (mg/dL) | 46 (37.8-53) | 48 (40-55) | 0.432 |
| LDL (mg/dL) | 108.5 (89-138.4) | 107 (88.5-137.4) | 0.793 |
| Platelets (109/L) | 184 ± 67.4 | 308 ± 1.4 | 0.053 |
| Ferritin (ng/mL) | 109 (56.75-267.5) | 69 (37.2-143.8) | 0.019 |
| D vitamin (ng/mL) | 21.1 (17.55-25.9) | 23.4 (19-28) | 0.086 |
| HOMA-IR | 5.26 (2.97-8.8) | 3.36 (1.77-4.7) | 0.330 |
A significantly higher proportion of subjects in the NASH group had T2DM or impaired fasting glucose (IFG) when compared to the SS group, but there was no difference regarding the frequency of arterial hypertension, dyslipidemia or BMI. AST, ALT and ferritin serum concentration levels were significantly higher in the NASH group than in the SS group (Table 2). Fibrosis was detected in 35 of the 65 biopsies: F1 = 18, F2 = 5, F3 = 5, and F4 = 7.
The PNPLA3 gene was successfully amplified by real-time PCR in all NAFLD patients and controls. The minor allele frequency (MAF) of PNPLA3 rs738409 in overall patients was 34%, and the genotype frequencies were CC 47%, CG 38% and GG 15%. The Hardy-Weinberg equilibrium was demonstrated for the selected SNP of PNPLA3 in the NAFLD (P = 0.26) and the control (P = 0.07) groups.
The C and G allele frequencies in the NAFLD patients were 59% and 41%, respectively, and were 74% and 26% among the controls. PNPLA3 genotypes are described in Table 3; their frequencies were significantly different between the NAFLD patients and the controls (P < 0.001).
| Genotype frequency % (n) | P value | |||
| CC | CG | GG | ||
| NAFLD | 37.2 (55) | 43.9 (65) | 18.9 (28) | < 0.001 |
| Controls | 58.4 (80) | 31.4 (43) | 10.2 (14) | |
The chance of NAFLD was increased by the presence of the G allele [odds ratio (OR) = 2.37, 95%CI: 1.47-3.82; P < 0.001], even after adjustment for age, gender, BMI and T2DM/IFG (OR = 1.69, 95%CI: 1.21-2.36; P = 0.002) (Table 4). This association was even stronger when CC homozygotes were compared with GG homozygotes (OR = 3.13, 95%CI: 1.49-6.57; P = 0.003).
| Genotypes | OR | 95%CI | P value |
| NAFLD vs Control | |||
| CC vs CG/GG | 1.69 | 1.21–2.36 | 0.002 |
| CC vs GG | 3.13 | 1.49–6.57 | 0.003 |
| NASH vs SS | |||
| CC vs CG/GG | 3.50 | 1.84–6.64 | < 0.001 |
| CC vs GG | 5.53 | 2.04-14.92 | 0.001 |
In the NAFLD group, no association was found between PNPLA3 CC vs CC + CG genotypes concerning BMI (P = 0.421), waist circumference (P = 0.641), FG (P = 0.795), high-density lipoprotein cholesterol (P = 0.723), low-density lipoprotein (P = 0.614) and TG (P = 0.269). On the other hand, serum AST (P < 0.001) and ALT (P = 0.002) were associated with the presence of rs738409 G allele at PNPLA3.
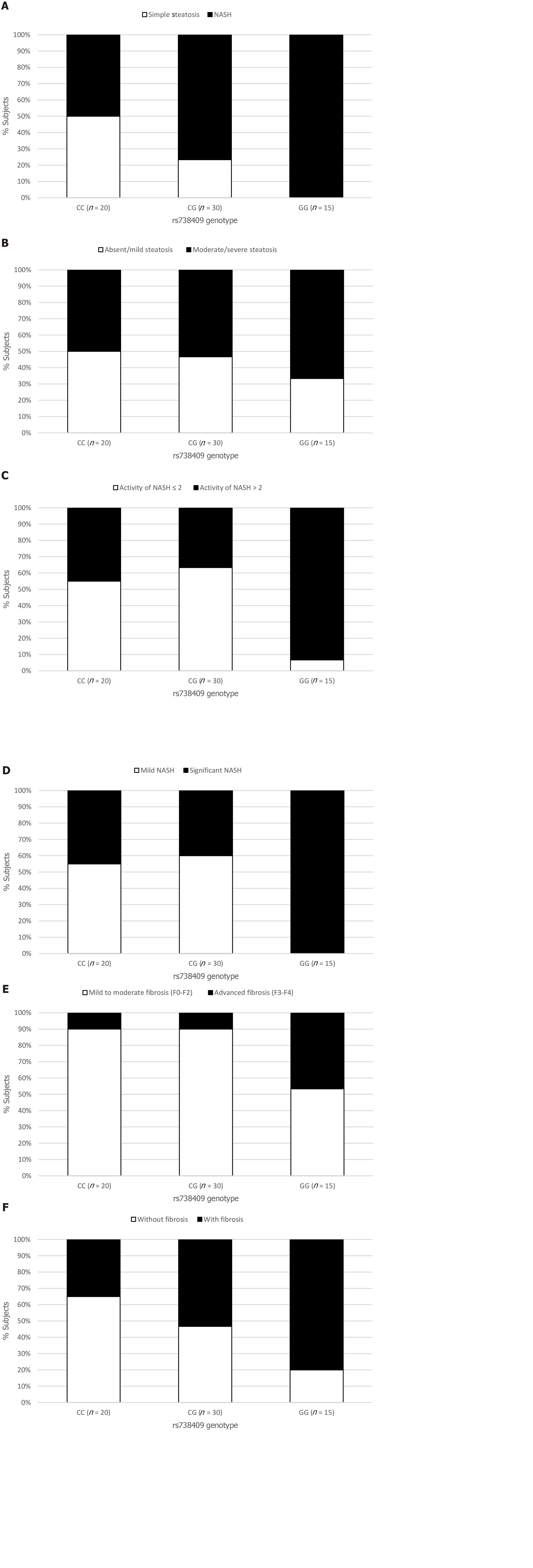
The NAFLD group was assessed for the association between histological features including steatosis, ballooning, inflammation and fibrosis, and the PNPLA3 genotype. Although the presence of hepatic steatosis on US was associated with the G variant rs738409, we found no association between the histological grade of steatosis and the presence of the G allele (Figure 1B). The severity of both lobular inflammation and hepatocellular ballooning was associated with the PNPLA3 variant G allele (P < 0.001).
The prevalence of NASH was 50% in the NAFLD subjects with the CC genotype (10/20), 77% in those with the CG genotype (23/30), and 100% in those with the GG genotype (15/15) (P < 0.001) (Figure 1A). G allele presence increased the chance of NASH (OR = 2.21, 95%CI: 1.04-4.71, P = 0.039), especially after adjusted for age, gender, BMI and T2DM (OR = 3.50, 95%CI: 1.84-6.64, P < 0.001). This chance was even higher when we analyzed homozygosis GG vs CC (OR = 6.07, 95%CI: 2.06-17.81, P = 0.001), even after the same adjusted model (OR = 5.53, 95%CI: 2.04-14.92, P = 0.001) (Table 4).
The chance of significant NASH activity (A > 2) was higher in homozygosis GG (OR = 17.11; 95%CI: 1.87-156.25; P = 0.012) in multivariate analysis. Based on the Steatosis, Activity, and Fibrosis score, significant NASH was associated with PNPLA3 GG genotype (Figure 1D).
The presence of fibrosis was observed in 7 of the 20 (35%) patients who underwent liver biopsy with the CC genotype; in 16 of 30 (53%) with the CG genotype, and in 12 of 15 (80%) with the GG genotype (P = 0.01) (Figure 1F). Presence of the G allele increased the chance of liver fibrosis in a model adjusted for age, gender, BMI and T2DM/IFG (OR = 3.05; 95%CI: 1.01-9.17; P = 0.04). The GG homozygosis increased the chance of fibrosis (OR = 7.42; 95%CI: 1.55-35.47; P = 0.01) after the same adjusted model (Table 5).
| Genotypes | OR | 95%CI | P value |
| Activity of NASH (A > 2 vs A < 2) | |||
| CC vs CG/GG | 0.65 | 0.22-1.88 | 0.433 |
| CC vs GG | 17.11 | 1.87 - 156.25 | 0.012 |
| Fibrosis (presence vs absence) | |||
| CC vs CG/GG | 3.05 | 1.01-9.17 | 0.046 |
| CC vs GG | 7.42 | 1.55-35.47 | 0.012 |
The TM6SF2 rs58542926 genotypes were confirmed to be in Hardy–Weinberg equilibrium with a MAF in the overall NAFLD cohort of 8%. The genotype frequencies were CC 84%; CT 15%; and TT 1%.
TM6SF2 genotype distribution among NAFLD and controls are described in Table 6. There was no significant difference in the allele distribution between cases and controls (P = 0.78). The frequencies of T alleles were 9% in the controls and 8% in NAFLD patients. The presence of T alleles was not associated with NAFLD or NASH in univariate or multivariate analysis. TM6SF2 T alleles were more frequent in the NASH group (22%) than in the SS group (12%), but without statistical significance (P = 0.89).
| Genotype frequency % (n) | P value | |||
| CC | CT | TT | ||
| NAFLD | 83.2 (125) | 14.9 (22) | 0.7 (1) | 0.78 |
| Controls | 84.5 (114) | 16.1 (22) | 0.7 (1) | |
Finally, there was no association between the gene TM6SF2 rs58542926 and liver steatosis (P = 0.62), ballooning (P = 0.14), lobular inflammation (P = 0.99) and fibrosis (P = 0.89).
This study demonstrated that the PNPLA3 genotype is associated with NAFLD susceptibility and different clinical forms. The presence of the G allele was associated with NAFLD occurrence when compared to the controls, and with NASH when compared to SS, in addition it was associated with higher NASH histological activity score and the presence of fibrosis in individuals with histopathological assessment. The TM6SF2 genotype frequency was analyzed for the first time in a Brazilian population, and we did not observe any significant difference in the variant gene distribution between NAFLD patients and controls, or between NASH and SS subjects.
The first GWA study in NAFLD identified a single highly significant association between increased hepatic TG content and the PNPLA3[17]. Subsequent studies demonstrated that this variant was also associated with the progression of NAFLD in different ethnic populations around the world[21,25,26,41,42].
In the Brazilian population, there are only two studies on NAFLD and PNPLA3 genotypes[30,43]. Machado et al[43] investigated the gene polymorphism in T2DM individuals and Mazo et al[30] evaluated the genetic variation in NAFLD subjects compared to controls. The TM6SF2 polymorphisms were assessed in the investigation by Mazo et al[30]; however, they were not in Hardy-Weinberg equilibrium in the NAFLD group, which precluded this analysis. Therefore, the present study contributes to the PNPLA3 genotype study in Brazil and is the first study to document the distribution of TM6SF2 alleles and genotypes in Brazilian NAFLD patients.
In this sample, the frequency of the minor (G) allele at PNPLA3 rs738409 was higher (34%) than the frequency reported in European (23%) and African (15%) individuals, lower than observed in American subjects (45%), and similar to that found in East Asian individuals (35%). The observed differences in the MAF for rs738409 between different racial groups are in agreement with the differences in the risk of hepatic steatosis[44].
Findings from this study confirmed the association between PNPLA3 and liver fat content, and showed that the rs738409 G allele increases the chance of NAFLD in Brazilian patients by about 1.6-fold, regardless of metabolic syndrome features. Furthermore, GG homozygosis seems to have an even stronger association with liver fat.
NAFLD is currently considered the hepatic manifestation of the metabolic syndrome[45]. In a recent meta-analysis, most of the included studies showed lack of association between PNPLA3 genotypes and BMI, fasting glucose levels and Homeostasis Model Assessment[21]. In the study by Machado et al[43], analysis of the associations between PNPLA3 genotypes and the metabolic syndrome components in a T2DM population demonstrated that the G allele was only associated with better glycemic control. In the current study, we found no association between anthropometric and metabolic parameters with the PNPLA3 gene variant, which confirms previous results.
The presence of the G allele was not only associated with NAFLD occurrence, but also with the NASH phenotype in patients who underwent liver biopsy. This finding was different to a previous Brazilian analysis[30], in which NASH occurrence was not associated with the presence of the G allele in NAFLD individuals. This could be attributed to the fact that the prior Brazilian study enrolled a small number of SS individuals (n = 34) and the SS: NASH proportion was 1.0: 6.3, whereas in our study it was 1.7: 1.0.
Interestingly, no significant association between rs738409 and steatosis grade was found in our study. However, the grade of NASH histological activity and the presence of fibrosis were associated with the PNPLA3 genotype. In fact, our data support that more aggressive disease with higher fibrosis scores was associated with rs738409 variation–subjects with higher activity scores (A > 2) were 17 times more likely to be GG homozygous than to be homozygous for the C allele; and subjects with liver fibrosis were 7.4-fold more likely to be GG homozygous than to be CC. Lack of significant association between histological steatosis grade and rs738409 genotype was observed in at least two other case-control studies[46,47]; and an association between PNPLA3 and inflammation activity and fibrosis in NAFLD has also been found in other studies[21,23,25]. Our study, however, was limited by the fact that only 44% of the patients underwent liver biopsy.
As stated, the TM6SF2 rs58542926 genotypes were described in this study for the first time in Brazilians. The MAF (8%) was similar to that observed in a Northern European sample (7%)[48], but lower than that observed in other European samples (12% and 13%)[28,29]. Different to that described in other studies[27,29,41,42], we did not find significant differences regarding the TM6SF2 genotypes between NAFLD patients and controls, or between NASH and SS individuals. This finding suggests that in the Brazilian population, the genetic variants of rs58542926 TM6SF2 may have a distinct influence on NAFLD than that observed in other populations, which is understandable, as Brazilian NAFLD patients have been reported as an admixed population presenting genetic ancestry contributions from European (48.8%), African (41.7%) and Amerindian (9.5%)[49]. Besides that, as TM6SF2 MAF is less frequent in the general population, larger samples may be required to confirm this finding.
In conclusion, we observed that PNPLA3 may be involved in the progression of NAFLD in the Brazilian population. Individuals who had histopathological assessment and more advanced liver disease were more likely to carry the G allele. The TM6SF2 genetic variants were not associated with NAFLD susceptibility and severity in the population studied, although further studies with larger samples are required to confirm these findings.
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease in Western countries and encompasses a spectrum ranging from simple steatosis to nonalcoholic steatohepatitis (NASH). Although highly prevalent, only a minority of NAFLD patients develop significant fibrosis. Thus, NAFLD is considered a complex disorder in which the disease phenotype results from an interaction between environmental exposure and a susceptible polygenic background. Polymorphisms of PNPLA3 and TM6SF2 genes have been associated with greater susceptibility to NAFLD development in previous studies.
There is only one genetic study in Brazilian NAFLD patients, and TM6SF2 polymorphism has not yet been analyzed in this population. As Brazilian NAFLD subjects have been reported as an admixed population presenting diverse genetic ancestry contributions, it is relevant to study the associations between various genotypes and fatty liver disease progression.
We aimed to investigate the association between PNPLA3 and TM6SF2 genotypes and clinical parameters of NAFLD, and analyzed the genotype variations as markers of liver histological features in adult Brazilian NAFLD patients. We also investigated the distribution of these genotype variations among Brazilians.
This cross-sectional study enrolled 285 individuals, of which 148 patients had features of NAFLD (case patients) and 137 were non-NAFLD control subjects. NAFLD was diagnosed based on hepatic steatosis by liver ultrasonography and exclusion of other causes of liver disease. Patients who had decompensated cirrhosis or were taking drugs that induce steatosis were excluded. From the total of NAFLD patients, 65 underwent liver biopsy according to the clinical protocol of increased risk for NASH and/or advanced fibrosis. DNA was obtained for genotyping PNPLA3 at rs738409 and TM6SF2 at rs58542926.
PNPLA3 CC, CG and GG genotype frequencies were 37%, 44% and 19%, respectively, in NAFLD patients and were 58%, 31% and 10% in controls (P < 0.001). In a model adjusted for gender, age, body mass index and type 2 diabetes mellitus, the G allele increased the chance of NAFLD (OR = 1.69, 95%CI: 1.21-2.36, P = 0.002) and NASH (OR = 3.50, 95%CI: 1.84-6.64, P < 0.001). The chance of NASH was even higher with GG homozygosis (OR = 5.53, 95%CI: 2.04-14.92, P = 0.001). No association was found between G allele and the features of metabolic syndrome. In the histological assessment, PNPLA3 genotype was not associated with steatosis grade, although GG homozygosis increased the chance of significant NASH activity (OR = 17.11, 95%CI: 1.87-156.25, P = 0.01) and fibrosis (OR = 7.42, 95%CI: 1.55-34.47, P = 0.01) in the same adjusted model. TM6SF2 CC, CT and TT genotype frequencies were 83%, 15% and 0.7%, respectively, in NAFLD patients and were 84%, 16% and 0.7% in controls (P = 0.78). Presence of the T allele was not associated with NAFLD or NASH, or with histological features.
PNPLA3 may be involved in the susceptibility and progression of NAFLD and NASH in the Brazilian population. More advanced histological liver disease was associated with the G allele. The TM6SF2 genetic variants were not associated with NAFLD susceptibility and progressive histological forms in the population studied.
The description of variant genotypes distribution in NAFLD Brazilian patients contributes to a better understanding of the disease clinical characteristics and atypical features in this population. As the TM6SF2 polymorphism is less frequent in the general population, investigations with larger sample are needed. Further studies may investigate additional particular components of fatty liver disease in Brazil. The role of genotyping assessment for risk stratification is still uncertain.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Han T S-Editor: Zhang H L-Editor: Webster JR P-Editor: Wang LL
| 1. | European Association for the Study of the Liver (EASL). EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J Hepatol. 2016;64:1388-1402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2290] [Cited by in RCA: 3179] [Article Influence: 353.2] [Reference Citation Analysis (4)] |
| 2. | Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, Harrison SA, Brunt EM, Sanyal AJ. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67:328-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3544] [Cited by in RCA: 4946] [Article Influence: 706.6] [Reference Citation Analysis (9)] |
| 3. | Takahashi Y, Fukusato T, Inui A, Fujisawa T. [Pediatric nonalcoholic fatty liver disease/nonalcoholic steatohepatitis]. Nihon Rinsho. 2012;70:1827-1834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 400] [Article Influence: 44.4] [Reference Citation Analysis (3)] |
| 4. | Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5322] [Cited by in RCA: 7533] [Article Influence: 837.0] [Reference Citation Analysis (0)] |
| 5. | Non-alcoholic Fatty Liver Disease Study Group, Lonardo A, Bellentani S, Argo CK, Ballestri S, Byrne CD, Caldwell SH, Cortez-Pinto H, Grieco A, Machado MV, Miele L, Targher G. Epidemiological modifiers of non-alcoholic fatty liver disease: Focus on high-risk groups. Dig Liver Dis. 2015;47:997-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 347] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 6. | Farrell GC, Larter CZ. Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology. 2006;43:S99-S112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1756] [Cited by in RCA: 1822] [Article Influence: 95.9] [Reference Citation Analysis (0)] |
| 7. | Younossi Z, Stepanova M, Ong JP, Jacobson IM, Bugianesi E, Duseja A, Eguchi Y, Wong VW, Negro F, Yilmaz Y, Romero-Gomez M, George J, Ahmed A, Wong R, Younossi I, Ziayee M, Afendy A; Global Nonalcoholic Steatohepatitis Council. Nonalcoholic Steatohepatitis Is the Fastest Growing Cause of Hepatocellular Carcinoma in Liver Transplant Candidates. Clin Gastroenterol Hepatol 2019; 17: 748-755. e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 599] [Cited by in RCA: 574] [Article Influence: 95.7] [Reference Citation Analysis (0)] |
| 8. | Kim D, Kim WR, Kim HJ, Therneau TM. Association between noninvasive fibrosis markers and mortality among adults with nonalcoholic fatty liver disease in the United States. Hepatology. 2013;57:1357-1365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 626] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 9. | Dulai PS, Singh S, Patel J, Soni M, Prokop LJ, Younossi Z, Sebastiani G, Ekstedt M, Hagstrom H, Nasr P, Stal P, Wong VW, Kechagias S, Hultcrantz R, Loomba R. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: Systematic review and meta-analysis. Hepatology. 2017;65:1557-1565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 984] [Cited by in RCA: 1430] [Article Influence: 178.8] [Reference Citation Analysis (0)] |
| 10. | Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol. 2013;10:330-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1066] [Cited by in RCA: 1318] [Article Influence: 109.8] [Reference Citation Analysis (0)] |
| 11. | Singh S, Allen AM, Wang Z, Prokop LJ, Murad MH, Loomba R. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: a systematic review and meta-analysis of paired-biopsy studies. Clin Gastroenterol Hepatol 2015; 13: 643-54. quiz e39-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 932] [Cited by in RCA: 1236] [Article Influence: 123.6] [Reference Citation Analysis (0)] |
| 12. | Dongiovanni P, Romeo S, Valenti L. Genetic Factors in the Pathogenesis of Nonalcoholic Fatty Liver and Steatohepatitis. Biomed Res Int. 2015;2015:460190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 13. | Anstee QM, Day CP. The genetics of NAFLD. Nat Rev Gastroenterol Hepatol. 2013;10:645-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 264] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 14. | Anstee QM, Day CP. The Genetics of Nonalcoholic Fatty Liver Disease: Spotlight on PNPLA3 and TM6SF2. Semin Liver Dis. 2015;35:270-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 15. | Wood KL, Miller MH, Dillon JF. Systematic review of genetic association studies involving histologically confirmed non-alcoholic fatty liver disease. BMJ Open Gastroenterol. 2015;2:e000019. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 16. | Severson TJ, Besur S, Bonkovsky HL. Genetic factors that affect nonalcoholic fatty liver disease: A systematic clinical review. World J Gastroenterol. 2016;22:6742-6756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 79] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 17. | Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461-1465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2701] [Cited by in RCA: 2596] [Article Influence: 152.7] [Reference Citation Analysis (0)] |
| 18. | Chung GE, Lee Y, Yim JY, Choe EK, Kwak MS, Yang JI, Park B, Lee JE, Kim JA, Kim JS. Genetic Polymorphisms of PNPLA3 and SAMM50 Are Associated with Nonalcoholic Fatty Liver Disease in a Korean Population. Gut Liver. 2018;12:316-323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 19. | Kitamoto T, Kitamoto A, Yoneda M, Hyogo H, Ochi H, Nakamura T, Teranishi H, Mizusawa S, Ueno T, Chayama K, Nakajima A, Nakao K, Sekine A, Hotta K. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Hum Genet. 2013;132:783-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 157] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 20. | Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, Gudnason V, Eiriksdottir G, Garcia ME, Launer LJ, Nalls MA, Clark JM, Mitchell BD, Shuldiner AR, Butler JL, Tomas M, Hoffmann U, Hwang SJ, Massaro JM, O'Donnell CJ, Sahani DV, Salomaa V, Schadt EE, Schwartz SM, Siscovick DS; NASH CRN; GIANT Consortium; MAGIC Investigators; Voight BF; Carr JJ; Feitosa MF; Harris TB; Fox CS; Smith AV; Kao WH; Hirschhorn JN; Borecki IB; GOLD Consortium. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 775] [Cited by in RCA: 761] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 21. | Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883-1894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 756] [Cited by in RCA: 739] [Article Influence: 52.8] [Reference Citation Analysis (1)] |
| 22. | Palmer ND, Musani SK, Yerges-Armstrong LM, Feitosa MF, Bielak LF, Hernaez R, Kahali B, Carr JJ, Harris TB, Jhun MA, Kardia SL, Langefeld CD, Mosley TH Jr, Norris JM, Smith AV, Taylor HA, Wagenknecht LE, Liu J, Borecki IB, Peyser PA, Speliotes EK. Characterization of European ancestry nonalcoholic fatty liver disease-associated variants in individuals of African and Hispanic descent. Hepatology. 2013;58:966-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 122] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 23. | Zhang L, You W, Zhang H, Peng R, Zhu Q, Yao A, Li X, Zhou Y, Wang X, Pu L, Wu J. PNPLA3 polymorphisms (rs738409) and non-alcoholic fatty liver disease risk and related phenotypes: a meta-analysis. J Gastroenterol Hepatol. 2015;30:821-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 24. | Xu R, Tao A, Zhang S, Deng Y, Chen G. Association between patatin-like phospholipase domain containing 3 gene (PNPLA3) polymorphisms and nonalcoholic fatty liver disease: a HuGE review and meta-analysis. Sci Rep. 2015;5:9284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 25. | Salameh H, Hanayneh MA, Masadeh M, Naseemuddin M, Matin T, Erwin A, Singal AK. PNPLA3 as a Genetic Determinant of Risk for and Severity of Non-alcoholic Fatty Liver Disease Spectrum. J Clin Transl Hepatol. 2016;4:175-191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 26. | Dai G, Liu P, Li X, Zhou X, He S. Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease (NAFLD) susceptibility and severity: A meta-analysis. Medicine (Baltimore). 2019;98:e14324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 27. | Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjærg-Hansen A, Vogt TF, Hobbs HH, Cohen JC. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352-356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 724] [Cited by in RCA: 930] [Article Influence: 84.5] [Reference Citation Analysis (0)] |
| 28. | Dongiovanni P, Petta S, Maglio C, Fracanzani AL, Pipitone R, Mozzi E, Motta BM, Kaminska D, Rametta R, Grimaudo S, Pelusi S, Montalcini T, Alisi A, Maggioni M, Kärjä V, Borén J, Käkelä P, Di Marco V, Xing C, Nobili V, Dallapiccola B, Craxi A, Pihlajamäki J, Fargion S, Sjöström L, Carlsson LM, Romeo S, Valenti L. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61:506-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 418] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 29. | Liu YL, Reeves HL, Burt AD, Tiniakos D, McPherson S, Leathart JB, Allison ME, Alexander GJ, Piguet AC, Anty R, Donaldson P, Aithal GP, Francque S, Van Gaal L, Clement K, Ratziu V, Dufour JF, Day CP, Daly AK, Anstee QM. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:4309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 362] [Cited by in RCA: 460] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 30. | Mazo DF, Malta FM, Stefano JT, Salles APM, Gomes-Gouvea MS, Nastri ACS, Almeida JR, Pinho JRR, Carrilho FJ, Oliveira CP. Validation of PNPLA3 polymorphisms as risk factor for NAFLD and liver fibrosis in an admixed population. Ann Hepatol. 2019;18:466-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 31. | Hamaguchi M, Kojima T, Itoh Y, Harano Y, Fujii K, Nakajima T, Kato T, Takeda N, Okuda J, Ida K, Kawahito Y, Yoshikawa T, Okanoue T. The severity of ultrasonographic findings in nonalcoholic fatty liver disease reflects the metabolic syndrome and visceral fat accumulation. Am J Gastroenterol. 2007;102:2708-2715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 514] [Cited by in RCA: 673] [Article Influence: 37.4] [Reference Citation Analysis (3)] |
| 32. | Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, Fruchart JC, James WP, Loria CM, Smith SC Jr; International Diabetes Federation Task Force on Epidemiology and Prevention; Hational Heart; Lung; and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; International Association for the Study of Obesity. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640-1645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8720] [Cited by in RCA: 10555] [Article Influence: 659.7] [Reference Citation Analysis (0)] |
| 33. | Angulo P, Hui JM, Marchesini G, Bugianesi E, George J, Farrell GC, Enders F, Saksena S, Burt AD, Bida JP, Lindor K, Sanderson SO, Lenzi M, Adams LA, Kench J, Therneau TM, Day CP. The NAFLD fibrosis score: a noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatology. 2007;45:846-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1917] [Cited by in RCA: 2283] [Article Influence: 126.8] [Reference Citation Analysis (1)] |
| 34. | Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, Yeh M, McCullough AJ, Sanyal AJ; Nonalcoholic Steatohepatitis Clinical Research Network. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6807] [Cited by in RCA: 8233] [Article Influence: 411.7] [Reference Citation Analysis (5)] |
| 35. | Bedossa P; FLIP Pathology Consortium. Utility and appropriateness of the fatty liver inhibition of progression (FLIP) algorithm and steatosis, activity, and fibrosis (SAF) score in the evaluation of biopsies of nonalcoholic fatty liver disease. Hepatology. 2014;60:565-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 458] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 36. | Alberti KG, Zimmet P, Shaw J; IDF Epidemiology Task Force Consensus Group. The metabolic syndrome--a new worldwide definition. Lancet. 2005;366:1059-1062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5130] [Cited by in RCA: 5325] [Article Influence: 266.3] [Reference Citation Analysis (0)] |
| 37. | Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22373] [Cited by in RCA: 24508] [Article Influence: 612.7] [Reference Citation Analysis (0)] |
| 38. | Lahiri DK, Nurnberger JI Jr. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991;19:5444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 42] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 39. | Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169:505-514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 804] [Cited by in RCA: 790] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 40. | Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 986] [Cited by in RCA: 1047] [Article Influence: 55.1] [Reference Citation Analysis (0)] |
| 41. | Krawczyk M, Rau M, Schattenberg JM, Bantel H, Pathil A, Demir M, Kluwe J, Boettler T, Lammert F, Geier A; NAFLD Clinical Study Group. Combined effects of the PNPLA3 rs738409, TM6SF2 rs58542926, and MBOAT7 rs641738 variants on NAFLD severity: a multicenter biopsy-based study. J Lipid Res. 2017;58:247-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 149] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 42. | Koo BK, Joo SK, Kim D, Bae JM, Park JH, Kim JH, Kim W. Additive effects of PNPLA3 and TM6SF2 on the histological severity of non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2018;33:1277-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 43. | Machado CM, Leite NC, França PH, Cardoso CR, Salles GF, Villela-Nogueira CA. PNPLA3 gene polymorphism in Brazilian patients with type 2 diabetes: A prognostic marker beyond liver disease? Nutr Metab Cardiovasc Dis. 2019;29:965-971. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 44. | Kalia HS, Gaglio PJ. The Prevalence and Pathobiology of Nonalcoholic Fatty Liver Disease in Patients of Different Races or Ethnicities. Clin Liver Dis. 2016;20:215-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (1)] |
| 45. | Socha P, Wierzbicka A, Neuhoff-Murawska J, Włodarek D, Podleśny J, Socha J. Nonalcoholic fatty liver disease as a feature of the metabolic syndrome. Rocz Panstw Zakl Hig. 2007;58:129-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1725] [Cited by in RCA: 1745] [Article Influence: 72.7] [Reference Citation Analysis (0)] |
| 46. | Hotta K, Yoneda M, Hyogo H, Ochi H, Mizusawa S, Ueno T, Chayama K, Nakajima A, Nakao K, Sekine A. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med Genet. 2010;11:172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 133] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 47. | Li X, Zhao Q, Wu K, Fan D. I148M variant of PNPLA3 confer increased risk for nonalcoholic fatty liver disease not only in European population, but also in Chinese population. Hepatology. 2011;54:2275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 48. | 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68-74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10217] [Cited by in RCA: 12011] [Article Influence: 1201.1] [Reference Citation Analysis (0)] |
| 49. | Cavalcante LN, Stefano JT, Machado MV, Mazo DF, Rabelo F, Sandes KA, Carrilho FJ, Cortez-Pinto H, Lyra AC, de Oliveira CP. Genetic ancestry analysis in non-alcoholic fatty liver disease patients from Brazil and Portugal. World J Hepatol. 2015;7:1433-1438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |