Published online Jan 27, 2018. doi: 10.4254/wjh.v10.i1.155
Peer-review started: November 5, 2017
First decision: November 15, 2017
Revised: November 17, 2017
Accepted: December 5, 2017
Article in press: December 7, 2017
Published online: January 27, 2018
Processing time: 82 Days and 6 Hours
To identify the key epigenetically modulated genes and pathways in HCC by performing an integrative meta-analysis of all major, well-annotated and publicly available methylation datasets using tools of network analysis.
PubMed and Gene Expression Omnibus were searched for genome-wide DNA methylation datasets. Patient clinical and demographic characteristics were obtained. DNA methylation data were integrated using the Ingenuity Pathway Analysis, a software package for visualizing and analyzing biological networks. Pathway enrichment analysis was performed using IPA, which also provides literature-driven and computationally-predicted annotations for significant association of genes to curated molecular pathways.
From an initial 928 potential abstracts, we identified and analyzed 11 eligible high-throughput methylation datasets representing 354 patients. A significant proportion of studies did not provide concomitant clinical data. In the promoter region, HIST1H2AJ and SPDYA were the most commonly methylated, whereas HRNBP3 gene was the most commonly hypomethylated. ESR1 and ERK were central genes in the principal networks. The pathways most associated with the frequently methylated genes were G-protein coupled receptor and cAMP-mediated signalling.
Using an integrative network-based analysis approach of genome-wide DNA methylation data of both the promoter and body of genes, we identified G-protein coupled receptor signalling as the most highly associated with HCC. This encompasses a diverse range of cancer pathways, such as the PI3K/Akt/mTOR and Ras/Raf/MAPK pathways, and is therefore supportive of previous literature on gene expression in HCC. However, there are novel targetable genes such as HIST1H2AJ that are epigenetically modified, suggesting their potential as biomarkers and for therapeutic targeting of the HCC epigenome.
Core tip: Hepatocellular carcinoma (HCC) is a high-fatality cancer with limited screening biomarkers and therapeutic options. It arises in the context of chronic liver disease, having accumulated epigenetic changes over time. The goal of this study was to perform an integrative network-based meta-analysis of all genome-wide DNA methylation data in HCC. Using bioinformatics tools, we identified the most important aberrantly methylated genes and associated pathways. G-protein receptor signaling was the most significantly associated with HCC based on differential methylation of involved genes, which is consistent with the implication of the Ras/Raf/MAPK and mTOR pathways. The identification of novel epigenetically modified genes such as HIST1H2AJ within known pathways suggests targeting of the epigenome as a potential therapeutic avenue for HCC.
- Citation: Bhat V, Srinathan S, Pasini E, Angeli M, Chen E, Baciu C, Bhat M. Epigenetic basis of hepatocellular carcinoma: A network-based integrative meta-analysis. World J Hepatol 2018; 10(1): 155-165
- URL: https://www.wjgnet.com/1948-5182/full/v10/i1/155.htm
- DOI: https://dx.doi.org/10.4254/wjh.v10.i1.155
Hepatocellular carcinoma (HCC) arises in the context of chronic liver disease, where there is ongoing injury over decades. HCC incidence in North America has been increasing in recent years, in the setting of a higher prevalence of cirrhosis secondary to hepatitis C and fatty liver disease[1]. It is the fifth most common cancer worldwide, and five-year survival is the second worst worldwide among all cancers at 8.9%. HCC is often diagnosed at later stages, and there is an inability to tolerate chemotherapy in patients with cirrhosis[1]. Curative treatment with resection, radiofrequency ablation or transplantation is only possible in early stage disease[2]. When diagnosed at a later stage, the first-line chemotherapeutic agent is sorafenib, which extends survival only by 3 mo[2]. Several other trials of chemotherapeutic regimens have been developed based on studies of genomic and transcriptomic data, with no further improvement in overall survival[3]. There is therefore a dire need to better understand HCC pathogenesis, elucidate screening biomarkers in patients at risk, and develop more optimal therapeutic agents.
Epigenetic changes are of significant interest to this malignancy, given that it results from mutations accumulating over time with exposure to various insults such as viral hepatitis, alcohol or fatty liver. Epigenetic modifications are heritable states of gene expression without altering DNA sequences[4]. They encompass processes such as DNA methylation, histone modifications, non-coding RNAs and nucleosome positioning. These changes are passed along faithfully to daughter cells during cell division[4]. Among these, DNA methylation has been the most studied, regulating gene expression through a stable silencing mechanism[5]. Covalent modification by DNA methyltransferases of cytosine residues with methyl groups in CpG dinucleotides occurs preferentially at the 5’ end in promoter regions. Transcriptional gene silencing results from this through two mechanisms: steric hindrance of transcription factors being able to access their cognate binding sites on gene promoters[5], and direct binding of methyl CpG binding domain containing proteins to the methylated DNA causing transcriptional repression[6]. The recent advent of genome-wide methylation analysis has enabled an appreciation of methylation status in genes of interest to cancer: Hypermethylation of tumor suppressor genes, hypomethylation of oncogenes, and methylation of repetitive elements[7]. The extensive reprogramming of the epigenome in cancer has led to a growing interest in epigenetic therapy. Specifically in HCC, aberrant DNA methylation of tumor suppressor gene promoters has been documented[8]. These epigenetic changes have been closely correlated with disease stage and clinical outcome[9]. There has been significant variability in the reported frequency of hypermethylated loci in HCC[10-12]. CDKN2A is methylated in 30%-70% of HCCs[10,12,13], RASSF1A in up to 85%[11,12], GSTP1 in 50-90%[14] and MGMT in 40%[15]. DNA methylation loci have also been reported as significantly enriched in the signaling networks of cellular development, gene expression, cell death, and cancer[16].
Our study represents the first comprehensive network-based attempt to integrate all relevant, publicly available, high-throughput genome-wide DNA methylation data to better understand the epigenetic landscape in HCC. Network and pathway analysis tools can enable identification of the most commonly methylated genes across studies and associated pathways, and propose novel treatment options using network-based analysis[17,18].
The goal of this study was to identify key epigenetically modulated genes and pathways in HCC by integrating all major, well-annotated and publicly available methylation datasets datasets using tools of network analysis.
Genome-wide methylation profiles related to HCC samples were downloaded from published datasets (PubMed, http://www.ncbi.nlm.nih.gov/PubMed) using the following MeSH terms: “{[“methylation”(MeSH Terms) or “methylation” (all fields)] and [“carcinoma, hepatocellular” (MeSH terms) or [“carcinoma” (all fields) and “hepatocellular” (all fields)] or “hepatocellular carcinoma” (All Fields) or ["hepatocellular" (all fields) and "carcinoma" (all fields)]} and ["humans" (MeSH Terms) and English (lang)]. All entries on PubMed since 2002, which represents the advent of high-throughput profiling, were considered for inclusion. A second search was performed using Gene Expression Omnibus (GEO), a public functional genomics data repository containing genome-wide methylation profile array data (https://http://www.ncbi.nlm.nih.gov/geo). This search was performed using the following MeSH terms {[“methylation” (MeSH Terms) or methylation (all fields)] and [“carcinoma, hepatocellular” (MeSH terms) or hepatocellular carcinoma (all fields) and “Homo sapiens” (porgn) and “Homo sapiens” (porgn) and “Homo sapiens” (porgn)] and “Homo sapiens” (porgn)}, covering all HCC high-throughput methylation profiling datasets comparing HCC to adjacent non-tumoral tissue.
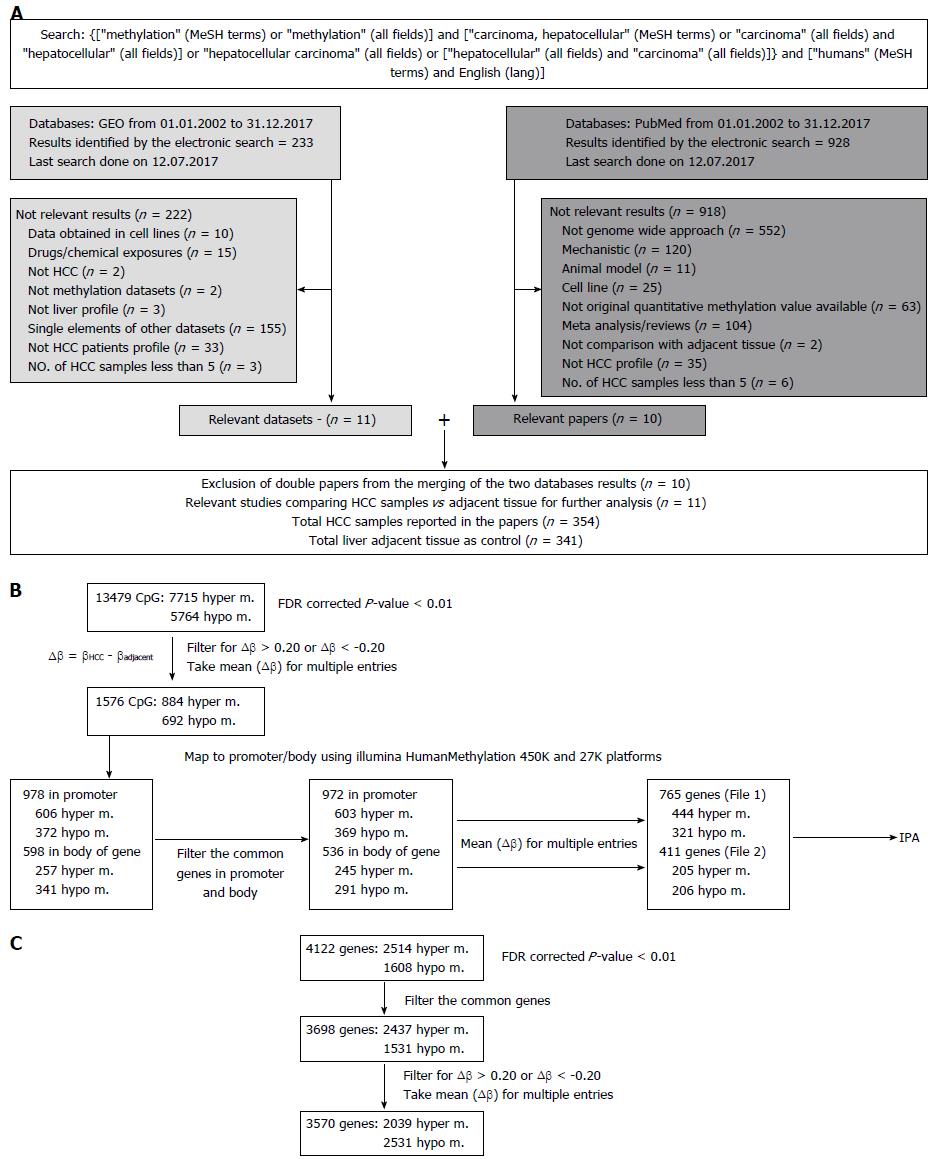
The study workflow is illustrated in Figure 1A. Results were retrieved from both databases: GEO and PubMed. The exclusion criteria listed in Figure 1A were applied to identify papers reporting quantitative results of methylation profile performed on HCC patients and the relative adjacent tissue as control.
Available patient data, including etiology of liver disease (HCV, HBV, alcohol, fatty liver disease) on the basis of which the HCC tumors developed, presence of cirrhosis, the Model for End-stage Liver Disease score (MELD score, an assessment of the severity of liver dysfunction), tumor histology, stage of cancer, alpha-fetoprotein (AFP) level, overall and recurrence-free survival following treatment were also documented.
We identified 928 abstracts retrieved by the search on PubMed and 233 results were obtained from GEO. The flow chart outlining the selection process is detailed in Figure 1A. Details regarding the 11 included studies[8,19-30], together with the information on number of samples per group, per study are provided in Table 1.
Demographic and clinical patient information pertaining to each dataset are presented in Tables 2 and 3.
| Clinical-pathological information | n = 11 | % |
| Cirrhosis status in HCC samples | 6 | 55 |
| Child-Pugh/MELD Score | 3 | 27 |
| HCC Etiology | 10 | 91 |
| Alphafetoprotein level | 5 | 45 |
| Tumor grade | 5 | 45 |
| Tumor stage | 5 | 45 |
| Survival data | 3 | 27 |
| Dataset | Year | PMID | GEO dataset | HCC (n) | Controls (n) | Liver cirrhosis in HCC samples | Etiology of liver disease (n) | Method |
| 1 | 2011 | 21500188 | 13 | 12 | Y (12) | HBV (3), HCV (4), alcoholic (6) | Human methylation 27 DNA analysis bead-chip | |
| 2 | 2014 | 24306662 | 45 | 45 | Y (120), N (34) | HBV (149), HCV (1), nonviral (4) | Illumina GoldenGate Methylation Beadarray Cancer Panel I | |
| 3 | 2014 | 25376292 | 22 | 22 | N/A | HBV (1), HCV (9), alcohol (4), other (8) | Infinium Humanmethylation 27 Beadchip | |
| 4 | 2015 | 25945129 | GSE59260 | 8 | 8 | N/A | HBV-HCV-(8) | Nimblegen Human DNA Methylation 3 x 720K CpG Island PI |
| 5 | 2011 | 21747116 | GSE29720 | 12 | 12 | N/A | N/A | Agilent-017075 human hg 18 promoter 800-200 |
| 6 | 2010 | 20165882 | GSE18081 | 20 | 20 | Y (20) | HCV (20) | Illumina Golden Gate Methylation Beadarray Cancer Panel I |
| 7 | 2012 | 22234943 | GSE37988 | 62 | 62 | N/A | HBV-HCV-(7), HBV + HCV-(36), HBV-HCV+(6), HBV + HCV+(13) | Illumina Human Methylation27 Beadchip |
| 8 | 2013 | 24012984 | GSE44970 | 20 | 8 | N/A | HCV (8) | Human Methylation27 Beadchip |
| 9 | 2013 | 23208076 | GSE54503 | 66 | 66 | Y (48), N (17), missing (1) | HBV-HCV-(19), HCV (19), HBV (13), HBV + HCV (4), missing (11) | Infinium Human Methylation 450K Beadchip |
| 10 | 2014 | 25093504 | GSE57956 | 59 | 59 | Y (37), N (21) | HBV+(36), HBV-(23) | Intinium Ilumanmethylation27 Beadchip |
| 11 | 2014 | 25294808 | GSE60753 | 27 | 27 | Y (26), N (1) | HBV (1), HCV (7), alcohol (9), other (10) | Iinfinium -450K Human Methy lation Beadchip |
Only 6 out of 11 papers (55%) included details regarding presence/absence of cirrhosis, and 10 out 11 papers (91%) provided details regarding the etiology of liver disease. MELD or Child-Pugh score were provided in 27% of papers. Less than half of the selected publications, 5 out of 11 (45%), included details regarding the stage of cancer, although all studies were performed using hepatectomy patient samples. The same trend is identified for information regarding the histologic grade of the tumor (well-, moderately-, or poorly-differentiated tumors). Only 5 out of 11 papers (45%) had alpha-fetoprotein levels available. Overall survival and HCC recurrence statistics as follow-up data were available in 3/11 studies (27%).
For the final network-based integrative analysis, we selected 11 datasets (Table 1). Out of these, raw data from eight studies were available on the GEO website (https://http://www.ncbi.nlm.nih.gov/geo/). Except for the GSE60753[25] dataset, for which we have performed our own analysis with R[31] (due to comparison between sample groups required being different from the main paper), we selected the CpG sites or genes reported to be hyper-or hypo- methylated in the corresponding publications. In 6/11 datasets, CpGs and the mapped genes were selected, whereas in the remaining 5/11 datasets, only the genes without CpG sites were found. Therefore, we separated our analysis into two parts: (1) Taking into account approximately 13500 CpG sites provided or obtained with R analysis (Figure 1B); and (2) considering only the 4122 differentially methylated genes without information on the corresponding CpG sites (Figure 1C). In both cases, the genomic region is considered differentially methylated between HCC tissue and the adjacent non-tumoral sample, if the FDR[32] corrected P-value < 0.01. Furthermore, we filtered out everything that did not satisfy the criteria: ∆β ≥ 0.20 or ∆β ≤ -0.20, where ∆β = βHCC - βadjacent was the difference in methylation between above specified groups. When the CpG sites were considered, the Illumina HumanMethylation450K and 27K platforms were used for mapping to the genes. When multiple sites or genes were found having the same sense of differential methylation, the mean value of ∆β was calculated. The CpGs in the 5’UTR, 1st Exon, TSS200, TSS1500 or in CpG islands were considered in the promoter and all other CpGs were considered to be in the body of the gene.
Two final lists of differentially methylated genes corresponding to CpGs in the promoter (n = 765, Supplementary Table 1) or to the body of the gene (n = 411, Supplementary Table 2) and their corresponding mean (∆β) were uploaded into IPA (Ingenuity Systems®, http://www.ingenuity.com). Based on the manually-curated Ingenuity Knowledge Base derived from experiments and findings published in top peer-reviewed journals, IPA identifies a series of canonical pathways, diseases and functions or networks associated with the molecules in the input list. For each of these, a P-value is calculated with the right-tailed Fisher’s exact test[33], which takes into account the number of focus molecules (input genes) in the network and the total number of molecules in the IPA database that could be included in the corresponding networks.
Based on datasets with known CpG sites, the most frequently hypermethylated genes in the promoter region included HIST1H2AJ, which is a histone protein and SPDYA, a cell cycle regulator known to trigger transition from G1 to S phase. The HRNBP3 gene, an RNA-binding protein, was the most commonly hypomethylated in the promoter region. Further details are provided in Supplementary Tables 1 and 2.
Canonical pathways: The most significantly associated pathways with our list of differentially methylated genes are given in Table 4. G-protein coupled receptor signaling, Transcriptional Regulatory Network in Embryonic Cells, cAMP-mediated signaling were the top hits.
| Ingenuity canonical pathways | -log (P-value) | Molecules |
| G-protein coupled receptor signaling | 3.84E+00 | DRD5, GNA11, VIPR2, ADCY5, ADRB1, CNR1, PIK3R5, NPY1R, FPR1, FFAR3, NFKBID, MC2R, PDPK1, GRM4, MC3R, CXCR2, PRKAR1B, DRD4, PDE6B, HCAR2, DUSP4, PTGDR |
| Transcriptional regulatory network in embryonic stem cells | 3.42E+00 | MYF5, SIX3, PAX6, GBX2, CDYL, FOXD3, ONECUT1, FOXC1 |
| cAMP-mediated signaling | 3.22E+00 | DRD5, VIPR2, ADCY5, ADRB1, CNR1, NPY1R, FPR1, FFAR3, MC2R, GRM4, MC3R, CXCR2, PRKAR1B, DRD4, PDE6B, HCAR2, DUSP4, PTGDR |
Diseases and functions: Not surprisingly Cancer, Organismal Injury and Abnormalities were identified as some of the top diseases and functions. Among different types of cancer, abdominal cancer (P = 1.7E-210), digestive system cancer (P = 7.88E-14), abdominal carcinoma and digestive organ tumor (P = 8.58E-13–1.76E-12) were listed. Cellular development (P-value=3.22E-03–2.66E-08), growth and proliferation (P = 3.22E-03–6.99E-06) were the most important molecular and cellular functions associated with HCC methylated data.
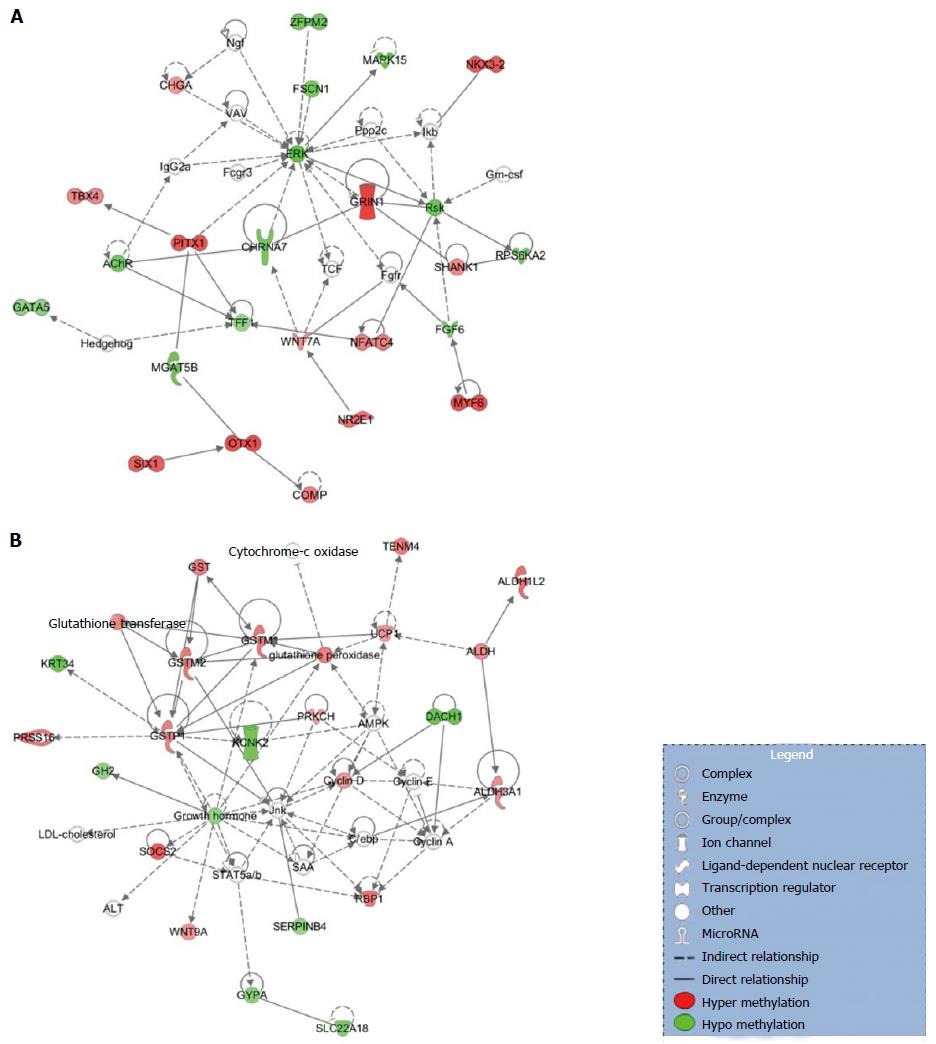
Networks: Among the most statistically and biologically significant networks associated with the genes differentially methylated in the promoter region, three of them captured our attention: Organismal Development, Organismal Injury and Abnormalities, Cellular Development (Figure 2A), Lipid Metabolism, Small Molecule Biochemistry, Cell Death and Survival (Figure 2B) and Cell-to-Cell Signaling and Interaction, Drug Metabolism, Small Molecule Biochemistry (Figure 2C). We noted that networks 1 and 3 were mainly formed by the hyper-methylated genes, whereas network 2 is constituted by both hyper- and hypo-methylated genes approximately equally.
Canonical pathways: G-protein coupled receptor signaling and cAMP-mediated signaling were among the top 20 hits (Table 5), which was similar to the pathways generated by the genes with methylated CpG sites in the promoter region.
| Ingenuity canonical pathways | -log (P-value) | Molecules |
| Aryl hydrocarbon receptor signaling | 2.48E+00 | GSTM1, CCND2, TFF1, ALDH1L2, GSTM2, TP73, GSTP1, ALDH3A1 |
| G-protein coupled receptor signaling | 2.03E+00 | CAMK2B, RGS7, GABBR1, PDE4D, ADCY2, PDE1C, ADRA1D, NPR3, PDE10A, GRM6, PRKCG |
| cAMP-mediated signaling | 1.73E+00 | CAMK2B, RGS7, GABBR1, PDE4D, ADCY2, PDE1C, NPR3, PDE10A, GRM6 |
Diseases and functions: Our study shows that DNA methylation in HCC patients leads to the same diseases and functions, regardless of the CpG site position (promoter or body), with cancer and, in particular, abdominal/digestive system cancer among the top listed by IPA.
Networks: From the top detected networks, the Cell-To-Cell Signaling and Interaction, Cellular Assembly and Organization, Cellular Function and Maintenance (Figure 3A) and Drug Metabolism, Glutathione Depletion in Liver, Small Molecule Biochemistry (Figure 3B) are closely related to HCC.
Having identified the differentially methylated genes corresponding to the CpG sites in the promoter or the body of the genes through this integrative meta-analysis, we wanted to verify how many of these overlapped with the reported differentially methylated genes in those studies that did not include information on CpG sites. The Venn diagram in Figure 4 shows 165 genes reported genes in common with CpG sites in the promoter and 82 genes in common with CpG sites in the body (Supplementary Table 3).
The literature on the epigenome in HCC has grown since the advent of tools permitting genome-wide methylation analysis. Epigenetic changes in HCC arise in the context of various etiologies of chronic liver disease, and have been revealed to contribute to tumorigenesis and cancer progression. Therapeutic targeting of HCC has not been as successful as in other malignancies, and requires exploration of a different approach[34,35]. Given that this cancer is driven by various known environmental factors, targeting epigenetic changes in HCC represents a potentially promising therapeutic avenue[36].
The current study is the largest network-based integrative meta-analysis of all publicly available genome-wide DNA methylation data in HCC, with 354 HCC samples represented. These HCCs had arisen mainly in the context of viral hepatitis B and C, with only a few occurring in patients with alcoholic cirrhosis. Therefore, the literature on methylation in HCC is heavily weighted towards epigenetic changes from viral infection, and the aberrantly methylated genes in our analysis will likewise be influenced by the greater proportion of hepatitis B and C. Clinical information regarding tumor grade, disease stage, and survival were only available in around half of the datasets, thereby limiting the ability to correlate with histopathological characteristics and disease outcome. The genome-wide DNA methylation datasets included in our integrative analysis were published from 2010 to 2015.
Network-based tools offer a different and unique perspective into the key genes and pathways implicated in disease pathogenesis and progression[37]. Network-based medicine is critical to a broader understanding of HCC, whose pathogenesis has been difficult to elucidate given the multiplicity of underlying liver disease etiologies[30]. Epigenetic changes impact genetic networks, and a network-based integrative meta-analysis is ideally suited to integrating and exploring effects of networks on disease pathogenesis[38]. Using IPA, we performed this integrative analysis in order to identify the most commonly aberrantly methylated genes and associated pathways. The most commonly hyper- and hypomethylated genes were identified. These included HIST1H2AJ, which is a histone-coding cell cycle gene previously also identified as hypermethylated in patient lung adenocarcinoma samples[37] and head and neck squamous cell carcinomas[39]. In a study investigating the genetic-and-epigenetic cell cycle network in HeLa cancer cells, methylation of HIST1H2AJ (among other genes) was found to result in cell proliferation and anti-apoptosis through NFκB, TGF-β, and PI3K/Akt/mTOR pathways[40]. HIST1H2AJ has not previously been highlighted as a gene of interest in HCC, which is a novel finding of our integrative analysis of methylation datasets. This illustrates the power of integrating all available high-throughput data to better understand important genes in cancer. SPDYA, a cell cycle regulator known to trigger transition from G1 to S phase, was differentially hypermethylated. The HRNBP3 gene, an RNA-binding protein, was the most commonly hypomethylated, as had been reported in the integrative analysis of epigenetic data by Song et al[16] in 2012. One would thereby anticipate increased gene expression of HRNBP3 in HCC. These genes with differential methylation have not previously been highlighted in the HCC literature, and serve as potential new biomarkers and therapeutic targets[41].
Using IPA, we then determined the most commonly affected networks in HCC.
G-protein coupled receptor signaling, Transcriptional Regulatory Network in Embryonic Cells, cAMP-mediated signaling were the top hits, which was in perfect agreement with the work of Song et al[16], wherein they used IPA to analyze methylation profiling for a set of 27 HCC tumors compared with 20 normal patients. G-protein coupled receptor signalling is common to various principal pathways known to be implicated in HCC, including the PI3K/Akt/mTOR and Ras/Raf/MAPK pathways based on genomic and gene expression analyses. Therefore, our results reinforce the biological rationale of targeting these pathways. We also elucidated the crosstalk between proteins within the networks of interest to HCC. This analysis revealed ESR1 and ERK to be proteins central to they key networks.
A unique aspect of our study was the analysis of methylation at CpG sites in both the promoter and body of genes. Whereas methylation in the gene promoter is known to cause transcriptional repression, methylation in the body has the opposite effect, promoting gene expression. A novel finding was that the genes with the greatest differential methylation in the promoter were the same as those with the greatest differential methylation in the body, further confirming the importance of these genes. We were also able to validate the identity of several genes with data on CpG sites within datasets without such data available.
Limitations of our study include the lack of methylation data on individual HCC samples. Given the relatively recent advent of genome-wide methylation analysis methods, with the earliest dataset in HCC being released in 2010, this analysis was representative of only 354 samples in comparison to a similar number of non-cancerous liver samples. Nonetheless, our study is the largest integrative network-based analysis of DNA methylation in HCC. Clinicopathological characteristics such as grade, stage and survival were available only for half of the datasets, thereby limiting the ability to correlate these data points with the most aberrantly methylated genes. Finally, these data were most representative of hepatitis B and C, as described above.
In conclusion, our integrative analysis of genome-wide DNA methylation represents the largest such study in HCC. By integrating all genome-wide DNA methylation data with network-based tools, we have systematically elucidated the landscape of epigenetic DNA modifications in HCC and identified novel potential biomarkers and targetable genes within known pathways of interest to HCC. Therapeutic targeting of the epigenome in HCC is a potential avenue to address this malignancy that arises in the context of various etiologies of chronic liver disease.
The advent of high-throughput technologies in epigenetics has led to improved characterization of methylation status and its impact on development of Hepatocellular carcinoma (HCC).
HCC is a malignancy that arises in the context of ongoing liver injury from various causes, such as hepatitis B, hepatitis C, alcoholic and non-alcoholic liver disease. Therefore, epigenetic changes are very likely to contribute to the pathogenesis of this malignancy.
We aimed to identify the key epigenetically modulated genes and pathways in HCC by performing an integrative meta-analysis of all major, well-annotated and publicly available methylation datasets using tools of network analysis.
PubMed and Gene Expression Omnibus were searched for genome-wide DNA methylation datasets. Patient clinical and demographic characteristics were obtained. DNA methylation data were integrated using the Ingenuity Pathway Analysis, a software package for visualizing and analyzing biological networks. Pathway enrichment analysis was performed using IPA, which also provides literature-driven and computationally-predicted annotations for significant association of genes to curated molecular pathways.
From an initial 928 potential abstracts, we identified and analyzed 11 eligible high-throughput methylation datasets representing 354 patients. A significant proportion of studies did not provide concomitant clinical data. In the promoter region, HIST1H2AJ and SPDYA were the most commonly methylated, whereas HRNBP3 gene was the most commonly hypomethylated. ESR1 and ERK were central genes in the principal networks. The pathways most associated with the frequently methylated genes were G-protein coupled receptor and cAMP-mediated signalling.
Using an integrative network-based analysis approach of genome-wide DNA methylation data of both the promoter and body of genes, we identified G-protein coupled receptor signalling as the most highly associated with HCC. This encompasses a diverse range of cancer pathways, such as the PI3K/Akt/mTOR and Ras/Raf/MAPK pathways, and is therefore supportive of previous literature on gene expression in HCC. However, there are novel targetable genes such as HIST1H2AJ that are epigenetically modified, suggesting their potential as biomarkers and for therapeutic targeting of the HCC epigenome.
Our integrative analysis of genome-wide DNA methylation represents the largest such study in HCC. By integrating all genome-wide DNA methylation data with network-based tools, we have systematically elucidated the landscape of epigenetic DNA modifications in HCC and identified novel potential biomarkers and targetable genes within known pathways of interest to HCC. Therapeutic targeting of the epigenome in HCC is a potential avenue to address this malignancy that arises in the context of various etiologies of chronic liver disease.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Canada
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: He S, Lin ZY, Mendez-Sanchez N S- Editor: Gong ZM L- Editor: A E- Editor: Li D
| 1. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4260] [Article Influence: 236.7] [Reference Citation Analysis (2)] |
| 2. | Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5972] [Cited by in RCA: 6560] [Article Influence: 468.6] [Reference Citation Analysis (1)] |
| 3. | Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12:436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 133] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 4. | Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2210] [Cited by in RCA: 2194] [Article Influence: 104.5] [Reference Citation Analysis (0)] |
| 5. | Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068-1070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1432] [Cited by in RCA: 1414] [Article Influence: 58.9] [Reference Citation Analysis (0)] |
| 6. | Karpf AR, Jones DA. Reactivating the expression of methylation silenced genes in human cancer. Oncogene. 2002;21:5496-5503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 203] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 7. | Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632-4642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 797] [Cited by in RCA: 869] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 8. | Yang B, Guo M, Herman JG, Clark DP. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol. 2003;163:1101-1107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 288] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 9. | Ma L, Chua MS, Andrisani O, So S. Epigenetics in hepatocellular carcinoma: an update and future therapy perspectives. World J Gastroenterol. 2014;20:333-345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 77] [Cited by in RCA: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 10. | Matsuda Y, Ichida T, Matsuzawa J, Sugimura K, Asakura H. p16(INK4) is inactivated by extensive CpG methylation in human hepatocellular carcinoma. Gastroenterology. 1999;116:394-400. [PubMed] |
| 11. | Yeo W, Wong N, Wong WL, Lai PB, Zhong S, Johnson PJ. High frequency of promoter hypermethylation of RASSF1A in tumor and plasma of patients with hepatocellular carcinoma. Liver Int. 2005;25:266-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 87] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 12. | Zhang YJ, Ahsan H, Chen Y, Lunn RM, Wang LY, Chen SY, Lee PH, Chen CJ, Santella RM. High frequency of promoter hypermethylation of RASSF1A and p16 and its relationship to aflatoxin B1-DNA adduct levels in human hepatocellular carcinoma. Mol Carcinog. 2002;35:85-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 92] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 13. | Zhang YJ, Rossner P Jr, Chen Y, Agrawal M, Wang Q, Wang L, Ahsan H, Yu MW, Lee PH, Santella RM. Aflatoxin B1 and polycyclic aromatic hydrocarbon adducts, p53 mutations and p16 methylation in liver tissue and plasma of hepatocellular carcinoma patients. Int J Cancer. 2006;119:985-991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 60] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 14. | Zhong S, Tang MW, Yeo W, Liu C, Lo YM, Johnson PJ. Silencing of GSTP1 gene by CpG island DNA hypermethylation in HBV-associated hepatocellular carcinomas. Clin Cancer Res. 2002;8:1087-1092. [PubMed] |
| 15. | Zhang YJ, Chen Y, Ahsan H, Lunn RM, Lee PH, Chen CJ, Santella RM. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation and its relationship to aflatoxin B1-DNA adducts and p53 mutation in hepatocellular carcinoma. Int J Cancer. 2003;103:440-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 57] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 16. | Song MA, Tiirikainen M, Kwee S, Okimoto G, Yu H, Wong LL. Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma. PLoS One. 2013;8:e55761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 17. | Shangguan H, Tan SY, Zhang JR. Bioinformatics analysis of gene expression profiles in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2015;19:2054-2061. [PubMed] |
| 18. | Fortney K, Griesman J, Kotlyar M, Pastrello C, Angeli M, Sound-Tsao M, Jurisica I. Prioritizing therapeutics for lung cancer: an integrative meta-analysis of cancer gene signatures and chemogenomic data. PLoS Comput Biol. 2015;11:e1004068. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 19. | Stefanska B, Huang J, Bhattacharyya B, Suderman M, Hallett M, Han ZG, Szyf M. Definition of the landscape of promoter DNA hypomethylation in liver cancer. Cancer Res. 2011;71:5891-5903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 142] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 20. | Archer KJ, Mas VR, Maluf DG, Fisher RA. High-throughput assessment of CpG site methylation for distinguishing between HCV-cirrhosis and HCV-associated hepatocellular carcinoma. Mol Genet Genomics. 2010;283:341-349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 21. | Shen J, Wang S, Zhang YJ, Kappil M, Wu HC, Kibriya MG, Wang Q, Jasmine F, Ahsan H, Lee PH. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012;55:1799-1808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 163] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 22. | Revill K, Wang T, Lachenmayer A, Kojima K, Harrington A, Li J, Hoshida Y, Llovet JM, Powers S. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology. 2013;145:1424-35.e1-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 174] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 23. | Shen J, Wang S, Zhang YJ, Wu HC, Kibriya MG, Jasmine F, Ahsan H, Wu DP, Siegel AB, Remotti H. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics. 2013;8:34-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 130] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 24. | Mah WC, Thurnherr T, Chow PK, Chung AY, Ooi LL, Toh HC, Teh BT, Saunthararajah Y, Lee CG. Methylation profiles reveal distinct subgroup of hepatocellular carcinoma patients with poor prognosis. PLoS One. 2014;9:e104158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 25. | Hlady RA, Tiedemann RL, Puszyk W, Zendejas I, Roberts LR, Choi JH, Liu C, Robertson KD. Epigenetic signatures of alcohol abuse and hepatitis infection during human hepatocarcinogenesis. Oncotarget. 2014;5:9425-9443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 26. | Ammerpohl O, Pratschke J, Schafmayer C, Haake A, Faber W, von Kampen O, Brosch M, Sipos B, von Schönfels W, Balschun K. Distinct DNA methylation patterns in cirrhotic liver and hepatocellular carcinoma. Int J Cancer. 2012;130:1319-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 27. | Udali S, Guarini P, Ruzzenente A, Ferrarini A, Guglielmi A, Lotto V, Tononi P, Pattini P, Moruzzi S, Campagnaro T. DNA methylation and gene expression profiles show novel regulatory pathways in hepatocellular carcinoma. Clin Epigenetics. 2015;7:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 80] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 28. | Hou X, Peng JX, Hao XY, Cai JP, Liang LJ, Zhai JM, Zhang KS, Lai JM, Yin XY. DNA methylation profiling identifies EYA4 gene as a prognostic molecular marker in hepatocellular carcinoma. Ann Surg Oncol. 2014;21:3891-3899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Nishida N, Chishina H, Arizumi T, Takita M, Kitai S, Yada N, Hagiwara S, Inoue T, Minami Y, Ueshima K. Identification of epigenetically inactivated genes in human hepatocellular carcinoma by integrative analyses of methylation profiling and pharmacological unmasking. Dig Dis. 2014;32:740-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 30. | Woo HG, Choi JH, Yoon S, Jee BA, Cho EJ, Lee JH, Yu SJ, Yoon JH, Yi NJ, Lee KW. Integrative analysis of genomic and epigenomic regulation of the transcriptome in liver cancer. Nat Commun. 2017;8:839. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 89] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 31. | R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing 2008. . |
| 32. | Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological). 1995;57:125-133. |
| 33. | Fisher RA. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. J Royal Stat Soc. 1922;85:87-94. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1674] [Cited by in RCA: 1727] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 34. | Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12:408-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 390] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 35. | Heimbach JK, Kulik LM, Finn R, Sirlin CB, Abecassis M, Roberts LR, Zhu A, Murad MH, Marrero J. Aasld guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2017;67:358-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2107] [Cited by in RCA: 3005] [Article Influence: 429.3] [Reference Citation Analysis (3)] |
| 36. | Wahid B, Ali A, Rafique S, Idrees M. New Insights into the Epigenetics of Hepatocellular Carcinoma. Biomed Res Int. 2017;2017:1609575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 37. | Barabási AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56-68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3846] [Cited by in RCA: 3024] [Article Influence: 216.0] [Reference Citation Analysis (0)] |
| 38. | Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317-330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5261] [Cited by in RCA: 4421] [Article Influence: 442.1] [Reference Citation Analysis (0)] |
| 39. | Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157-1160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2034] [Cited by in RCA: 1964] [Article Influence: 140.3] [Reference Citation Analysis (0)] |
| 40. | Li CW, Chen BS. Investigating core genetic-and-epigenetic cell cycle networks for stemness and carcinogenic mechanisms, and cancer drug design using big database mining and genome-wide next-generation sequencing data. Cell Cycle. 2016;15:2593-2607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 41. | Mani S, Herceg Z. DNA demethylating agents and epigenetic therapy of cancer. Adv Genet. 2010;70:327-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |