Published online Jan 27, 2018. doi: 10.4254/wjh.v10.i1.22
Peer-review started: October 7, 2017
First decision: November 23, 2017
Revised: November 28, 2017
Accepted: December 28, 2017
Article in press: December 29, 2017
Published online: January 27, 2018
Processing time: 111 Days and 21.8 Hours
To develop appropriate humanized three-dimensional ex-vivo model system for drug testing.
Bioengineered humanized livers were developed in this study using human hepatic stem cells repopulation within the acellularized liver scaffolds which mimics with the natural organ anatomy and physiology. Six cytochrome P-450 probes were used to enable efficient identification of drug metabolism in bioengineered humanized livers. The drug metabolism study in bioengineered livers was evaluated to identify the absorption, distribution, metabolism, excretion and toxicity responses.
The bioengineered humanized livers showed cellular and molecular characteristics of human livers. The bioengineered liver showed three-dimensional natural architecture with intact vasculature and extra-cellular matrix. Human hepatic cells were engrafted similar to the human liver. Drug metabolism studies provided a suitable platform alternative to available ex-vivo and in vivo models for identifying cellular and molecular dynamics of pharmacological drugs.
The present study paves a way towards the development of suitable humanized preclinical model systems for pharmacological testing. This approach may reduce the cost and time duration of preclinical drug testing and further overcomes on the anatomical and physiological variations in xenogeneic systems.
Core tip: Liver is the central organ for absorption, distribution, metabolism, excretion and toxicity (ADMET) of pharmacological drugs and molecules. Available in vitro and in vivo preclinical models deals with several limitations including xenogeneic barrier, lack of natural humanized liver architecture and functional responses. Bioengineered humanized livers developed in present study can overcome on such limitations. This humanized liver model system provides better platform which could be used more efficiently to screen the ADMET of several pipeline drugs and other pharmacological molecules. This approach could reduce the time and cost of the total drug screening experiments as compared to the animal models. It provides enhanced dose response relationship by using drug concentrations relative to human exposure. Ease of ex-vivo access of cellular and molecular responses in humanized liver model system during pharmacological screening also offers high-throughput studies to determine the cellular response networks and toxicity pathways.
- Citation: Vishwakarma SK, Bardia A, Lakkireddy C, Nagarapu R, Habeeb MA, Khan AA. Bioengineered humanized livers as better three-dimensional drug testing model system. World J Hepatol 2018; 10(1): 22-33
- URL: https://www.wjgnet.com/1948-5182/full/v10/i1/22.htm
- DOI: https://dx.doi.org/10.4254/wjh.v10.i1.22
Drug testing has been one of the most critical challenges faced by the pharmaceutical companies with approximately 90% failure due to unpredictable adverse events which remain unidentified in preclinical phase[1]. The average time to introduce one drug to market is approximately 8.5 years from the time of clinical testing to Food and Drug Administration (FDA) approval which has 21.5% of clinical success rate imposing about $2 billion cost per drug[2]. Since many years, animal models have been gold standard and the most preferred choice of drug testing to understand the underlying mechanisms of various human pathologies. However, the marked biochemical variations, anatomical complexities and physiological responses limit the bio-mimetic outcomes of drug testing. To overcome these hurdles, patient specific stem cells have been employed to recapitulate the human pathologies in vitro to evaluate cellular process which is also termed as “disease-in-a-dish”[3].
Human primary hepatocytes culture system gives closest representation of human liver physiology[4]. However, the source of tissue along with phenotypic variations represent major limitations[5]. In addition, suspension culture of primary human hepatocytes offer the maximum drug incubation time for 4-6 h thereby requiring high dose of drugs to identify the cellular toxicity. Whereas, the monolayer cultures of human primary hepatocytes allows drug toxicity study for 4-72 h, but the drug metabolism capacity of such cultures represent severe downregulation which negatively impact the correlation with clinical outcomes[6]. In addition, the conventional two-dimensional (2D) cell culture systems do not complement the higher order processes which further neglect the crucial stimuli for cellular organization and function. These drawbacks of conventional cultures have been because tissue specific functions are dependent on several crucial factors other than only cell autonomous system which includes extracellular microenvironment with soluble factors, physical strength and extra-cellular matrix (ECM)[7]. Conventional cultures lack these crucial factors for proper cell to cell and cell to microenvironment interactions.
The leveraging tissue-engineering strategies to stabilize the functions of primary human hepatocytes within the xenogenic liver scaffolds provides unique model which can be utilized for better predicting human drug responses, pharmacokinetics and metabolic synchronization similar to human system[8,9]. Hence, the present study was designed to bioengineer humanized livers using more efficient technology of whole xenogenic liver acellularization and human hepatic stem/progenitor cells repopulation. This technology provides biomimetic natural organ scaffold with highly intact native ECM, vascular networks and mechanical strength. Repopulated cells in these acellularized whole liver scaffolds are organized in natural manner and perform high level of bio-mimetic liver functions better than conventional 2D culture systems.
In present study, the structural and functional advantage of humanized livers has been evaluated by testing the metabolism of six cytochrome P-450 (CYP) probe substrates (phenacetin, diclofenac, S-mephenytoin, dextromethorphan, nifedipine and testosterone). CYP has been considered the most common drug metabolizing enzymes which are profusely expressed in liver apart from lungs, kidney, intestine and brain etc. The expression level of these CYPs changes according to the physiological conditions and disease status[10]. Hence studying the behavior of these CYPs against different kinds of substrates in humanized liver could provide better choice for identifying the pharmacokinetics and pharmacodynamics of drugs. This technology offers tremendous potential option for pre-clinical pharmacological drug testing which can reduce the cost, time and unpredictable adverse events.
Spontaneously aborted 10 wk gestation aged human fetuses (n = 2) were collected from local maternity hospitals after taking written informed consent from their parents. The study was approved by the Institutional Ethics Committee of Deccan College of Medical Sciences, Hyderabad. The study was conducted according to the ethical and regulatory guidelines of Indian Council of Medical Research (ICMR), India.
The whole liver was harvested by laparotomy from male Wister rats (n = 10, average body weight = 180-200 g) having intact hepatic artery and portal vein. The rats were obtained from National Institute of Nutrition (NIN), Hyderabad, Telangana, India. Harvested rat liver was initially perfused with heparinized phosphate buffered saline (100 U/mL) through portal vein with the help of 22G (gauge) intravenous catheter. Following to this, 3.8% of Sodium Citrate solution was infused to completely remove the red blood cells from liver. Afterwards a sequential perfusion was performed through main hepatic artery using different concentration gradients of Sodium dodecyl sulphate (SDS) at 30 Hg pressure and with flow rate of 1 mL/min for 16 h. After obtaining the complete acellularized whole liver scaffold, distilled water was run for 10min followed by Triton-X-100 (1.0%) perfusion. Completely acellularized rat liver scaffolds (ALS) were preserved in distilled water containing antibiotic and antimycotic solutions and stored at 4 °C until further use.
Identifying the residual nucleic acids: Acellularized xenogenic liver scaffolds were first characterized for the absence of nucleic acid contents in tissue lysate and flow through after 16 h of acellularization. Briefly; the lysate of acellularized and native liver was prepared by digestion with 0.1% papain solution, 1 mmol/L EDTA, 7.0 mmol/L cysteine and 1 mol/L NaCl in 1 × PBS at 60 °C for 48 h in an incubator shaker and residual nucleic acid content was quantified using spectrophotometric analysis at 260 and 280 nm. The ratio of 260/280 nm sample optical density was calculated to compare the presence of nucleic acid content.
Immunohistochemical staining: Further immunohistological staining was performed for the ALS using H and E staining. The presence of intact ECM components within ALS was determined using immunofluorescence staining for collagen, fibronectin and laminin. Briefly; ALS was fixed in 4% paraformaldehyde (PFA) and further used for the preparation of 3-5 μm thin sections which were stained using specific primary and secondary antibodies and analyzed under the microscope. Parallel analysis was also performed using native rat liver sections for comparison.
Ultra-structure analysis: The ultra-structure analysis of ALS was performed using scanning electron microscopy (SEM). Section were prepared and fixed in 2.5% (v/v) Gluteraldehyde in 1 × PBS and further subjected to dehydration using graded series of ethanol (50%, 75%, 80%, 95% and 100%) for 15 min each and dried in a HCP-2 critical-point dryer using CO2. The cross sections were mounted and subjected for SEM analysis using JOEL-JSM 5600 SEM at RUSKA Lab’s College of Veterinary Science, SVVU, Rajendranagar, Hyderabad, India[11].
Sterilization of ALS: Sterilization of ALS was performed to preserve the functional homology of liver matrix. Briefly; the ALS were perfused with 0.1% per-acetic acid (PAA) for 30 min in laminar chamber at room temperature and further exposed to ultra-violet (UV) light for 30 min.
Mechanical strength: ALS was subjected to mechanical strength analysis following to sterilization procedure using mixed tensile strength, suture retention strength and compressive strength assays[12]. All the mechanical properties of ALS were compared with the native liver without acellularization.
Methylene blue dye was infused into the acellularized liver through main hepatic artery to check the integrity of liver vascular system. The microvasculature and the surface capsule integrity was evaluated further by increasing the infusion rate using peristaltic pump.
Human hepatic cells were isolated from 10 wk gestation aged spontaneously aborted human fetal livers following the protocol as described in our earlier studies[13,14]. The enrichment of human hepatic progenitor cells (hHPCs) was performed by magnetic activated cell sorting (MACS) using epithelial cell adhesion molecular (EpCAM) antibody tagged with the iron nanoparticles (MiltenyBiotec)[15]. EpCAM+ve enriched cells were termed as hHPCs which were further tested for their viability and counted using hemocytometer. Human HPCs were further characterized for the expression of other liver cell specific pluripotent markers using immunofluorescence and molecular analysis. Human HPCs with more than 90% viability were used for repopulating the ALS.
Post-sterilization ALS was transferred to a perfusion chamber kept in a CO2 incubator having inlet and outlet for the flow of culture media. Initially, Dulbecco’s Modified Eagle’s Medium (DMEM)-F12 medium supplemented with 10% Fetal Calf Serum (FCS), 0.0036 μg/mL insulin, 10 ng/mL Epidermal Growth Factor (EGF), 1 × antibiotics and antimycotics was perfused through cannula connected with the main hepatic artery. Following to this, 12 × 106 EpCAM+ve enriched hHPCs were resuspended in 5 mL of human hepatic maturation medium and infused into ALF through hepatic artery at flow rate of 1 mL/min. Recellularized liver was incubated for three hours in static culture. The flow through before and after incubation was collected to determine the cells repopulation efficiency. After static culture, continuous fluidic culture was established by supplying culture media to the repopulated liver at flow rate of less than 0.5 mL/min with the help of a peristaltic pump. The perfusate was collected after 24, 48 and 72 h of culture and used for DNA quantification and release of lactate dehydrogenase (LDH).
SEM: SEM analysis of repopulated liver scaffold (RLS) was performed using the standard protocol described by Bozzola and Russell (1998)[11]. The humanized liver tissues were fixed in 2.5% (v/v) gluteraldehyde in 1 × PBS and the ultra-structures of these tissues were documented using JOEL-JSM 5600 SEM at RUSKA Lab’s College of Veterinary Science, SVVU, Rajendranagar, Hyderabad, India.
Immunofluorescence staining: The immunofluorescence staining of repopulated hHPCs in ALS was identified using specific antibodies for Glucose-6-phosphatase catalytic subunit (G6PC) and albumin (ALB). 4’,6-diamino-2-phenylindole (DAPI, Sigma) was used as counterstain for nuclear components. Stained sections were imaged using inverted fluorescence microscope (Carl Zeiss, Germany).
Histology: Histological analysis of RLS was performed using H and E staining of liver tissue microsections and compared with the native liver.
Functional analysis: The functional response of humanized livers was identified by quantification of albumin and glucose-6-phosphatase catalytic subunit (G6PC) liver enzyme in culture supernatant at different time point’s post-repopulation.
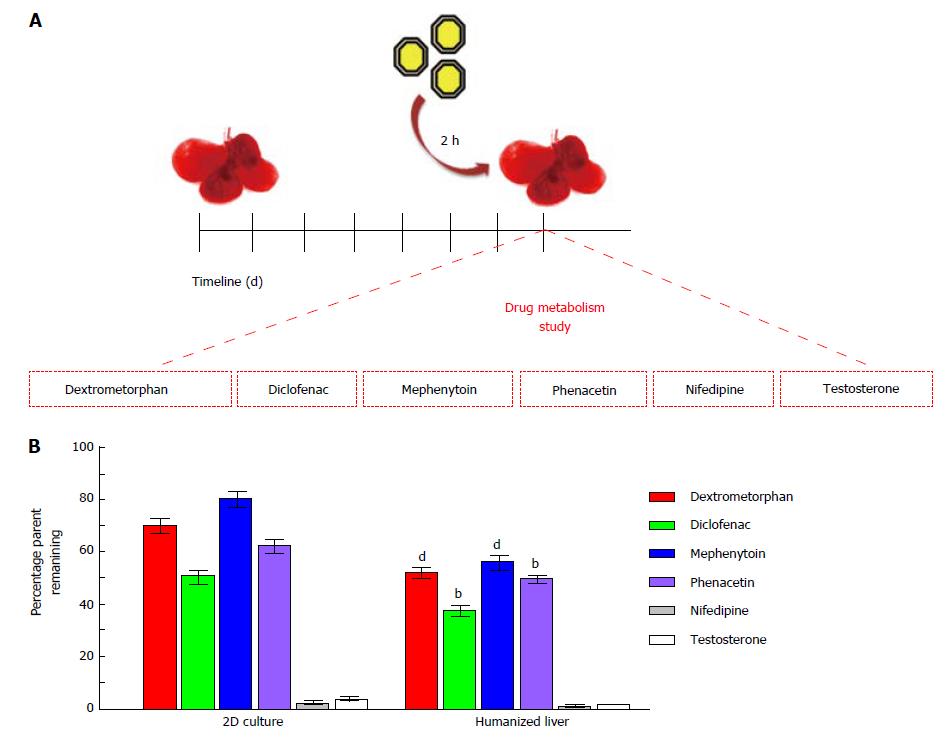
Six of the commonly used CYP probe substrates such asphenacetin (100 μmol/L) specific to CYP1A2[16], diclofenac (25 μmol/L) specific to CYP2C9[17], S-mephenytoin (5 μmol/L) specific to CYP2C19[18], dextromethorphan (50 μmol/L) as a substrate of CYP2D6[19], nifedipine (5 μmol/L) as a substrate of CYP3A4 and testosterone (120 μmol/L) as a substrate of CYP3A4[20] were used to evaluate cellular metabolism in humanized livers in comparison to 2D-cultures. One hundred microlitres of each test compound (diluted in DMSO water) was infused in repopulated humanized livers to maintain final volume of DMSO below 0.2% (v/v). The drug metabolism time was set for two hour post-treatment in CO2 incubator at 37 °C temperature, 5% CO2 and 95% humid atmospheres. Each of the treatment condition was performed in triplicates in two separate cohort studies. The reaction was terminated using 2 mL of acetonitrile containing 1 mg/mL celecoxib as internal control. The supernatant from each treatment group was transferred in sterile glass tubes and the contents were dried using steam of nitrogen with the help of multivap evaporator set at 40 °C (N-evap, Orginomation, Berlin, MA, United States). The residue was reconstituted in 200 μL mobile phase (A:B, 1:1). One hundred microlitres of this reconstituted solution was injected in High-performance liquid chromatography (HPLC) for further analysis.
HPLC analysis of drug metabolism was performed as earlier described by Rao et al[21] 2003. Briefly; HPLC system containing water alliance separation module attached with a water photodynamic array detector was set at detection range of 190-400 nm. A C18, 3V column (GL Sciences, Inc, Japan) was used for the analysis. A tertiary mobile phase gradient system containing three different types of solutions (solution A: 0.01 mol/L ammonium acetate having pH 5 and acetonitrile 90:10, solution B: 0.01 mol/L ammonium acetate having pH 5 and acetonitrile 5:95, and solution C: 0.01 mol/L ammonium acetate having pH 5 and methanol 5:95). The total run time was 40 min with gradient flow. Analysis was conducted by estimating the peak area at individual UV-spectra with the integration of the peak area counts obtained from the internal standards. The area counts of each test compound were divided by the area counts of internal standard within the same analytical run to find the area ratio. This calculated area ratio was used to determine the percentage depletion of the parent compound after 2 h of metabolism in 3D-humanized liver as compared to 2D-culture system.
The data were expressed as mean ± SEM. Each experiment was performed in triplicate in two separate cohort studies to maintain the reproducibility. During metabolism studies, area of the drug was divided by the area of internal standard to calculate the area ratio. The area ratio obtained at 0 h was considered as 100% and at 2 h was calculated to get the metabolic stability of test compounds. Drug metabolism in each group was estimated using the substrate depletion approach[22] with the formula: Percentage substrate remained in test sample = (ratio of substrate in test sample/ratio of substrate in control sample) × 100. One way and two way ANOVA was performed using Graph Pad Prism (version V) to identify the statistical significance among multiple groups. P < 0.05 was set as statistical significance for all the variables in different groups.
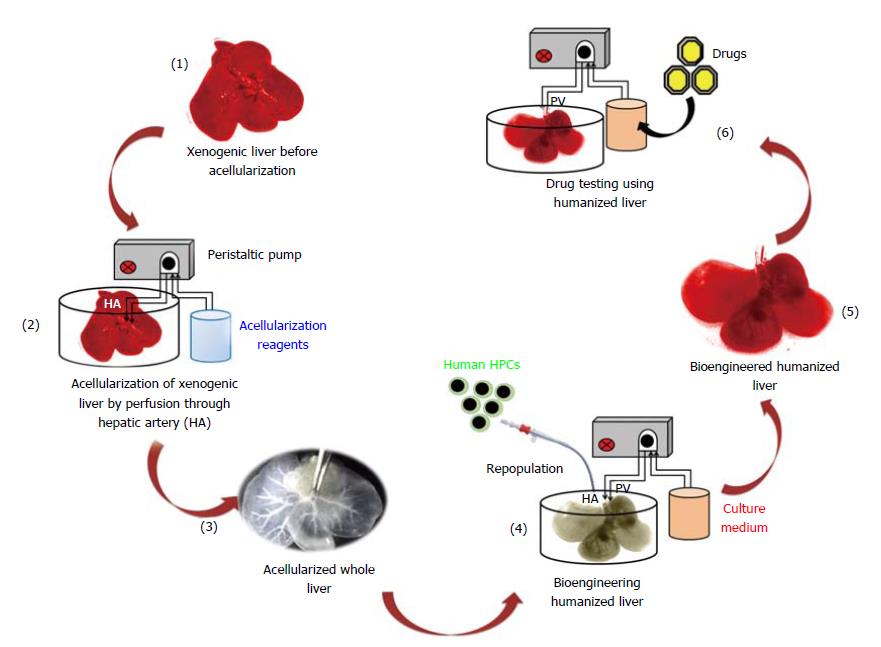
Humanized livers were bioengineered based on the acellularization and human HPCs repopulation technology as demonstrated in Figure 1. This strategy involves the compete removal of cellular components from the total liver through perfusion with acellularization reagents. The continuous flow of acellularization reagents at fixed speed and pressure is maintained with the help of peristaltic pump.
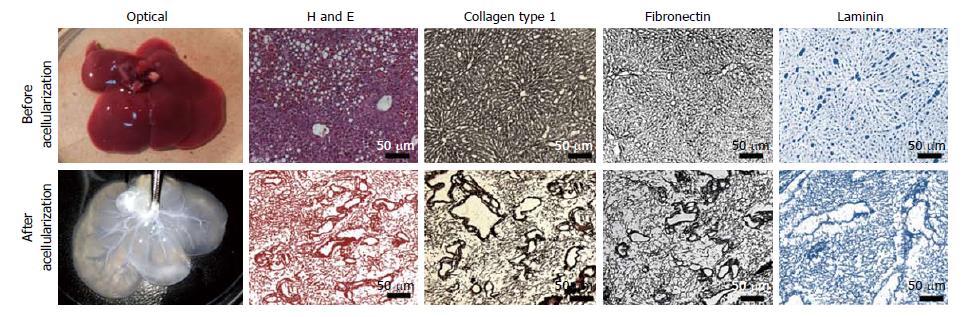
The optical characterization of liver tissues post-acellularization showed absence of liver parenchyma and non-parenchyma cells. The solid and red color whole liver became translucent post-acellularization while retaining intact vascular networks (Figure 2). Further expression and distribution analysis before and after acellularization of whole liver showed intact ECM proteins. More specifically, the immunohistochemistry of liver key ECM proteins collagen type 1 and fibronectins showed complete preservation of liver ECM components post-acellularization. In addition, laminin staining showed intact lining of liver vasculature representing the intact network of vascular tree within the liver post-acellularization.
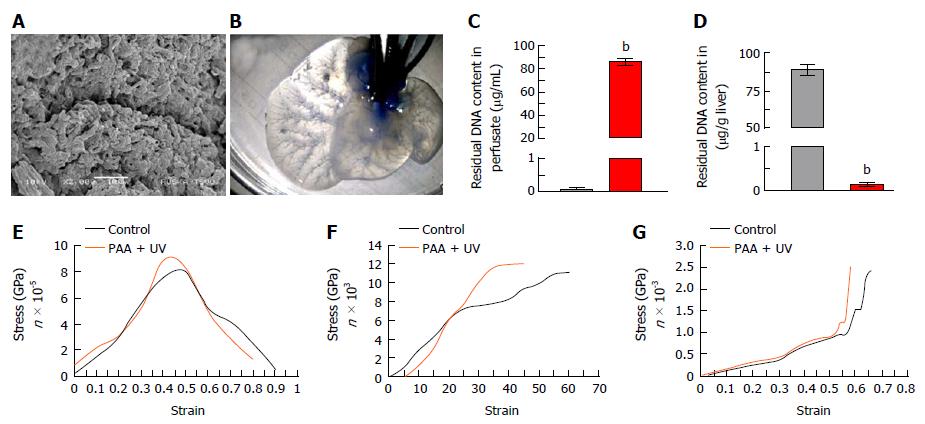
Ultra-structural characterization of ALS: SEM analysis of ALS showed intact 3D-architecture and retention of micro-structures of liver specific ECM proteins. The key structural components such as organ vasculatures were well maintained and distributed throughout the scaffold. The prints of liver cells could be easily recognized in parenchyma region of the liver scaffolds which were surrounded by the network of ECM proteins (Figure 3A). Overall, these observations confirmed the intact three-dimensional anatomy and ultra-structures of liver post-acellularization.
Vascular-tree imaging: Methylene blue dye infusion through main hepatic artery in ALS confirmed the intactness of liver vasculature through gradual distribution from major artery to distant smaller arteries. The vasculature dying also demonstrated the intactness of all three vascular systems named portal, arterial and biliary. The dye infusion first colored the liver parenchyma and finally reached to the central venous system which showed the complete retention of intact perfusion polarity within the ALS (Figure 3B).
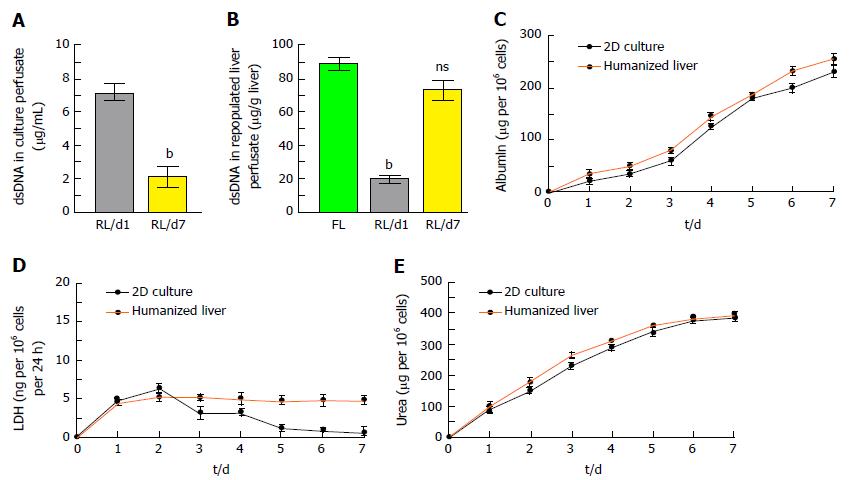
Residual DNA content in ALS: The liver perfusate showed high quantity of dsDNA (Figure 3C) whereas complete reduction of dsDNA in ALS was identified (Figure 3D).
Mechanical properties of ALS post-sterilization: The retention of preserved mechanical properties of ALS was determined post-sterilization with PAA and UV and further compared with the fresh liver as control. Three different types of mechanical characterization using tensile strength test, suture retention test and compressive strength analysis revealed that all three mechanical properties of ALS were preserved similar to the control (Figure 3E-G).
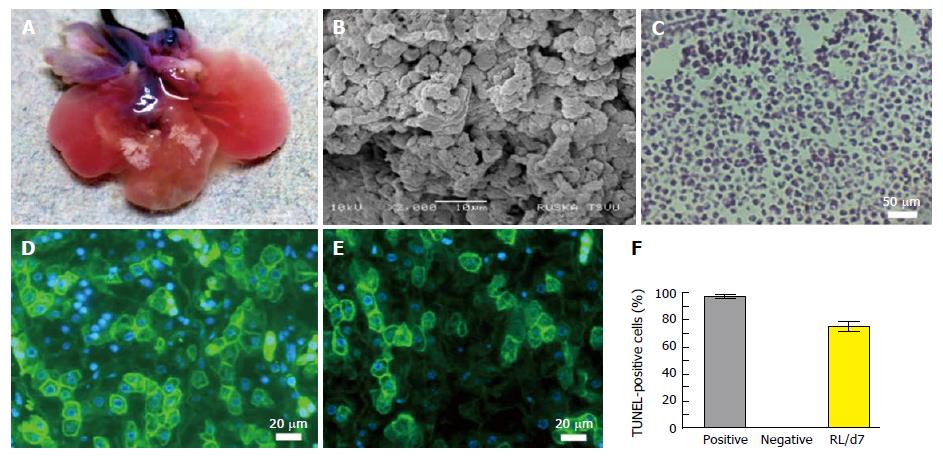
The intact vasculature and liver capsule post-acellularization offers the development of neo-humanized liver system. Herein, the humanized liver was achieved through repopulation of human HPCs into completely ALS at day 7. Optical images of humanized liver showed revival of liver tissues with well intact capsule and liver architecture (Figure 4A). The repopulation efficiency was calculated to be > 80% at day 7 post-repopulation.
Ultra-structural analysis of humanized liver using SEM showed that human liver cells are well engrafted and proliferated in parenchyma and around the vascular spaces. Liver parenchyma was completely surrounded by the human liver cells at day 7 which suggests that the cells were migrated beyond the ECM barrier to reach the acellularized sinusoidal spaces (Figure 4B).
H and E staining was performed to characteize the cellular arrangement in humanized liver tissue at day 7 post-repopulation (Figure 4C). The staining revealed proper arrangement and distribution of cells at defined locations as observed by SEM analysis. Further the functional analysis of cells engrafted within the humanized liver was identified using immunocytochemuical staining for albumin (Figure 4D) and key liver cell enzyme G6PC (Figure 4E). These investigations confirmed that the repopulated human liver cells are viable and functional. The percentage cell apoptosis post-repopulation was determined by the TUNEL staining of humanized liver tissue microsections before (negative) and after humanization at day 7 (RL/d7). The analysis revealed < 10% cells were apoptotic after 7 d of repopulation (Figure 4F).
The quantity of nuclear contents in humanized liver showed that < 25% dsDNA were present in liver perfusate during repopulation at day 7 which may include 10% of apoptotic DNA as observed by TUNEL assay (Figure 5A). Humanized liver tissue extract also showed almost similar quantity of dsDNA per gram of humanized liver tissue as compared to the fresh liver tissue (Figure 5B).
The integrated cellular function of humanized liver was identified by estimating the albumin secretion and LDH released from human hepatic cells at different time points of repopulation and compared with the 2D-culture system. Albumin estimation in liver perfusate revealed extensively increased albumin secretion by the liver cells in humanized liver along with the time and was comparatively higher than the 2D-cultured cells (Figure 5C). LDH released from the cells in humanized liver was quite stable as compared to 2D-culture system which represents the stability and well established synchronization in liver cells within the scaffold (Figure 5D). Furthermore, urea synthesis, one of the functional characteristic of mature liver cells also demonstrated progressively higher degree response in humanized livers as compared to 2D-culture system (Figure 5E).
The drug metabolism study performed in humanized livers for six well-known CYP substrates (Figure 6A) using substrate depletion assay showed that humanized liver metabolites the CYP substrates better than the 2D-cultured cells. The percentage retention of examined six CYP substrates was significantly lower in humanized livers than the 2D-culture system. Complete depletion was observed for nifedipine and testosterone in both humanized liver as well as 2D-culture system. Whereas, the depletion rate was quite high for dextromethorphan (> 20%, P < 0.001), diclofenac (> 10%, P < 0.01), mephenytoin (> 25%, P < 0.001) and phenacetin (> 10%, P < 0.01) in humanized liver as compared to the 2D-culture system (Figure 6B). These results clearly suggest that humanized liver could be better in vitro three-dimensional drug testing model system to optimize the dose for safety evaluations prior to clinical applications.
Human liver play significant role in drug metabolism and toxicological response. Therefore drug-induced liver toxicity has been a major concern for the development of acute liver failure and post-market drug withdrawal due to the absence of suitable humanized preclinical model system. Animal models have been the gold standard platform to identify the toxicological effects of pharmacological drugs/molecules. However, species difference always does not allow predictive outcome similar to human system[23]. Hence, several in vitro models of human livers have been developed to complement the animal model system. The most widely used in vitro models include human liver specific cell lines such as HepG2, Hep 3B and SNU-398. However, these cell lines lack expression of several molecular cues for drug targeting. Currently, human stem cells have been considered the most suitable cell types for such studies.
Among the various choices of stem cells, induced pluripotent stem cells (iPSCs) have been proposed as the best choice for in vitro drug testing. These cells are generated by reprogramming of somatic cells into pluripotent nature by inducing OSKM Yamanaka transcription factors[24]. The major advantages of iPSCs are its highly proliferative nature, ease of accessibility and less/or no ethical constraints[25]. However, the preclinical and clinical applicability of iPSCs has been limited due to reprogramming obstacles, financial hurdles, reprogramming inefficiencies, and genetic instability[26,27]. One of the examples of failing iPSCs pre-clinical and clinical applicability was demonstrated by ophthalmologist Masayo Takahashi in collaboration with Shinya Yamanaka where they claimed for the regeneration and improvement in vision post-transplantation of iPSCs-derived retinal pigment epithelial (RPE) sheets in patient suffering with age related muscular degeneration. This trial was halted due to unexplained mutations in transplanted RPE and the patient’s iPSCs which concluded that several crucial safety assays need to be established before considering the pre-clinical applicability of iPSCs[28,29].
Alternative to iPSCs, human HPCs have been proposed as better cell type for drug testing model development. However, the source and isolation technique has been challenging to obtain enriched homogenous population of primary hHPCs. Our group has reported well established approach to isolate human fetal hepatic progenitor cells[30,31]. However, due to the ethical concerns alternative adult sources are needed to be identified. Our earlier studies have demonstrated tremendous clinical beneficial effects in the field of stem cells transplantation specifically in patients with acute[32,33] and chronic liver failure[13,34-36] and metabolic syndrome[37]. In addition to this, various other groups have also demonstrated significant role of stem cells in liver regeneration[38,39].
Development of humanized organs has always been a challenging area in regenerative biology. Discovery of stem cells has given a potential hope to regenerate the diseased organs or tissues in human body. Since then, various strategies have been tried to evolve humanized organs and/or tissue using stem cell technology for in vitro discoveries and in vivo transplantation studies. However, very limited success has been achieved as far as ex-vivo development of whole functional humanized organs/tissues is concerned. Due to the enormous potential of humanized tissues/organs in pharmacological studies, several investigations have been focused to generate biomimetic humanized organs which is an urgent need to replace the conventional 2D/or 3D ex-vivo systems and animal models to reduce the investigatory and economic burdens towards the preclinical evaluation of drugs.
Over the past decades, micro culture technology has emerged to probe the biomedical mechanisms and functions[40]. However, the natural 3D system is critical to bridge the preclinical and clinical studies more effectively. The most popular 3D-model system named organoid culture exhibit more complexity in structure and function than the 2D cultures which results in several challenges in systemic assessment of pharmacological interventions. Furthermore, the batch to batch diversity in complexity, size, morphology, 3D-arrangement of cells and more importantly the protocol variability represent major challenges to overcome.
The very recent preclinical technology named “human organs-on-chip” do not mimic with the full complexity of human liver functions and show limited pharmacokinetic recapitulation and can’t exhibit the clinically relevant processes[41-43]. Hence, identifying a more clinically relevant 3D-humanized model system can streamline and expedite the drug evaluation process. Given the limitations of currently available preclinical models, human metabolites and their downstream effects often go undetectable until the human clinical trials which is the most costly and risky phase of drug development[44]. Despite these significant advances, several crucial issues related to drug absorption, distribution, metabolism, excretion and toxicity (ADMET) indicates lack of sufficient predictability in drug evaluation models. To avoid such higher failure rate in late-stages of drug testing processes, more appropriate humanized platform is highly desirable to generate better preclinical outcome. Our earlier effort was to generate such platforms using various bioengineering technologies in different organs[45-47]. However, drug metabolism studies in humanized liver remain to be studied.
The bioengineered humanized model developed in this study provides natural system for above described assumptions which could be more practical approach to replace the earlier developed models including animals. In addition to the natural architecture, presence of human primary hepatocytes provides activities of human liver metabolic enzymes to identify the real pharmacokinetics of drugs. As CYP is the most common group of enzymes found in liver for the clearance of drugs, it has been proposed better pathway to study the drug metabolism[48]. Another important role of CYPs has been its quantitative variability during the drug metabolism. Hence identifying CYP mapping could provide important information about the drug metabolism either by a single or multiple isoforms of CYPs. FDA guidance requires more than 25% clearance from the CYP mediated liver metabolism prior to conduct human trial on a particular drug[49]. The metabolism of six CYP substrates in present study using bioengineered liver system could provide better platform for future drug metabolism studies as a replacement of animal models as unique pre-clinical model system.
Humanized liver model system could be ideal choice for drug metabolism studies using tissue specific 3D-architecture, proper cell to cell and cell to ECM interactions which make them one of the best model systems to predict the drug responses. The 3D-architecture of this model provides in vivo like context and also eliminates the species differences. This system allows biomimetic humanized preclinical outcomes by allowing natural drug delivery and distribution. In summary, bioengineered humanized livers could be more suitable option for determining drug safety and efficacy in human mimetic preclinical model system. This unique biomimetic platform can produce better outcome during disease modeling and ADMET studies.
The present study offers a new platform for drug metabolism studies in 3D-biomimetic humanized model system.
This approach can provide more realistic outcome of drug metabolism in human cells under organ specific biological and mechanical cues.
The real-time pharmacokinetics of drug absorption, distribution, metabolism, excretion and toxicity can be identified in natural humanized system using cytochrome P-450 probes.
This unique system offers several advantages over the conventional models of drug metabolism studies such as comparatively less cost and time is required for the maintenance and care of the cultures than animal studies.
Smaller quantities of chemicals are required for ex-vivo drug testing.
The cellular response networks and toxicity pathways can be easily determined against drug exposure.
Enhanced dose-responsive relationships can be identified relative to human exposure.
All authors thank to CLRD, DCMS for providing platform and valuable support during the whole study duration. Authors also thank to Mr. Shaik Iqbal (CLRD) for his technical support to conduct the study. We also thank to Dr. Syed Ameer Basha Paspala (CLRD) for his encouraging support and valuable inputs.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: India
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Ding MX, Liaskou E, Parvez MK, Qi X, Zhu YY S- Editor: Ji FF L- Editor: A E- Editor: Li D
| 1. | Khanna S, Bajaj R, Khurana B, Srivastava K. Pharmacotherapeutic Principles of Ungual Drug Delivery System. Int J Drug Dev Res. 2012;3:9-18. |
| 2. | Kaitin KI. Obstacles and opportunities in new drug development. Clin Pharmacol Ther. 2008;83:210-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 3. | Tiscornia G, Monserrat N, Izpisua Belmonte JC. Modelling long QT syndrome with iPS cells: be still, my beating heart. Circ Res. 2011;108:648-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 4. | LeCluyse EL, Witek RP, Andersen ME, Powers MJ. Organotypic liver culture models: meeting current challenges in toxicity testing. Crit Rev Toxicol. 2012;42:501-548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 277] [Cited by in RCA: 255] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 5. | Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, Bode JG, Bolleyn J, Borner C, Böttger J. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol. 2013;87:1315-1530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1026] [Cited by in RCA: 972] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 6. | Xu JJ, Henstock PV, Dunn MC, Smith AR, Chabot JR, de Graaf D. Cellular imaging predictions of clinical drug-induced liver injury. Toxicol Sci. 2008;105:97-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 379] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 7. | Garreta E, Oria R, Tarantino C, Pla-Roca M, Prado P, Fernandez-Aviles F, Campistol JM, Samitier J, Montserrat N. Tissue engineering by decellularization and 3D bioprinting. Materials Today. 2017;20:166-178. [DOI] [Full Text] |
| 8. | Baptista PM, Siddiqui MM, Lozier G, Rodriguez SR, Atala A, Soker S. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology. 2011;53:604-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 493] [Cited by in RCA: 473] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 9. | Mazza G, Rombouts K, Rennie Hall A, Urbani L, Vinh Luong T, Al-Akkad W, Longato L, Brown D, Maghsoudlou P, Dhillon AP. Decellularized human liver as a natural 3D-scaffold for liver bioengineering and transplantation. Sci Rep. 2015;5:13079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 273] [Cited by in RCA: 296] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 10. | Shaik AN, Vishwakarma SK, Khan AA. Metabolism of six CYP probe substrates in fetal hepatocytes. ADMET. 2016;4:84-90. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 11. | Bozzola JJ, Russell LD. In: Electron microscopy principles and techniques for biologists. 2nd Edition. Sudbury, Massachusettes. Jones and Bartlett Publishers, 1998: 19-24, 54-55, 63-67. . |
| 12. | Kajbafzadeh AM, Javan-Farazmand N, Monajemzadeh M, Baghayee A. Determining the optimal decellularization and sterilization protocol for preparing a tissue scaffold of a human-sized liver tissue. Tissue Eng Part C Methods. 2013;19:642-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 13. | Habibullah CM, Syed IH, Qamar A, Taher-Uz Z. Human fetal hepatocyte transplantation in patients with fulminant hepatic failure. Transplantation. 1994;58:951-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 200] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 14. | Vishwakarma SK, Rahamathulla S, Bardia A, Tiwari SK, Srinivas G, Raj A, Tripura C, Sandhya A, Habeeb MA, Khan AA. In vitro quantitative and relative gene expression analysis of pancreatic transcription factors Pdx-1, Ngn-3, Isl-1, Pax-4, Pax-6 and Nkx-6.1 in trans-differentiated human hepatic progenitors. J Diabetes Investig. 2014;5:492-500. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Khan AA, Sivaram G, Vishwakarma SK, Lakki Reddy C, Srinivas G, Raj A, Nallari P, Habeeb MA, Venkateswarlu J. Transplantation of Epcam+Ve Human Hepatic Stem Cells in Liver Cirrhosis Patient and Cellular Immune Response. J J Transplant Technol Res. 2015;5:1-4. |
| 16. | Hoshino K, Inouye H, Unokuchi T, Ito M, Tamaoki N, Tsuji K. HLA and diseases in Japanese patients [proceedings]. Diabete Metab. 1976;2:157-158. [PubMed] |
| 17. | Yasar U, Eliasson E, Forslund-Bergengren C, Tybring G, Gadd M, Sjöqvist F, Dahl ML. The role of CYP2C9 genotype in the metabolism of diclofenac in vivo and in vitro. Eur J Clin Pharmacol. 2001;57:729-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 75] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem. 1994;269:15419-15422. [PubMed] |
| 19. | Kerry NL, Somogyi AA, Bochner F, Mikus G. The role of CYP2D6 in primary and secondary oxidative metabolism of dextromethorphan: in vitro studies using human liver microsomes. Br J Clin Pharmacol. 1994;38:243-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 70] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 20. | Patki KC, Von Moltke LL, Greenblatt DJ. In vitro metabolism of midazolam, triazolam, nifedipine, and testosterone by human liver microsomes and recombinant cytochromes p450: role of cyp3a4 and cyp3a5. Drug Metab Dispos. 2003;31:938-944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 204] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 21. | Rao MN, Biju B, Ansar AK, Mujeeb S, Ramesh M, Srinivas NR. ‘Open access’ generic method for continuous determination of major human CYP450 probe substrates/metabolites and its application in drug metabolism studies. Xenobiotica. 2003;33:1233-1245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 22. | Jones HM, Houston JB. Substrate depletion approach for determining in vitro metabolic clearance: time dependencies in hepatocyte and microsomal incubations. Drug Metab Dispos. 2004;32:973-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 138] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 23. | Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul Toxicol Pharmacol. 2000;32:56-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1359] [Cited by in RCA: 1228] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 24. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14327] [Cited by in RCA: 14278] [Article Influence: 839.9] [Reference Citation Analysis (0)] |
| 25. | Medvedev SP, Shevchenko AI, Zakian SM. Induced Pluripotent Stem Cells: Problems and Advantages when Applying them in Regenerative Medicine. Acta Naturae. 2010;2:18-28. [PubMed] |
| 26. | Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63-67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1035] [Cited by in RCA: 946] [Article Influence: 67.6] [Reference Citation Analysis (0)] |
| 27. | Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 722] [Cited by in RCA: 697] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 28. | Scudellari M. How iPS cells changed the world. Nature. 2016;534:310-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 29. | Fields M, Cai H, Gong J, Del Priore L. Potential of Induced Pluripotent Stem Cells (iPSCs) for Treating Age-Related Macular Degeneration (AMD). Cells. 2016;5:pii: E44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 30. | Rao MS, Khan AA, Parveen N, Habeeb MA, Habibullah CM, Pande G. Characterization of hepatic progenitors from human fetal liver during second trimester. World J Gastroenterol. 2008;14:5730-5737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Vali SM, Vishwakarma SK, Bardia A, Tiwari SK, Srinivas G, Raj A, Tripura C, Habeeb MA, Khan AA, Pande G. Isolation and characterization of stem cells sub population within the human fetal liver. Cell Biol Res Ther. 2014;S1:1-6. |
| 32. | Khan AA, Habeeb A, Parveen N, Naseem B, Babu RP, Capoor AK, Habibullah CM. Peritoneal transplantation of human fetal hepatocytes for the treatment of acute fatty liver of pregnancy: a case report. Trop Gastroenterol. 2004;25:141-143. [PubMed] |
| 33. | Khan AA, Parveen N, Habeeb MA, Paspala S, Rajendraprasad A, Mahaboob Vali S, Khaja M, Lakshmi N, Pramila R, Habibullah C. Cell Therapy for Acute Liver Failure - Ideal source of cell. J Stem Cells Regen Med. 2008;4:2-8. [PubMed] |
| 34. | Khan AA, Parveen N, Mahaboob VS, Rajendraprasad A, Ravindraprakash HR, Venkateswarlu J, Rao SG, Narusu ML, Khaja MN, Pramila R. Safety and efficacy of autologous bone marrow stem cell transplantation through hepatic artery for the treatment of chronic liver failure: a preliminary study. Transplant Proc. 2008;40:1140-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 56] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 35. | Khan AA, Shaik MV, Parveen N, Rajendraprasad A, Aleem MA, Habeeb MA, Srinivas G, Raj TA, Tiwari SK, Kumaresan K. Human fetal liver-derived stem cell transplantation as supportive modality in the management of end-stage decompensated liver cirrhosis. Cell Transplant. 2010;19:409-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 58] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 36. | Habeeb MA, Vishwakarma SK, Bardia A, Khan AA. Hepatic stem cells: A viable approach for the treatment of liver cirrhosis. World J Stem Cells. 2015;7:859-865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 37. | Khan AA, Parveen N, Mahaboob VS, Rajendraprasad A, Ravindraprakash HR, Venkateswarlu J, Rao P, Pande G, Narusu ML, Khaja MN. Treatment of Crigler-Najjar Syndrome type 1 by hepatic progenitor cell transplantation: a simple procedure for management of hyperbilirubinemia. Transplant Proc. 2008;40:1148-1150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 38. | Strom SC, Fisher RA, Thompson MT, Sanyal AJ, Cole PE, Ham JM, Posner MP. Hepatocyte transplantation as a bridge to orthotopic liver transplantation in terminal liver failure. Transplantation. 1997;63:559-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 364] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 39. | Fox IJ, Chowdhury JR, Kaufman SS, Goertzen TC, Chowdhury NR, Warkentin PI, Dorko K, Sauter BV, Strom SC. Treatment of the Crigler-Najjar syndrome type I with hepatocyte transplantation. N Engl J Med. 1998;338:1422-1426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 826] [Cited by in RCA: 716] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 40. | Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26:120-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 949] [Cited by in RCA: 895] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 41. | Vernetti LA, Senutovitch N, Boltz R, DeBiasio R, Shun TY, Gough A, Taylor DL. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp Biol Med (Maywood). 2016;241:101-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 172] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 42. | Wang Z, Samanipour R, Kim K. Organ-on-a-Chip Platforms for Drug Screeningand Tissue Engineering. Biomedical Engineering: Frontier Research and Converging Technologies 2016; 209-233. |
| 43. | Low LA, Tagle DA. Microphysiological Systems (“Organs-on-Chips”) for Drug Efficacy and Toxicity Testing. Clin Transl Sci. 2017;10:237-239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 44. | Leclercq L, Cuyckens F, Mannens GS, de Vries R, Timmerman P, Evans DC. Which human metabolites have we MIST? Retrospective analysis, practical aspects, and perspectives for metabolite identification and quantification in pharmaceutical development. Chem Res Toxicol. 2009;22:280-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 95] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 45. | Rout S, Vishwakarma SK, Khan AA. Decellularized heart: A step towards creating personalized bioengineered organs. Cur Sci. 2014;107:1. |
| 46. | Vishwakarma SK, Bhavani PG, Bardia A, Abkari A, Murthy GS, Venkateshwarulu J, Khan AA. Preparation of natural three-dimensional goat kidney scaffold for the development of bioartificial organ. Indian J Nephrol. 2014;24:372-375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 47. | Khan AA. Emerging technologies for development of humanized bio-artificial organs. J Med Allied Sci. 2016;6:01-02. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 48. | Jia L, Liu X. The conduct of drug metabolism studies considered good practice (II): in vitro experiments. Curr Drug Metab. 2007;8:822-829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 223] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 49. | FDA. Guidance for industry drug interaction studies-study design, data analysis, implications for dosing, and labeling recommendations. 2017; Available from: https://www.fda.gov/downloads/drugs/guidances/ucm292362.pdf. |