Published online Feb 27, 2019. doi: 10.4254/wjh.v11.i2.186
Peer-review started: December 10, 2018
First decision: January 5, 2019
Revised: January 17, 2019
Accepted: January 26, 2019
Article in press: January 26, 2019
Published online: February 27, 2019
Processing time: 80 Days and 16.8 Hours
Patients with hepatitis C virus (HCV) and hepatocellular carcinoma (HCC) may or not develop iron overload (IO), which is associated with worst prognosis, because can cause serious damage to organs. HFE gene controls the iron uptake from gut, particularly in patients with hereditary hemochromatosis (HH).
To identify associations between HFE coding region in patients exhibiting hereditary hemochromatosis and in diseases associated with acquired IO.
We sequenced exons 2 to 5 and boundary introns of HFE gene, evaluating all polymorphic sites in patients presenting hereditary (hemochromatosis) or acquired iron overload HCV and HCC) and in healthy controls, using Sanger sequencing. We also determined the ensemble of extended haplotype in healthy control individuals, including several major histocompatibility complex loci, using sequence specific probes. Haplotype reconstruction was performed using the Arlequin and Phase softwares, and linkage disequilibrium (LD) between histocompatibility loci and HFE gene was performed using the Haploview software.
The HFE*003 allele was overrepresented (f = 71%) and HFE*001 allele was underrepresented (f = 14%) in HH patients compared to all groups. A strong linkage disequilibrium was observed among the H63D-G, IVS2(+4)-C and C282Y-G gene variants, particularly in HH; however, the mutation IVS2(+4)T>C was not directly associated with HH susceptibility. The HFE*001/HFE*002 genotype conferred susceptibility to HCC in HCV patients exhibiting IO (P = 0.02, OR = 14.14). Although HFE is telomeric to other histocompatibility genes, the H63D-G/IVS2(+4)-C (P ≤ 0.00001/P ≤ 0.0057) combination was in LD with HLA-B*44 allele group in healthy controls. No LD was observed between HFE alleles and other major histocompatibility loci.
A differential HFE association was observed for HH and for diseases associated with acquired IO (HCV, HCC). Since HFE is very distant from other histocompatibility loci, only weak associations were observed with these alleles.
Core tip: Patients with hepatitis C virus (HCV) and hepatocellular carcinoma (HCC) may or not develop iron overload (IO), which is associated with worset prognosis. The sequencing of the HFE gene permitted to assemble the previously described variation sites (H63DC>G-, S65CA>T and C282YG>A) associated with hereditary hemochromatosis into HFE haplotypes, under the standardized HLA nomenclature. A differential association of HFE alleles was observed for hereditary and acquired IO (HCV, HCC). In addition to the HFE gene, we also typed other major histocompatibility loci (HLA-A/-B/-C/ DRB1/-DQB1, and HLA-G 14bp INDEL and TNFa-d microsatellites) in the healthy population to understand how the HFE gene variability is associated with these loci.
- Citation: de Campos WN, Massaro JD, Cançado ELR, Wiezel CEV, Simões AL, Teixeira AC, Souza FF, Mendes-Junior CT, Martinelli ALC, Donadi EA. Comprehensive analysis of HFE gene in hereditary hemochromatosis and in diseases associated with acquired iron overload. World J Hepatol 2019; 11(2): 186-198
- URL: https://www.wjgnet.com/1948-5182/full/v11/i2/186.htm
- DOI: https://dx.doi.org/10.4254/wjh.v11.i2.186
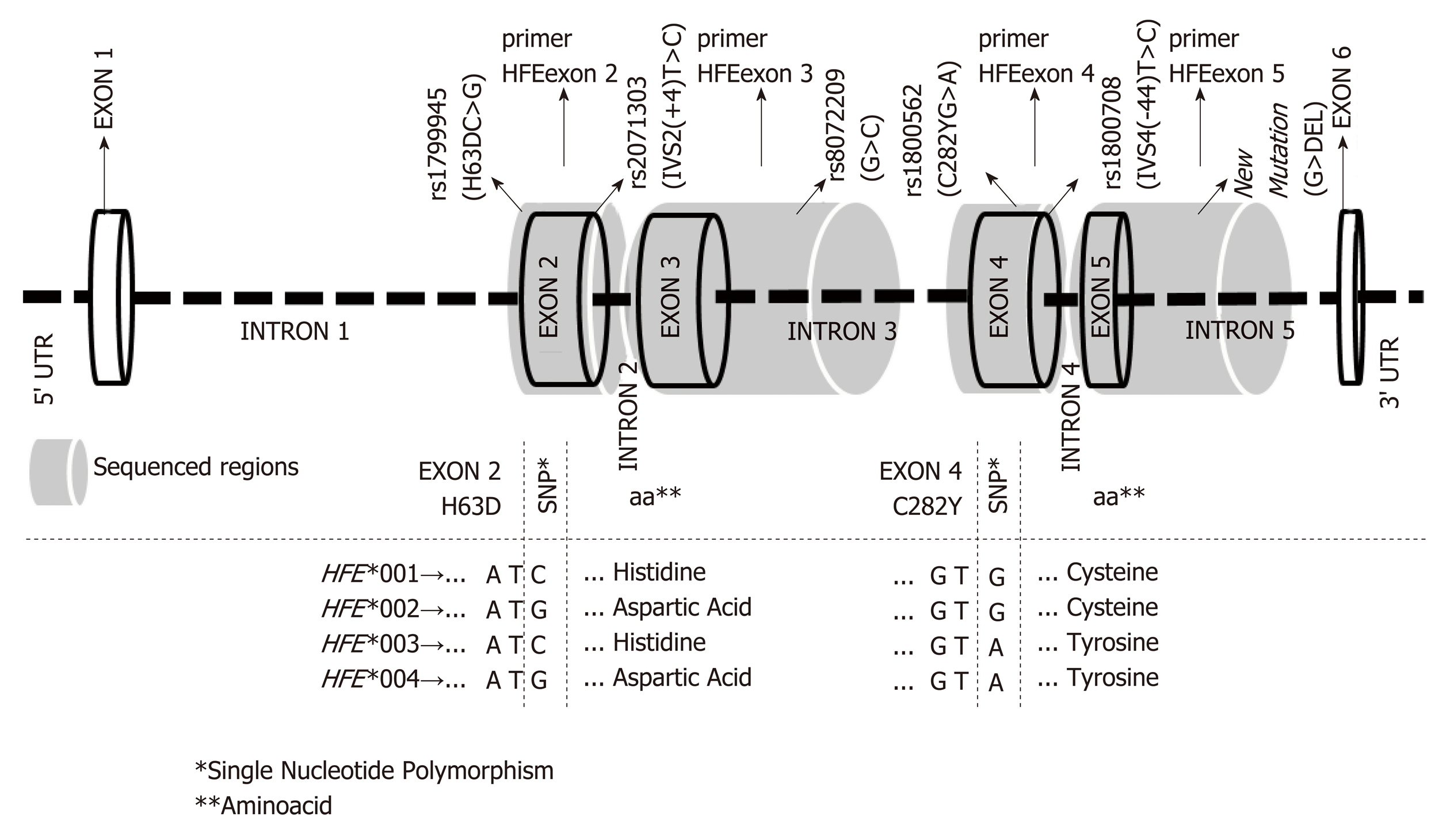
The HFE gene has seven exons and five introns, which code the α-heavy chain of the molecule. Exon 1 codes the signal peptide, exons 2-4 encode the α1, α2 and α3 domains, exons 5 the transmembrane domains, and the 5’ portion of exon 6 the cytoplasmic tail[1]. Considering that HFE gene controls the iron uptake from gut, defects of the encoded molecule have been associated with iron overload (IO), particularly in hemochromatosis hereditary (HH). Major variation sites observed at exons 2 to 4 have been associated with HH, including the H63DC>G (exon 2), S65CA>T (exon 2) and C282YG>A (exon 4) variants[2]. However, not all HH patients exhibit these mutations[1].
Besides HH, some acquired liver disorders have been associated with IO and fibrosis, including chronic hepatitis C virus (HCV), cirrhosis and hepatocellular carcinoma (HCC)[3]. The C282Y-A allele is associated with high iron serum levels, increased hepatic iron content and advanced fibrosis in HCV patients. Increased frequency of the classical HFE mutations has also been reported for HCC patients[4].
We sequenced exons 2 to 5 and boundary introns in HH patients, HCV patients presenting or not IO, and HCC patients exhibiting or not chronic HCV infection to associate with iron overload. We also evaluated the linkage disequilibrium (LD) between the HFE and HLA-A, HLA-B, HLA-C, HLA-DRB1 and DQB1 genes, as well as HLA-G 14bp INDEL and TNFa-d microsatellites to understand the association between HFE alleles and other major histocompatibility genes.
This study was approved by the local Ethics Research Committee (Process HCRP-FMRP, USP nº 4822/2011), and informed consent was obtained from all participants.
A total of 204 patients followed-up at Gastroenterology Units of University Hospitals of the University of São Paulo (USP) were studied: (1) 14 patients (9 men) aged 32-81 years (55.35 ± 15.16) exhibited HH, defined by high transferrin saturation (≥ 45%) and liver IO in the absence of secondary causes; (2) 130 patients with HCV (93 men) aged 19-69 years (42.60 ± 10.98), exhibiting (71 patients, 57 men) or not IO (59 patients, 36 men) (HCV-IO+ and HCV-IO-, respectively) in the absence of chronic alcohol ingestion (> 60 g/d). All patients exhibited IgG antibody against recombinant HCV antigens by second-generation ELISA (Abbott, Chicago, IL) for at least 6 mo and positive serum HCV RNA (Roche Diagnostic Systems, Branchburg, NJ). Serum levels of liver enzymes, iron, ferritin, and transferrin saturation were also determined. Liver specimens were scored for necroinflammatory activity, as previously described by Desmet et al[4]. Iron deposits were assessed and scored on the basis of the amount and cellular/lobular location[4,5]; and (3) 60 patients (43 men) aged 14-78 years (57 ± 14) exhibiting HCC, of whom 24 (18 men) presented IO and chronic hepatitis C (HCC HCV-IO+), and 36 (25 men) presented several underlying disorders including cryptogenic hepatitis, hepatitis B, non-alcoholic steatohepatitis and other co-morbidities. Since there is no need for liver biopsy for HCC diagnosis, liver iron was not screened in these patients (HCC-IO?). The diagnosis of HCC was performed according to Bruix and Sherman[6].
Iron overload was defined when iron deposits were detected in liver biopsy using Perl’s iron staining[7,8] and/or when serum transferrin saturation was higher than or equal to 45% with or without elevated ferritin. Patients presenting other types of congenital, virus or autoimmune liver disorders were excluded.
A total of 100 healthy unrelated blood donors (CTL), 80 men, and aged 20-52 years (33.31 ± 8.18) was also studied.
Exons 2 to 5 and boundary introns were evaluated using Sanger sequencing[9] (Figure 1). HFE nucleotide variations were retrieved from the NCBI (NC_000006.12) and Ensembl (ENSG00000010704) databases. Primer sequences, amplification conditions and allele nomenclature were defined as previously reported[10]. Sequencing was performed using an ABI 3500 sequencer (Applied Biosystems, Foster City, CA).
HLA-A/-B/-C/-DRB1 and -DQB1 typing was performed using commercial kits (One-Lambda, Canoga Park, CA). HLA-G 14bp INDEL[11] and TNFa-d microsatellites[12] were typed as previously described. Haplotype inferences combining major histocompatibility genes were performed only for healthy controls.
Allelic and genotype frequencies (f), Hardy Weinberg Equilibrium (HWE), Fisher exact test, and linkage disequilibrium (LD) were performed using the GENEPOP v.4.2 and ARLEQUIN v.3.1 softwares. Image map of the pairwise LD parameters [Log of the Odds (LOD) and Linkage Disequilibrium Coefficient (D')] was generated using the HAPLOVIEW v.3.32 software.
Extended major histocompatibility alleles were reconstructed by means of the EM (ARLEQUIN) and PHASE v.2 algorithms. For all situations, P values ≤ 0.05 were considered to be significant.
The results regarding HFE alleles are presented in two forms: (1) as previously reported in the literature, including the single nucleotide polymorphism (SNP) reference number (rs), the usual SNP names (H63DC>G, C282YG>A, IVS2(+4)T>C and IVS4(-44)T>C) and new variation sites (Table 1); and (2) as the newly described official HFE allele nomenclature (Table 2)[10]. The location of the previously reported variation sites with respect to the nucleotide sequence that defined the new HFE nomenclature is illustrated in Figure 1.
| SNPs | Allele/genotype | HH | HCV-IO+ | HCV-IO- | HCC HCV-IO+ | HCC-IO? | CTL |
| H63DC>G (rs1799945) [+3511] | C | 0.893 | 0.859 | 0.864 | 0.805 | 0.921 | 0.825 |
| G | 0.107 | 0.141 | 0.136 | 0.195 | 0.079 | 0.175 | |
| GG | 0.000 | 0.042 | 0.000 | 0.024 | 0.000 | 0.030 | |
| CG | 0.214 | 0.197 | 0.271 | 0.341 | 0.158 | 0.290 | |
| CC | 0.786 | 0.761 | 0.729 | 0.634 | 0.842 | 0.680 | |
| HW P value | 1.000 | 1.000 | 0.580 | 1.000 | 1.000 | 1.000 | |
| IVS2(+4)T>C (rs2071303) [+3668] | T | 0.857 | 0.641 | 0.669 | 0.585 | 0.684 | 0.610 |
| C | 0.143 | 0.359 | 0.331 | 0.415 | 0.316 | 0.390 | |
| TT | 0.7141 | 0.408 | 0.424 | 0.390 | 0.526 | 0.390 | |
| TC | 0.286 | 0.465 | 0.492 | 0.390 | 0.316 | 0.440 | |
| CC | 0.000 | 0.127 | 0.085 | 0.220 | 0.158 | 0.170 | |
| HW P value | 0.528 | 1.000 | 0.556 | 0.212 | 0.295 | 0.528 | |
| G>C (rs807209) [+5197] | G | 0.000 | 0.007 | 0.017 | 0.000 | 0.053 | 0.035 |
| C | 1.000 | 0.993 | 0.983 | 1.000 | 0.947 | 0.965 | |
| GG | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.010 | |
| GC | 0.000 | 0.014 | 0.034 | 0.000 | 0.105 | 0.050 | |
| CC | 1.000 | 0.986 | 0.966 | 1.000 | 0.895 | 0.940 | |
| HW P value | - | - | 1.000 | - | 1.000 | 0.103 | |
| C282YG>A (rs1800562) [+5473] | G | 0.2862 | 0.979 | 0.983 | 0.902 | 1.000 | 0.955 |
| A | 0.7142 | 0.021 | 0.017 | 0.098 | 0.000 | 0.045 | |
| GG | 0.0712 | 0.958 | 0.966 | 0.854 | 1.000 | 0.910 | |
| GA | 0.429 | 0.042 | 0.034 | 0.098 | 0.000 | 0.090 | |
| AA | 0.5002 | 0.000 | 0.000 | 0.049 | 0.000 | 0.000 | |
| HW P value | 1.000 | 1.000 | 1.000 | 0.031 | - | 1.000 | |
| IVS4(–44)T>C (rs1800708) [+5635] | T | 1.000 | 0.880 | 0.907 | 0.817 | 0.842 | 0.925 |
| C | 0.000 | 0.120 | 0.093 | 0.183 | 0.158 | 0.075 | |
| CC | 0.000 | 0.014 | 0.000 | 0.024 | 0.000 | 0.000 | |
| TC | 0.000 | 0.211 | 0.186 | 0.317 | 0.316 | 0.150 | |
| TT | 1.000 | 0.775 | 0.814 | 0.659 | 0.684 | 0.850 | |
| HW P value | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | |
| New mutation (G>Del) at intron 5 [+5811] | G | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 0.995 |
| Del | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.005 | |
| GG | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 0.990 | |
| G Del | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.010 | |
| Del Del | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| HW P value | - | - | - | - | - | - |
| Allele | SNPs sequences | Population frequencies (f) | |||||||||||||||||
| IO- | IO+ | IO? | Whole | ||||||||||||||||
| 1-2-3-4-5-63 | CTL | HCV-IO- | HCV-IO+ | HCC HCV-IO+ | HH | TOTAL | HCC-IO? | ||||||||||||
| n = 100 | % | n = 59 | % | n = 71 | % | n = 24 | % | n = 28 | % | n = 109 | % | n = 36 | % | n = 304 | % | ||||
| HFE*0014 | C-T-C-G-T-G | 107 | 0.54 | 74 | 0.63 | 86 | 0.61 | 23 | 0.48 | 42 | 0.14 | 113 | 0.52 | 40 | 0.56 | 334 | 0.55 | ||
| HFE*001:unofficial:025 | C-C-C-G-T-G | 28 | 0.14 | 12 | 0.10 | 15 | 0.11 | 3 | 0.06 | 1 | 0.04 | 19 | 0.09 | 4 | 0.06 | 63 | 0.10 | ||
| HFE*001:unofficial:035 | C-T-G-G-T-G | 8 | 0.04 | 2 | 0.02 | 1 | 0.01 | 0 | 0.00 | 0 | 0.00 | 1 | 0.00 | 2 | 0.03 | 12 | 0.02 | ||
| HFE*001:unofficial:045 | C-T-C-G-C-G | 0 | 0.00 | 0 | 0.00 | 1 | 0.01 | 1 | 0.02 | 0 | 0.00 | 2 | 0.01 | 1 | 0.01 | 3 | < 0.01 | ||
| HFE*001:unofficial:055 | C-C-C-G-C-G | 13 | 0.07 | 11 | 0.09 | 16 | 0.11 | 71 | 0.15 | 0 | 0.00 | 23 | 0.11 | 12 | 0.17 | 59 | 0.10 | ||
| HFE*001:unofficial:065 | C-C-G-G-T-Del | 1 | 0.01 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 1 | < 0.01 | ||
| HFE*0024 | G-C-C-G-T-G | 35 | 0.17 | 16 | 0.13 | 20 | 0.14 | 91 | 0.19 | 3 | 0.11 | 32 | 0.15 | 8 | 0.11 | 92 | 0.15 | ||
| HFE*0034 | C-T-C-A-T-G | 8 | 0.04 | 3 | 0.02 | 3 | 0.02 | 3 | 0.06 | 202 | 0.71 | 26 | 0.12 | 4 | 0.06 | 40 | 0.06 | ||
| HFE*0044 | G-C-C-A-T-G | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 1 | 0.02 | 0 | 0.00 | 1 | 0.00 | 0 | 0.00 | 1 | < 0.01 | ||
| Number of alleles | 200 | 118 | 142 | 48 | 28 | 218 | 72 | 608 | |||||||||||
All population samples adhered to the HWE, except HCC patients (IO+) at the C282YG>A variation site (P = 0.031). Overall, patients and healthy controls shared the same most frequent alleles at each SNP, except when HH patients were compared to healthy controls, for whom the C282Y-A (ƒ = 0.714) allele was the most frequently observed, significantly associated with susceptibility to HH (P < 0.001; OR = 53.06; 95%CI: 18.41-152.90). The C282Y-G allele was protective against HH (P < 0.001; OR = 0.01; 95%CI: 0.006-0.05). On the other hand, when the genotype frequencies were compared between HH patients and healthy controls several differences were observed. The IVS2(+4)-TT genotype was associated with susceptibility to HH (P = 0.04, OR = 3.91; 95%CI: 1.14-13.34). The C282Y-GG genotype was associated with protection against HH (P < 0.001; OR = 0.007; 95%CI: 0.0008-0.065), while the C282Y-AA genotype was associated with susceptibility to HH (P < 0.001; OR = 201.00; 95%CI: 10.44-3,871) (Table 1).
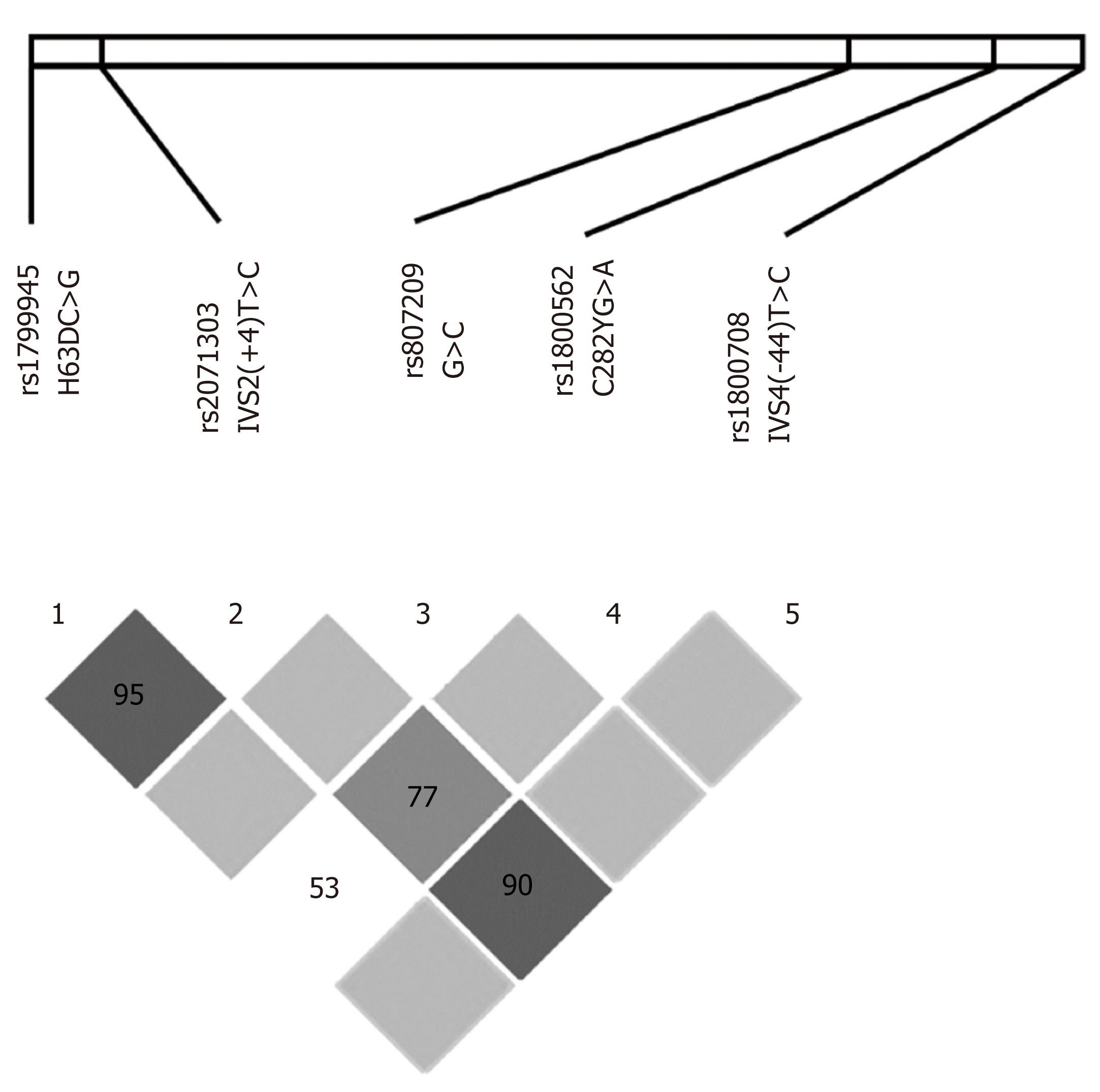
The most remarkable LD among these loci included: (1) H63DC>G and IVS2(+4)T<C in almost all groups analyzed separately and in the whole population; (2) IVS2(+4)T>C and IVS4(-44)T>C in most patient samples; and (3) IVS2(+4)T>C and C282YG>A in the HH population (Table S1). The Haploview software was used to analyze and visualize the patterns of linkage disequilibrium observed in these data and confirmed the strong LD between H63DC>G and IVS2(+4)T>C (D’ = 95) and IVS2(+4)T>C and IVS4(-44)T>C (D’ = 90), and a less strong linkage between IVS2(+4)T>C and C282YG>A (D’ = 77) (Figure 2). Therefore, the most relevant SNPs in LD with each other were H63DC>G, IVS2(+4)T>C, IVS4(-44)T>C and C282YG>A. Considering that: (1) H63DC>G and IVS2(+4)T>C were in LD in almost all analyses; (2) H63DC>G and C282YG>A presented LD only in HH patients; and (3) H63DC>G and C282YG>A polymorphic sites were frequently associated with susceptibility to HH in the literature, a third LD approach was performed, analyzing only HH and healthy control individuals to evaluate specifically-linked alleles and the strength of these associations. Accordingly, in both healthy controls and HH populations, a remarkable LD between the H63DC>G and IVS2(+4)T>C was observed (D' = 1.000 in both analyses, and r2 = 0.3318 and 0.7200, respectively). In both samples, a complete LD of H63D-G and IVS2(+4)-C was detected as well as an absence of the recombinant H63D-G in linkage with the IVS2(+4)-T. Another relevant result was the linkage of both mutant H63D-G and IVS(+4)-C mutants with the C282Y-G (D' = 1.000 in both analyses, and r2 = 0.3000 and 0.4267, respectively) (Table S2).
The reconstruction of the meiotic phase generated nine alleles, included into four major allele groups (HFE*001 to *004), as standardized by IMGT[10]. These allele groups encoded four distinct proteins (HFE*001 to *004) on the basis of polymorphic sites along the coding region, encompassing the H63DC>G (exon 2), IVS2(+4)T>C (intron 2), rs807209 (G>C intron 3), C282YG>A (exon 4) IVS4(-44)T>C (intron 4) and the new mutation (G>DEL at intron 5) (Table 2).
The HFE*001:01:01 was the most frequently observed allele in all studied populations (f varying from 48-63%), except in HH patients (f = 14%). In contrast, the HFE*003 allele was underrepresented in all studied populations (f varying from 2%-12%), except in HH patients (f = 71%). Therefore, the HFE*001, containing the H63D-C; IVS2(+4)-T; rs807209-C; C282Y-G; IVS4(–44)-T variation sites (from 5’ to 3’), conferred protection against the development of HH (P < 0.0001, OR = 0.14) and the HFE*003 allele, containing the H63D-C; IVS2(+4)-T; rs807209-C; C282Y-A; IVS4(–44)-T (from 5’ to 3’), conferred a high risk for HH development (P < 0.0001, OR = 60.00). The HFE*001/HFE*003 (P = 0.03, OR = 7.2) and HFE*003/HFE*003 (P < 0.001, OR = 174.20) genotypes, both containing the HFE*003 allele, were also overrepresented in HH patients. On the other hand, the HFE*001/HFE*002 genotype was associated with the development of HCC (P = 0.02, OR = 14.14) in patients exhibiting the underlying HCV infection and iron overload (HCC HCV-IO+).
The major histocompatibility complex (MHC) LD analysis was performed using two approaches: (1) considering HFE alleles (Table S3); and (2) considering separately the two HFE SNPs most frequently reported in association with HH (H63DC>G and C282YG>A) (Table 3). Considering the first approach, no LD was observed between HFE alleles and MHC alleles, except for H63DC>G and HLA-B locus (P = 0.03), showing a weak association between H63DC>G and HLA-B*44 (Table 3). We also observed an absence of LD between the classical C282YG>A SNP and HLA-A, HLA-B, HLA-C, 14bp HLA-G, TNFa-d microsatellites. Since the variation site IVS2(+4)T>C is located only 157bp downstream from the H63DC>G site and since these loci are in LD, the IVS2(+4)T>C would be a good candidate to be analyzed regarding the disequilibrium between HFE and HLA-B genes (Table 4). The weak HLA-B associations were confirmed.
| Observed frequency | Expected frequency | Standardized value of disequilibrium(D’) | Standardized value of correlation(r2) | QUI2value | P value of QUI2 | |||||
| HLA-B | HFE H63DC>G | |||||||||
| C | G | C | G | C | G | |||||
| 07 | 16 | 2 | 14.83 | 3.17 | 0.3683 | -0.3683 | 0.0029 | 0.5728 | 0.4492 | |
| 08 | 12 | 1 | 10.71 | 2.29 | 0.5626 | -0.5626 | 0.0047 | 0.9396 | 0.3324 | |
| 13 | 3 | 1 | 3.30 | 0.70 | -0.0899 | 0.0899 | 0.0008 | 0.1547 | 0.6941 | |
| 14 | 10 | 0 | 8.24 | 1.76 | 1.0000 | -1.0000 | 0.0113 | 2.2471 | 0.1339 | |
| 15 | 17 | 0 | 14.01 | 2.99 | 1.0000 | -1.0000 | 0.0199 | 3.9669 | 0.0464 | |
| 18 | 10 | 0 | 8.24 | 1.76 | 1.0000 | -1.0000 | 0.0113 | 2.2471 | 0.1339 | |
| 27 | 2 | 2 | 3.30 | 0.70 | -0.3933 | 0.3933 | 0.0149 | 2.9586 | 0.0854 | |
| 35 | 17 | 4 | 17.31 | 3.69 | -0.0177 | 0.0177 | 0.0002 | 0.0345 | 0.8526 | |
| 37 | 1 | 2 | 2.47 | 0.53 | -0.5955 | 0.5955 | 0.0254 | 5.0617 | 0.0245 | |
| 38 | 3 | 0 | 2.47 | 0.53 | 1.0000 | -1.0000 | 0.0033 | 0.6500 | 0.4201 | |
| 39 | 4 | 2 | 4.94 | 1.06 | -0.1911 | 0.1911 | 0.0053 | 1.0582 | 0.3036 | |
| 40 | 3 | 0 | 2.47 | 0.53 | 1.0000 | -1.0000 | 0.0033 | 0.6500 | 0.4201 | |
| 41 | 3 | 0 | 2.47 | 0.53 | 1.0000 | -1.0000 | 0.0033 | 0.6500 | 0.4201 | |
| 42 | 1 | 0 | 0.82 | 0.18 | 1.0000 | -1.0000 | 0.0011 | 0.2145 | 0.6433 | |
| 44 | 8 | 11 | 15.66 | 3.34 | -0.4891 | 0.4891 | 0.11831 | 23.5443 | < 0.00001 | |
| 45 | 6 | 0 | 4.94 | 1.06 | 1.0000 | -1.0000 | 0.0066 | 1.3203 | 0.2505 | |
| 48 | 2 | 0 | 1.65 | 0.35 | 1.0000 | -1.0000 | 0.0022 | 0.4312 | 0.5114 | |
| 49 | 8 | 0 | 6.59 | 1.41 | 1.0000 | -1.0000 | 0.0089 | 1.7788 | 0.1823 | |
| 50 | 2 | 0 | 1.65 | 0.35 | 1.0000 | -1.0000 | 0.0022 | 0.4312 | 0.5114 | |
| 51 | 9 | 5 | 11.54 | 2.46 | -0.2199 | 0.2199 | 0.0172 | 3.4137 | 0.0647 | |
| 52 | 7 | 0 | 5.77 | 1.23 | 1.0000 | -1.0000 | 0.0078 | 1.5484 | 0.2134 | |
| 53 | 5 | 0 | 4.12 | 0.88 | 1.0000 | -1.0000 | 0.0055 | 1.0946 | 0.2955 | |
| 55 | 2 | 0 | 1.65 | 0.35 | 1.0000 | -1.0000 | 0.0022 | 0.4312 | 0.5114 | |
| 56 | 0 | 1 | 0.82 | 0.18 | -1.0000 | 1.0000 | 0.0237 | 4.7094 | 0.0300 | |
| 57 | 8 | 2 | 8.24 | 1.76 | -0.0293 | 0.0293 | 0.0002 | 0.0423 | 0.8371 | |
| 58 | 2 | 4.94 | 1.06 | -0.1911 | 0.1911 | 0.0053 | 1.0582 | 0.3036 | ||
| 67 | 1 | 0 | 0.82 | 0.18 | 1.0000 | -1.0000 | 0.0011 | 0.2145 | 0.6433 | |
| Observed frequency | Expected frequency | Standardized value of disequilibrium(D’) | Standardized value of correlation(r2) | QUI2value | P value of QUI2 | ||||
| HLA-B | HFE IVS2(+4)T>C | ||||||||
| T | C | T | C | T | C | ||||
| 07 | 13 | 5 | 10.98 | 7.02 | 0.2877 | -0.2877 | 0.0052 | 1.0471 | 0.3062 |
| 08 | 8 | 5 | 7.93 | 5.07 | 0.0138 | -0.0138 | 0.0000 | 0.0017 | 0.9672 |
| 13 | 4 | 0 | 2.44 | 1.56 | 1.0000 | -1.0000 | 0.0130 | 2.6096 | 0.1062 |
| 14 | 4 | 6 | 6.10 | 3.90 | -0.3443 | 0.3443 | 0.0098 | 1.9513 | 0.1624 |
| 15 | 11 | 6 | 10.37 | 6.63 | 0.0950 | -0.0950 | 0.0005 | 0.1073 | 0.7433 |
| 18 | 8 | 2 | 6.10 | 3.90 | 0.4872 | -0.4872 | 0.0080 | 1.5973 | 0.2063 |
| 27 | 1 | 3 | 2.44 | 1.56 | -0.5902 | 0.5902 | 0.0111 | 2.2235 | 0.1359 |
| 35 | 17 | 4 | 12.81 | 8.19 | 0.5116 | -0.5116 | 0.0196 | 3.9264 | 0.0475 |
| 37 | 0 | 3 | 1.83 | 1.17 | -1.0000 | 1.0000 | 0.0238 | 4.7638 | 0.0291 |
| 38 | 2 | 1 | 1.83 | 1.17 | 0.1453 | -0.1453 | 0.0002 | 0.0411 | 0.8393 |
| 39 | 1 | 5 | 3.66 | 2.34 | -0.7268 | 0.7268 | 0.0256 | 5.1103 | 0.0238 |
| 40 | 3 | 0 | 1.83 | 1.17 | 1.0000 | -1.0000 | 0.0097 | 1.9472 | 0.1629 |
| 41 | 2 | 1 | 1.83 | 1.17 | 0.1453 | -0.1453 | 0.0002 | 0.0411 | 0.8393 |
| 42 | 1 | 0 | 0.61 | 0.39 | 1.0000 | -1.0000 | 0.0032 | 0.6426 | 0.4228 |
| 44 | 6 | 13 | 11.59 | 7.41 | -0.4823 | 0.4823 | 0.03821 | 7.6388 | 0.0057 |
| 45 | 3 | 3 | 3.66 | 2.34 | -0.1803 | 0.1803 | 0.0016 | 0.3146 | 0.5749 |
| 48 | 2 | 0 | 1.22 | 0.78 | 1.0000 | -1.0000 | 0.0065 | 1.2916 | 0.2558 |
| 49 | 8 | 0 | 4.88 | 3.12 | 1.0000 | -1.0000 | 0.0266 | 5.3279 | 0.0210 |
| 50 | 2 | 0 | 1.22 | 0.78 | 1.0000 | -1.0000 | 0.0065 | 1.2916 | 0.2558 |
| 51 | 10 | 4 | 8.54 | 5.46 | 0.2674 | -0.2674 | 0.0034 | 0.6882 | 0.4068 |
| 52 | 3 | 4 | 4.27 | 2.73 | -0.2974 | 0.2974 | 0.0050 | 1.0037 | 0.3164 |
| 53 | 5 | 0 | 3.05 | 1.95 | 1.0000 | -1.0000 | 0.0164 | 3.2787 | 0.0702 |
| 55 | 0 | 2 | 1.22 | 0.78 | -1.0000 | 1.0000 | 0.0158 | 3.1598 | 0.0755 |
| 56 | 1 | 0 | 0.61 | 0.39 | 1.0000 | -1.0000 | 0.0032 | 0.6426 | 0.4228 |
| 57 | 4 | 6 | 6.10 | 3.90 | -0.3443 | 0.3443 | 0.0098 | 1.9513 | 0.1624 |
| 58 | 1 | 5 | 3.66 | 2.34 | -0.7268 | 0.7268 | 0.0256 | 5.1103 | 0.0238 |
| 67 | 1 | 0 | 0.61 | 0.39 | 1.0000 | -1.0000 | 0.0032 | 0.6426 | 0.4228 |
The frequency of the H63D-G allele in healthy controls varies from 7.9% to 17.5% in worldwide populations, exhibiting high frequencies in Netherlands and Iberian Peninsula (around 20%)[13,14]. The frequency of the C282Y-A allele decreases from North (4%-10%) to South Europe (0%-3%)[15], and in populations without a high European genetic ancestry, the frequency of this allele is negligible. The frequency of the C282Y-A allele in our healthy control series, as well as in other Southern Brazilian samples[16-18], is closely similar to South European populations, indicating the European ancestry influenced on the Brazilian gene pool. The mutant S65C-T allele is observed at low frequency (0-1%) in European populations[19-21], as well as in the Brazilian population[22,23] (absent in our samples – data not shown).
Although the IVS2(+4)T>C SNP does not change protein sequence, it is in LD with H63DC>G, C282YG>A and IVS2(+4)-T alleles. Considering that IVS2(+4)-T allele is increased in HH population, and considering that this allele is only 157bp distant from the H63D-G allele, this association probably reflects a hitch-hiking effect, and possibly does not present biological significance in the susceptibility to HH. Indeed, de Lucas et al[24] reported that HH patients presenting homozygosis for the C282Y-A allele did not exhibit the IVS2(+4)-C allele, indicating that the presence of the C282Y-A allele excludes the presence of IVS2(+4)-C allele in the same haplotype. Therefore, the sole analysis of the allelic frequency of the IVS2(+4)T>C SNP is not adequate to evaluate HH susceptibility, since the frequency of the C282Y-A allele is high in HH patients, and consequently, there is a high frequency of IVS2(+4)-T allele in the same sample (Table 1). The C282Y-A allele and the AA genotype have been associated with susceptibility to HH patients[24,25], including the HH patients of this study and other Brazilian HH populations[23]. Although the HH cohort is small, the mutated AA genotype appeared in high frequency in patients and was not observed in the healthy control group. The C282Y-G allele and the GG genotype have been associated with protection against HH development in various worldwide populations[21]. The H63D-G allele and the GG genotype have been associated with HH in European and North American patients[1,25]. However, these associations were not observed in ours nor in other HH Brazilian samples[26].
The role of H63DC>G and C282YG>A variation sites in acquired IO disorders is controversial. Apart from HH, no other association involving such polymorphisms was observed in the present study. A previous study evaluating chronic hepatitis C patients reported an association between HFE mutations (H63DC>G and C282YG>A) and elevated serum transferrin saturation, but not with liver iron deposits[5]. On the other hand, some authors have observed an increased prevalence of C282YG>A mutation in hepatitis C patients from North England[27], Austria[28], and North America[29]. These studies have shown an association between HFE mutations and higher serum iron indices and liver iron deposits, especially for C282Y homozygotes. In contrast, another study did not show association between HFE mutations and liver iron deposits[30].
The association between the C282YG>A mutation and the HCC risk is also still controversial. HH is a condition characterized by hepatic IO, leading to higher cancer incidence[31]. However, the role of moderate liver IO and of the carriage of HFE mutations on the HCC risk remains unclear. Some studies have shown higher prevalence of the C282YG>A mutation in patients with HCC compared with cirrhotic patients without HCC[32], whereas other studies found no association between HFE and HCC[33]. Additionally, another study reported an association between liver IO and C282YG>A with a higher risk of HCC in patients with alcoholic but not with HCV-related cirrhosis[34].
The HFE*001 allele was underrepresented, while the HFE*003 was overrepresented in HH patients of this series. These findings corroborate the importance of the C282YG>A SNP on the susceptibility to HH, since only the HFE*003 allele has an Adenine at this position (C282Y-A), which is the unique difference between both alleles. In addition, the HFE*001/HFE*003 and HFE*003/HFE*003 genotypes were also significantly associated with high risk for HH development. The homozygosis for the HFE*003 allele group, which was not observed in the healthy control population, drastically increased the susceptibility to HH. Indeed, the HFE*003 allele was present in 13 out of 14 patients and its presence in double doses was observed in 7 out of 14 HH patients.
In relation to acquired diseases exhibiting IO, the HFE*001/HFE*002 genotype was overrepresented in HCC patients exhibiting HCV infection and IO. When the HFE SNPs were analyzed separately, no significant differences were observed. Noteworthy, these results indicate that these populations are heterogeneous and in some circumstances represented small groups.
HH was initially associated with the HLA-A3, HLA-A14 and HLA-B14 antigens[35]. Microsatellite evaluations pointed out a susceptibility locus for HH. This locus was initially named as HLA-H[25], which is the same name of a pseudogene, located close to HLA-A, stressing the disequilibrium concept between HLA-A/B genes and the HH locus. Later, this HH locus was renamed HFE to put an end on this ambiguity[36]. Considering the great distance between the HFE and HLA-A, -B and -C loci, strong LD between these genes are not expected; however, some studies reported LD between H63DC>G and C282YG>A SNPs with HLA-A and HLA-B alleles. Taking advantage of the fact that our healthy control population was typed for ten additional MHC loci, LD between HFE and all these loci was evaluated.
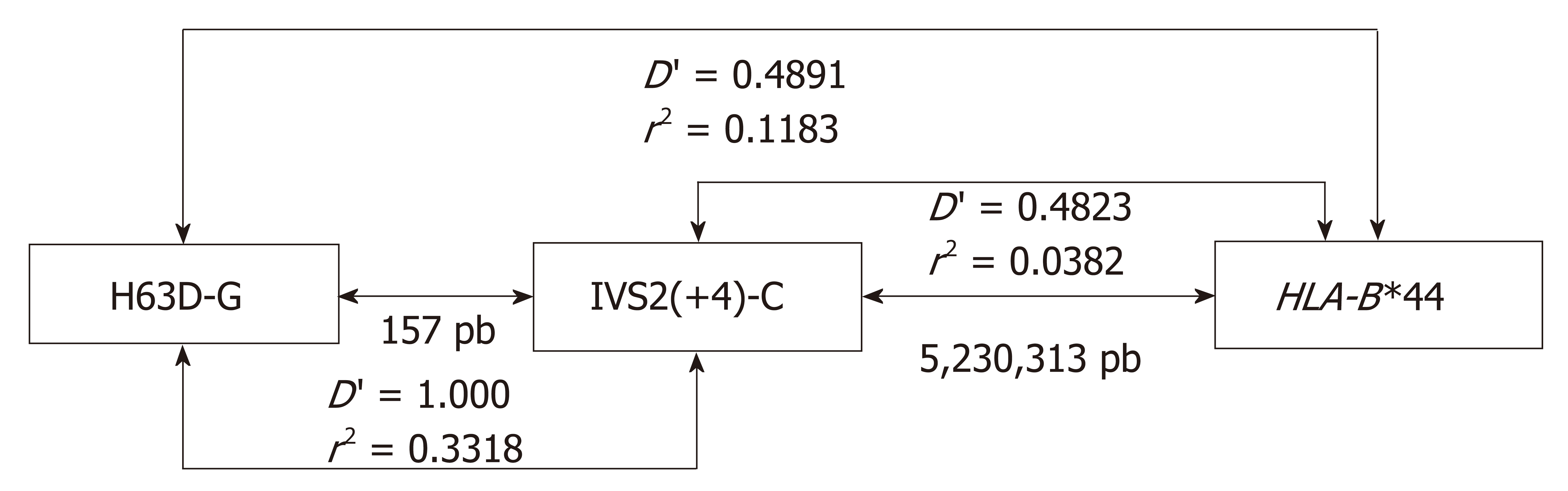
The pairwise test detected no disequilibrium between the HFE alleles and other MHC loci (Table S3), which is in agreement with the argument that the HFE gene is far from the other loci tested. When LD analyses were performed evaluating the H63DC>G and C282YG>A SNPs, a significant disequilibrium between the H63DC>G and HLA-B (P = 0.03) was observed, encompassing HLA-B*15/H63D-C, HLA-B*37/H63D-G, HLA-B*44/H63D-G and HLA-B*56/H63D-G alleles (Table 3), being stronger for HLA-B*15/H63D-C and HLA-B*56/H63D-G alleles (D' = 1). Since HLA-B locus is multiallelic, H63DC>G is biallelic, and H63D-G is rare, it is possible that not all H63DC>G/HLA-B haplotypes were represented in our CTL. In addition, the recombination coefficient, which indicates the power of the correlation between alleles, was weak for all these combinations, except for the HLA-B*44/H63D-G (r2 = 0.11) (Figure 3 and Table 3) which was much stronger than in the other combinations (r2 = 0.01-0.02). Most likely, this HLA-B*44/H63D-G disequilibrium has a historical origin.
Since the IVS2(+4)T>C SNP exhibited a significant LD with the H63DC>G SNP, as we discussed before, and considering that both SNPs are located at a relatively short distance, we further evaluated the LD between this SNP and HLA-B, which showed similar results: HLA-B*35/IVS2(+4)-T; HLA-B*37/IVS2(+4)-C; HLA-B*44/IVS2(+4)-C; HLA-B*49/IVS2(+4)-T and HLA-B*58/IVS2(+4)-C. The analyses of LD between HLA-B alleles and IVS2(+4)T>C and H63DC>G showed that HLA-B*37 and B*44 exhibited weaker correlations in relation to H63DC>G (r2 = 0.02 and 0.03, respectively) (Table 4). This analysis resulted on the identification of the extended H63D-G/IVS2(+4)-C/HLA-B*44 haplotype (Figure 3).
Regarding genetic studies in patients with IO, HLA-B*44 and C282Y-A alleles are reported to be overrepresented in patients with HH[1] or in patients with acquired diseases associated with IO[37], however, without reaching significance. Since haplotypes containing HLA-B*44 are common in Europe, West and North Africa, and in North-American Caucasians[38], there is a high probability of overrepresentation of the H63D-G/HLA-B*44 haplotype in these populations. Although the present study revealed that C282Y-A is not a part of this extended haplotype, the mentioned associations suggest an independent role of H63D-G and C282Y-A on HH susceptibility.
In conclusion, this study systematically reports variation sites along the HFE gene using HFE allelic official nomenclature, previously described by our group. The HFE*003 was frequently observed in HH patients, whereas the HFE*001 was frequently observed in healthy controls. The HFE*001/HFE*002 genotype was identified as a risk factor for HCC HCV patients exhibiting IO. Even if a strong LD has been observed among the H63D-G, IVS2(+4)-C and C282Y-G alleles, particularly in HH patients, the mutation IVS2(+4)T>C was not directly associated with HH susceptibility. Although the HFE gene is distant from other MHC genes, the HFE H63D-G/IVS2(+4)-C alleles were in weak LD with the HLA-B*44 allele.
HFE gene controls the iron uptake from gut, and defects of the encoded molecule have been associated with iron overload (IO), particularly in hemochromatosis hereditary (HH), which can cause serious damage to the liver. Besides HH, patients with hepatitis C virus (HCV) and hepatocellular carcinoma (HCC) may or not develop IO.
The search for markers associated with IO may be very useful for the early diagnosis of these patients, which is essential for their survival.
The main objectives of this work is to identify associations between HFE coding region variable sites in patients exhibiting HH and in diseases associated with acquired IO.
We sequenced exons 2 to 5 and boundary introns of the HFE gene to evaluate all polymorphic sites in patients presenting HH or acquired IO (HCV and HCC), and in healthy controls, using Sanger sequencing. We also determined the extended haplotype in healthy controls, including other major histocompatibility genes (HLA-A/-B/-C/-DRB1/-DQB1 alleles, and HLA-G 14bp INDEL and TNFa-d microsatellites). Haplotype reconstruction was performed using the Arlequin and Phase softwares, and linkage disequilibrium (LD) between histocompatibility loci and HFE gene was performed using the Haploview software.
The HFE*003 allele was overrepresented (f = 71%) and HFE*001 allele was underrepresented (f = 14%) in HH patients compared to all groups. A strong LD was observed among the previously reported H63D-G, IVS2(+4)-C and C282Y-G gene variants, particularly in HH; however, the mutation IVS2(+4)T>C was not associated with HH susceptibility. The HFE*001/HFE*002 genotype conferred susceptibility to HCC in HCV patients exhibiting IO (P = 0.02, OR = 14.14). Although HFE is telomeric to other histocompatibility genes, the H63D-G/IVS2(+4)-C (P ≤ 0.00001/P ≤ 0.0057) combination was in LD with HLA-B*44 allele group in healthy controls.
This study systematically evaluated variation sites along the HFE gene using the HLA official nomenclature, previously described by our group. The HFE*003 allele that was overrepresented in HH patients encompasses major variation sites previously described in association with HH in several worldwide populations, in contrast with the HFE*001 allele which does not present HH-associated variation sites and predominates among healthy controls. On the other hand, the HFE*001/HFE*002 genotype was identified as a risk factor for HCC and HCV patients exhibiting IO. Although the HFE gene is distant from other histocompatibility genes, the HFE H63D-G/IVS2(+4)-C alleles were in weak LD with the HLA-B*44 allele. Thus, a differential HFE association was observed for HH and for diseases associated with acquired IO (HCV, HCC).
Besides the identification of markers associated with IO, which may permit an early detection of patients prone to develop iron deposits, the knowledgement of the major gene associated with iron uptake may help on the understanding of the IO pathogenesis.
We thank Flavia Tremeschin de Almeida Vieira and Sandra Silva Rodrigues dos Santos for technical support.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and Hepatology
Country of origin: Brazil
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Luo GH, Mekky MA S- Editor: Ma YJ L- Editor: A E- Editor: Zhang YL
| 1. | Barton JC, Acton RT. HLA-A and -B alleles and haplotypes in hemochromatosis probands with HFE C282Y homozygosity in central Alabama. BMC Med Genet. 2002;3:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 2. | Anderson GJ, Ramm GA, Subramaniam VN, Powell LW. HFE gene and hemochromatosis. J Gastroenterol Hepatol. 2004;19:712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 3. | Racchi O, Mangerini R, Rapezzi D, Gaetani GF, Nobile MT, Picciotto A, Ferraris AM. Mutations of the HFE gene and the risk of hepatocellular carcinoma. Blood Cells Mol Dis. 1999;25:350-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 4. | Desmet VJ, Gerber M, Hoofnagle JH, Manns M, Scheuer PJ. Classification of chronic hepatitis: diagnosis, grading and staging. Hepatology. 1994;19:1513-1520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1582] [Cited by in RCA: 1506] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 5. | Martinelli AL, Franco RF, Villanova MG, Figueiredo JF, Secaf M, Tavella MH, Ramalho LN, Zucoloto S, Zago MA. Are haemochromatosis mutations related to the severity of liver disease in hepatitis C virus infection? Acta Haematol. 2000;102:152-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 6. | Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5972] [Cited by in RCA: 6569] [Article Influence: 469.2] [Reference Citation Analysis (1)] |
| 7. | Brissot P, Bourel M, Herry D, Verger JP, Messner M, Beaumont C, Regnouard F, Ferrand B, Simon M. Assessment of liver iron content in 271 patients: a reevaluation of direct and indirect methods. Gastroenterology. 1981;80:557-565. [PubMed] |
| 8. | Sciot R, van Eyken P, Facchetti F, Callea F, van der Steen K, van Dijck H, van Parys G, Desmet VJ. Hepatocellular transferrin receptor expression in secondary siderosis. Liver. 1989;9:52-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 9. | Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463-5467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40145] [Cited by in RCA: 46842] [Article Influence: 975.9] [Reference Citation Analysis (0)] |
| 10. | Campos WN, Massaro JD, Martinelli ALC, Halliwell JA, Marsh SGE, Mendes-Junior CT, Donadi EA. HFE gene polymorphism defined by sequence-based typing of the Brazilian population and a standardized nomenclature for HFE allele sequences. HLA. 2017;90:238-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (1)] |
| 11. | Castelli EC, Mendes-Junior CT, Deghaide NH, de Albuquerque RS, Muniz YC, Simões RT, Carosella ED, Moreau P, Donadi EA. The genetic structure of 3'untranslated region of the HLA-G gene: polymorphisms and haplotypes. Genes Immun. 2010;11:134-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 170] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 12. | Udalova IA, Nedospasov SA, Webb GC, Chaplin DD, Turetskaya RL. Highly informative typing of the human TNF locus using six adjacent polymorphic markers. Genomics. 1993;16:180-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 201] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 13. | Cardoso CS, de Sousa M. HFE, the MHC and hemochromatosis: paradigm for an extended function for MHC class I. Tissue Antigens. 2003;61:263-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | Porto G, de Sousa M; Variation of hemochromatosis prevalence and genotype in national groups. Hemochromatosis: Genetics, pathophysiology, diagnosis and treatment . Cambridge, 2000:51-62. Available from: URL: https://max.book118.com/html/2018/0213/152915262.shtm. |
| 15. | Trifa AP, Popp RA, Militaru MS, Farcaş MF, Crişan TO, Gana I, Cucuianu A, Pop IV. HFE gene C282Y, H63D and S65C mutations frequency in the Transylvania region, Romania. J Gastrointestin Liver Dis. 2012;21:177-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 16. | Bueno S, Duch CR, Figueiredo MS. Mutations in the HFE gene (C282Y, H63D, S65C) in a Brazilian population. Rev Bras Hematol Hemoter. 2006;28:293–295. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 17. | Santos PC, Cançado RD, Terada CT, Rostelato S, Gonzales I, Hirata RD, Hirata MH, Chiattone CS, Guerra-Shinohara EM. HFE gene mutations and iron status of Brazilian blood donors. Braz J Med Biol Res. 2010;43:107-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 18. | de Lima Santos PC. Hemocromatose hereditária: associação entre as mutações no gene HFE e o estado de ferro em doadores de sangue e pesquisa de mutações nos genes HFE, HJV, HAMP, TFR2 e SLC40A1 em pacientes com sobrecarga de ferro primária. São Paulo: Universidade de São Paulo 2010; . [DOI] [Full Text] |
| 19. | Torres FR, Souza-Neiras WC, D'Almeida Couto AA, D'Almeida Couto VS, Cavasini CE, Rossit AR, Machado RL, Bonini-Domingos CR. Frequency of the HFE C282Y and H63D polymorphisms in Brazilian malaria patients and blood donors from the Amazon region. Genet Mol Res. 2008;7:60-64. [PubMed] |
| 20. | Mura C, Raguenes O, Férec C. HFE mutations analysis in 711 hemochromatosis probands: evidence for S65C implication in mild form of hemochromatosis. Blood. 1999;93:2502-2505. [PubMed] |
| 21. | Merryweather-Clarke AT, Pointon JJ, Jouanolle AM, Rochette J, Robson KJ. Geography of HFE C282Y and H63D mutations. Genet Test. 2000;4:183-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 217] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 22. | Cançado RD, Guglielmi ACO, Vergueiro CS V, Rolim EG, Figueiredo MS, Chiattone CS. Estudo das mutações C282Y, H63D e S65C do gene HFE em doentes brasileiros com sobrecarga de ferro. Rev Bras Hematol Hemoter. 2007;29:351–360. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 23. | Cançado RD, Guglielmi AC, Vergueiro CS, Rolim EG, Figueiredo MS, Chiattone CS. Analysis of HFE gene mutations and HLA-A alleles in Brazilian patients with iron overload. Sao Paulo Med J. 2006;124:55-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 24. | de Lucas AP, Fulgencio MG, Robles JM, Sierra EM, del Rey Cerros MJ, Perez PM. Is the IVS2+4T>C variant of the HFE gene a splicing mutation or a polymorphism? A study in the Spanish population. Genet Med. 2005;7:212-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Prass CE, Quintana L, Starnes SM, Schatzman RC, Brunke KJ, Drayna DT, Risch NJ, Bacon BR, Wolff RK. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2676] [Cited by in RCA: 2550] [Article Influence: 87.9] [Reference Citation Analysis (0)] |
| 26. | Lok CY, Merryweather-Clarke AT, Viprakasit V, Chinthammitr Y, Srichairatanakool S, Limwongse C, Oleesky D, Robins AJ, Hudson J, Wai P, Premawardhena A, de Silva HJ, Dassanayake A, McKeown C, Jackson M, Gama R, Khan N, Newman W, Banait G, Chilton A, Wilson-Morkeh I, Weatherall DJ, Robson KJ. Iron overload in the Asian community. Blood. 2009;114:20-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 66] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 27. | Smith BC, Gorve J, Guzail MA, Day CP, Daly AK, Burt AD, Bassendine MF. Heterozygosity for hereditary hemochromatosis is associated with more fibrosis in chronic hepatitis C. Hepatology. 1998;27:1695-1699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 144] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 28. | Kazemi-Shirazi L, Datz C, Maier-Dobersberger T, Kaserer K, Hackl F, Polli C, Steindl PE, Penner E, Ferenci P. The relation of iron status and hemochromatosis gene mutations in patients with chronic hepatitis C. Gastroenterology. 1999;116:127-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 100] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 29. | Bonkovsky HL, Troy N, McNeal K, Banner BF, Sharma A, Obando J, Mehta S, Koff RS, Liu Q, Hsieh CC. Iron and HFE or TfR1 mutations as comorbid factors for development and progression of chronic hepatitis C. J Hepatol. 2002;37:848-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 67] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 30. | Höhler T, Leininger S, Köhler HH, Schirmacher P, Galle PR. Heterozygosity for the hemochromatosis gene in liver diseases--prevalence and effects on liver histology. Liver. 2000;20:482-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 31. | Fargion S, Mandelli C, Piperno A, Cesana B, Fracanzani AL, Fraquelli M, Bianchi PA, Fiorelli G, Conte D. Survival and prognostic factors in 212 Italian patients with genetic hemochromatosis. Hepatology. 1992;15:655-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 126] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 32. | Hellerbrand C, Pöppl A, Hartmann A, Schölmerich J, Lock G. HFE C282Y heterozygosity in hepatocellular carcinoma: evidence for an increased prevalence. Clin Gastroenterol Hepatol. 2003;1:279-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | Boige V, Castéra L, de Roux N, Ganne-Carrié N, Ducot B, Pelletier G, Beaugrand M, Buffet C. Lack of association between HFE gene mutations and hepatocellular carcinoma in patients with cirrhosis. Gut. 2003;52:1178-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 34. | Nahon P, Sutton A, Rufat P, Ziol M, Thabut G, Schischmanoff PO, Vidaud D, Charnaux N, Couvert P, Ganne-Carrie N, Trinchet JC, Gattegno L, Beaugrand M. Liver iron, HFE gene mutations, and hepatocellular carcinoma occurrence in patients with cirrhosis. Gastroenterology. 2008;134:102-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 92] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 35. | Simon M, Bourel M, Fauchet R, Genetet B. Association of HLA-A3 and HLA-B14 antigens with idiopathic haemochromatosis. Gut. 1976;17:332-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 270] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 36. | Wain HM, White JA, Bruford E, Povey S. Hemochromatosis gene nomenclature. Am J Med Genet. 2000;93:77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 37. | Porto G, Alves H, Rodrigues P, Cabeda JM, Portal C, Ruivo A, Justiça B, Wolff R, De Sousa M. Major histocompatibility complex class I associations in iron overload: evidence for a new link between the HFE H63D mutation, HLA-A29, and non-classical forms of hemochromatosis. Immunogenetics. 1998;47:404-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 43] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 38. | Arnaiz-Villena A, Martínez-Laso J, Gómez-Casado E, Díaz-Campos N, Santos P, Martinho A, Breda-Coimbra H. Relatedness among Basques, Portuguese, Spaniards, and Algerians studied by HLA allelic frequencies and haplotypes. Immunogenetics. 1997;47:37-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 85] [Article Influence: 3.1] [Reference Citation Analysis (0)] |